Abstract
Protein glycosylation modulates a wide variety of intracellular events and dysfunction of the glycosylation pathway has been reported in a variety of human pathologies. Endo-apyrases have been suggested to have critical roles in protein glycosylation and sugar metabolism. However, deciphering the physiological relevance of Endo-apyrases activity has actually proved difficult, owing to their complexity and the functional redundancy within the family. We report here that a UDP/GDPase, homologous to the human apyrase Scan-1, is present in the membranes of Caenorhabditis elegans, encoded by the ORF F08C6.6 and hereinafter-named APY-1. We showed that ER stress induced by tunicamycin or high temperature resulted in increased transcription of apy-1. This increase was not observed in C. elegans mutants defective in ire-1 or atf-6, demonstrating the requirement of both ER stress sensors for up-regulation of apy-1. Depletion of APY-1 resulted in constitutively activated unfolded protein response. Defects in the pharynx and impaired organization of thin fibers in muscle cells were observed in adult worms depleted of APY-1. Some of the apy-1(RNAi) phenotypes are suggestive of premature aging, because these animals also showed accumulation of lipofuscin and reduced lifespan that was not dependent on the functioning of DAF-2, the receptor of the insulin/IGF-1 signaling pathway.
INTRODUCTION
From observations on human diseases and mutant mice, it has become clear that glycosylation plays a major role in metazoan development (Schachter, 2004). Given the numerous physiological roles of glycoproteins, which encompass many aspects of normal cellular function and survival, the structures of glycan moieties vary markedly providing the chemical diversity required to fulfill the vast range of their biological tasks. The diversity of natural sugar structures is ultimately derived from the activities of the enzymes responsible for sugar attachment, namely glycosyltransferases, which determine the feasibility of natural product enzymatic glycodiversification. Lumenal ecto-nucleoside tri- and diphosphohydrolases (E-NTPDases) of the secretory pathway of eukaryotes hydrolyzes the nucleoside diphosphates (NDPs) generated by glycosyltransferases. The resulting nucleoside monophosphates (NMPs) are weaker inhibitors of glycosyltransferases than NDPs. NMPs also serve as antiporters in the transport of nucleotide-sugars from the cytosol to the lumen of the endoplasmic reticulum (ER) and Golgi apparatus. Glycosylation plays an important role in the biogenesis of secreted and transmembrane proteins and an elaborate mechanism ensure that only properly folded and assembled proteins exit the ER, a process termed “quality control.”
However, The ER's capacity to process proteins is relatively limited and the stress caused by accumulation of unfolded and misfolded proteins (ER stress) contributes to a number of important human diseases (Zhang and Kaufman, 2006). Cells respond to ER stress by activating the unfolded protein response (UPR), which limits new protein synthesis and promotes the expression of genes that enhance the organelle's capacity to process unfolded proteins (Patil and Walter, 2001). During this process UDP-glucose is transported from the cytosol into the lumen of the ER, where it serves as a substrate for reglucosylation of incompletely folded glycoproteins. A byproduct of this reaction is UDP, which, upon accumulation, can inhibit further progress of reglucosylation reactions (Trombetta and Helenius, 1999). Nucleoside diphosphatases (NDPases) are therefore though to play an essential role for efficient UPR process.
E-NTPases and E-NDPases or apyrases have been traditionally grouped into one of two families, with each family being distinguished from one another on the basis of sequence similarity. The first family of extracellular nucleotidases is the E-NTPDases (Zimmermann, 2000). At least six different members of the human E-NTPDases have been discovered (Kaczmarek et al., 1996; Smith and Kirley, 1998; Wang et al., 1998; Mateo et al., 1999), each of which possesses different enzymatic properties and different physiological localizations. Each member of this family possesses amino acid sequence similarities in the extracellular region of the proteins, with five “apyrase-conserved regions” (ACR; Handa and Guidotti, 1996; Schulte et al., 1997) shown to be essential for enzymatic activity (Smith and Kirley, 1998; Drosopoulos et al., 2000). All reported members of the E-NTPDases possess these conserved phosphate-binding motifs consisting of invariant amino acids comprising the nucleotide-binding and hydrolysis sites. The second family of extracellular apyrases is relatively new in formation and consists of those apyrases cloned from a variety of hematophagous arthropods (Valenzuela et al., 1998, 2001). Although analogous to the E-NTPDases in their enzymatic action, these enzymes are not similar to the E-NTPDase family with respect to their amino acid sequences, and it appears that these two families of apyrases are evolutionarily unrelated (Valenzuela et al., 2001). Sequences related to the Ca2+-dependent secreted apyrases from bloodsucking arthropods were also found in other metazoans from Xenopus to mammalian species (Failer et al., 2002; Smith et al., 2002; Devader et al., 2006). The cDNA isolated from rat brain encodes a membrane-bound Ca2+-dependent NDPase (Ca2+-NDPase) that is targeted to the ER after heterologous expression.
C. elegans is an attractive model to study the relevance of intracellular E-NTPDases in alleviating ER stress and regulating protein and lipid glycosylation. Its genome encodes at least three proteins belonging to the former family of E-NTPDases with sequence similarity to the Saccharomyces cerevisiae Golgi apparatus (GA) Gda1p: UDA-1 related, in substrate specificity, to the yeast Gda1p, NTP-1 an apyrase related to the yeast Ynd1p (Abeijon et al., 1993; Xiao-Dong et al., 1999; Zhong and Guidotti, 1999; Uccelletti et al., 2004) and a nucleotide diphosphatase functionally homologous to the yeast Ynd1p, encoded by the mig-23 gene. Loss of MIG-23 function results in altered gonad morphogenesis, demonstrating the importance of this enzyme (Nishiwaki et al., 2004). Transcription of uda-1 but not ntp-1 and mig-23 was up-regulated by conditions causing ER stress and the accumulation of unfolded proteins such as tunicamycin, or high temperature; however, no relevant phenotypes were associated with loss-of-function mutation in uda-1 obtained by RNA interference (RNAi; Uccelletti et al., 2004).
We report here the characterization of the C. elegans open reading frame (ORF) F08C6.6 coding for an NDPase similar to the apyrases of blood-sucking insects. The gene transcription was up-regulated by ER stress conditions under ire-1 and atf-6 control. Depletion of the enzyme by RNAi induced the activation of the UPR pathway and significantly shortened the lifespan of wild-type and daf-2 mutant animals; multiple premature aging-like phenotypes were also present, including altered organization of actin-containing thin filaments in muscle cells and accumulation of lipofuscin.
MATERIALS AND METHODS
Media Strains and Reagents
Worms were cultured as described previously (Brenner, 1974) and grown at 16°C, unless otherwise indicated. The following C. elegans strains were used in this study: Bristol strain N2 as standard wild-type strain, SJ30 [ire-1(zcI4) II; zcIs4 V], RB772 [atf6 (ok551) X], RB545 [pek-1 (ok275) X], SJ4005 [zcIs4 [hsp-4::GFP] V], SU93 (jcIs1) IV, and CB1370 [daf-2 (e1370) III], kindly provided by the C. elegans Genetics Center (CGC). The S. cerevisiae yeast strain Scgda1Δ G2-11 (MATα, ura3-52, lys2-801 am, ade2-101 oc, trp1-Δ1, his3-Δ200, leu2-Δ1, gda1::LEU2) was used for heterologous expression and was described previously (Abeijon et al., 1993). Yeast cells were grown at 30°C in yeast extract/peptone/dextrose or SD medium supplemented with amino acids as needed. Transformations with plasmids were done by electroporation (Sherman et al.,1986). Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA) was grown in LB medium with 100 μg/ml ampicillin when needed. Reagents for yeast media were obtained from Difco Laboratories (Detroit, MI). Unless otherwise stated, all other reagents were from Sigma (St. Louis, MO).
Characterization and Heterologous Expression of apy-1
Based on the nucleotide sequence of F08C6.6 (Chromosome X: 7568212-7569664; WormBase), forward (CEf 5′-CAGAAGATCATGACACAAGAAAGTAACTC-3′) and reverse (Cer 5′-GGTTAGGCAAATGCAATTCCTTCCTTC-3′) primers were designed to amplify a apy-1 cDNA fragment by PCR, which was performed with C. elegans mixed-stage cDNAs generated from total RNAs. The former ORF was sequenced and inserted appropriately into the L4440 double promoter vector (Timmons et al., 2003) to generate a apy-1 double-strand RNA (dsRNA) expression plasmid (L4440-apy-1) for RNAi assays and into the plasmid pRS426 (Mumberg et al., 1995) to obtain p426-apy-1 for heterologous expression into yeast gda1 mutant cells.
Drug Treatments
Mixed stage nematodes grown in liquid culture were treated with 5 μg/ml tunicamycin (Sigma) or 5% EtOH for 6 h and then collected.
Preparation of Membrane Fraction
Packed mixed-stage worms, grown in liquid culture, were suspended in equal volume of membrane buffer (0.8 M sorbitol, 1 mM EDTA, 10 mM triethanolamine/acetic acid, pH 7.2) supplemented with Protease Inhibitors mixture (Sigma) and disrupted several times (1 min each time) with 0.5-mm glass beads. The suspension was centrifuged at 700 × g for 3 min and then transferred to a Dounce homogenizer to disrupt the cells. The homogenate was centrifuged at 1500 × g for 10 min, and the resulting supernatant was centrifuged at 100,000 × g for 40 min at 4°C. The pellet was suspended in membrane buffer supplemented with protease inhibitor and stored in aliquots at −70°C.
Measurement of Nucleotidase Activities
Nucleotide phosphatase activity and substrate specificity were determined by incubating membrane fractions in solution containing 2 mM NDP/NTP/NMP, 2 mM CaCl2 or MnCl2 or MgCl2, 0.1% Triton X-100, and 0.2 M Tris-HCl. The optimal pH range was determined using a Tris-HCl buffer ranging from pH 6.0 to pH 12. For measurement of Pi, samples were then assayed as previously described (Yanagisawa et al., 1990).
Northern blot Analysis
Total RNA from mixed-stage worms, cultured on different media was isolated with Trizol reagent (Invitrogen), resolved on a 1% formaldehyde-containing gel, transferred to nylon membrane, and hybridized with 32P-labeled cDNA probes using random priming kit (Roche Applied Science, Indianapolis, IN). The cDNA probes were obtained by amplification of cDNA template from RT-PCR reaction.
RNAi and Brood Size
Synchronized worms at L4 larval stage were placed onto IPTG-containing NGM plates seeded with bacteria (E. coli HT115[DE3] carrying the empty vector L4440 (pPD129.36) or the construct for RNAi by feeding of apy-1). Worms were allowed to lay eggs at 16°C, and all progeny was observed daily and counted with a Zeiss Axiovert 25 microscope (Thornwood, NY).
Life Span Assays
All life span assays were performed at 16°C starting when the L4 wild-type worms were fed on bacteria expressing apy-1 RNAi or the empty vector as a control. Animals were transferred to fresh plates, monitored daily, and scored as dead when they no longer responded to gentle prodding with a platinum wire. Animals that crawled off the plates were not included in the analysis.
Pharyngeal Pumping Assay
Pharyngeal pumping was analyzed at 16°C under Zeiss Axiovert 25 microscope by counting the number of contractions (defined as backward grinder movements in the terminal bulb) on 40 animals for each treatment, during five periods of 30 s, starting when the L4 wild-type worms were fed on bacteria expressing apy-1 RNAi or the empty vector as a control. The experiment was performed from the first day of adulthood and last for 14 consecutive days.
Body Bends Assay
We counted the body bends of worms moving forward continuously on an undisturbed plate or immediately after prodding with platinum pick (Koelle and Horvitz, 1996). For each animal, the number of body bends was counted by direct observation for a total of 9 min in 3-min blocks with 30 min between each block. A body bend was scored each time a bend reached a maximum just posterior to the pharynx, either on the dorsal or ventral side of the animal. The experiment was performed when the L4 wild-type worms were fed on bacteria expressing apy-1 RNAi or the empty vector as a control.
Transgenic Animals Overexpressing apy-1::gfp
The reporter gene construct apy-1::gfp was obtained by a PCR fusion-based approach (Hobert, 2002). Genomic apy-1, with 2.86 kb immediately upstream of the start codon, was PCR-amplified from wild-type genomic DNA using Expand High Fidelity Taq (Roche Diagnostics, Penzberg, Germany). This product was then coamplified with a 1.8-kb PCR fragment encoding green fluorescent protein (GFP) from plasmid pPD95.75 (kindly provided by A. Fire, Stanford University, Stanford, CA). The resulting apy-1::gfp fragment was initially microinjected at 15–30 ng/μl into the syncytial gonad of 30 young wild-type adult hermaphrodites together with 130 ng/μl of the plasmid pRF4 (Mello and Fire, 1995) containing the dominant marker rol-6(su1006) (Mello and Fire, 1995). Because no transgenic progeny was obtained, two other microinjection experiments were performed decreasing the concentration of apy-1::gfp construct to 0.6–1 ng/μl.
Phalloidin Staining
Phalloidin staining was performed as described by Costa et al. (1997).
Electron Transmission Microscopy
Adults worms were used for ultrastructural analysis as reviewed by Hall (1985), with some minor differences. Briefly, worms were washed with M9 buffer and anesthetized in 8% alcohol in M9, fixed in 2.5% glutaraldehyde, 1% paraformaldehyde in 0.1 M sucrose, 0.05 M cacodylate on ice for 2 h. After three rinses in 0.2 M cacodylate on ice, worms were postfixed with 1% osmium tetroxide in 0.1 M cacodylate for 2 h on ice. After further rinses, worms were stained in 1% uranyl acetate for 1 h at room temperature, rinsed in distilled water, and pre-embedded in a thin 10% gelatin gel overnight. Gelatin small blocks were cut to have worms close to each other. Small pieces were dehydrated in an ethanol series and propylene oxide, infiltrated with a mixture of propylene oxide and Epon 812, embedded in pure Epon 812, and followed to polymerize at 60°C for 3 d. Transverse thin sections were cut, collected, and stained with uranyl acetate and lead citrate before viewing with a Philips CM10 electron microscope (Mahwah, NJ).
RESULTS
The ORF F08C6.6 Encodes for a Uridine Diphosphatase Up-regulated by ER Stress Conditions
We have previously identified and characterized the uridine diphosphatase (UDPase)/guanine diphosphatase (GDPase) UDA-1, involved in ER “quality control”; UDA-1 showed high similarity of sequence and of substrate specificity with S. cerevisiae Golgi Gda1p and with a human ER NDPase (Abeijon et al., 1993; Trombetta and Helenius, 1999; Uccelletti et al., 2004).
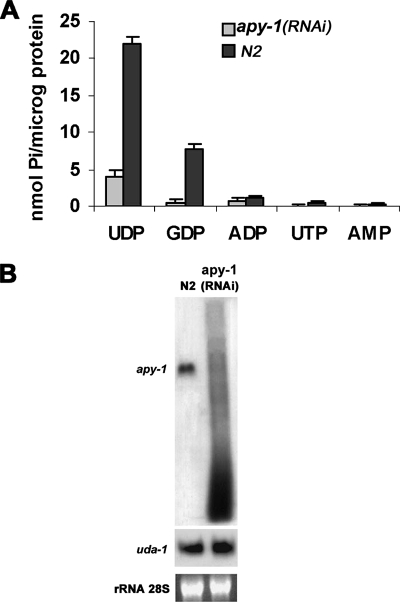
The presence in C. elegans ORFeome Database of the ORF F08C6.6 (hereinafter referred to as C. elegans apy-1), similar to a rat brain ER Ca2+-NDPase (Failer et al., 2002) and to human UDPase SCAN-1 (Smith et al., 2002), prompted us to ask whether APY-1 was an NDPase too. To answer this point, we carried out a nucleotidase activity assay on membrane fractions from apy-1(RNAi) mixed-stage worms. Fractions derived from RNAi individuals showed a drastic reduction of UDPase activity with respect to control ones; GDP was hydrolyzed to a smaller extent but the activity inhibition was still evident. No significant differences were observed in the very low catalytic activity with ADP as substrate, and no activity was detected with NMPs (Figure 1A); these results are consistent with the hypothesis that APY-1 has NDPase activity.
Figure 1.
Results of apy-1 RNAi on nucleotidase activity and gene expression in N2 strain. (A) Substrate specificity of membrane fractions from interfered (feeding) N2 worms. (B) Northern blot analysis of total RNA from N2 and N2 exposed to RNAi of apy-1 animals. The values reported are averages of three experiments; error bars, SD.
To further confirm these data a heterologous approach was followed by expressing apy-1 in a S. cerevisiae gda1Δ mutant strain, which has a low background in nucleotide phosphatase activities (Abeijon et al., 1993). UDPase and GDPase activities were significantly increased in membranes from gda1Δ cells transformed with the vector containing apy-1 cDNA compared with those transformed with the empty vector (Supplementary Figure S1).
To verify the apy-1 mRNA effective degradation in RNAi experiments, a Northern blot analysis of apy-1 transcript was performed on apy-1 RNAi worms and, as a control, on worms fed with bacteria carrying the empty vector. As shown in Figure 1B, the apy-1 mRNA from interfered individuals resulted completely degraded with respect to mock-treated, which had a normal profile. To verify the integrity of the mRNAs extracted from worms, the same samples as before were probed against uda-1, which showed an identical level of transcript between the mock- and the RNAi-treated animals (Figure 1B).
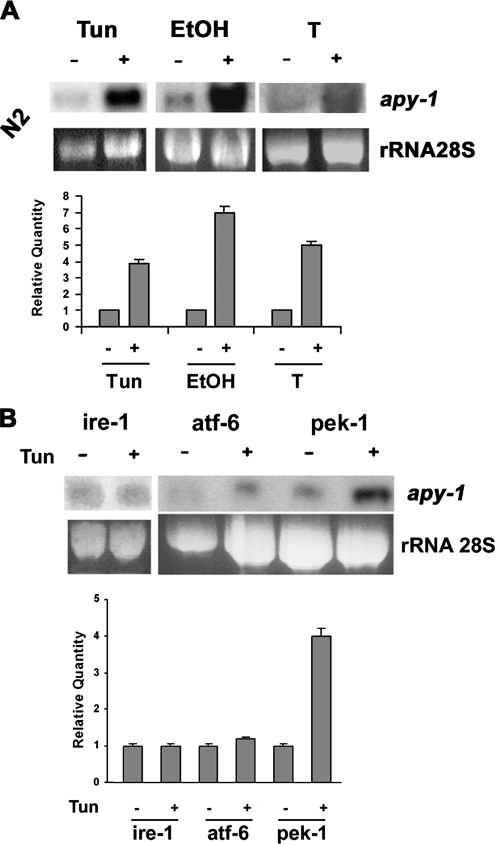
The expression of two different proteins involved in proteins folding and ER quality control with partially redundant catalytic activities may represent a safety margin. To verify a possible overlapping function of UDA-1 and APY-1 and to study transcriptional regulation of second one, we investigated if apy-1 expression could be induced by ER homeostasis–altered conditions. Total RNA from N2 mixed-stage worms, untreated and treated with 5 μg/ml tunicamycin, 6% EtOH, or high temperature was isolated and hybridized with radiolabeled full-length cDNA of apy-1 gene as a probe. Northern blot analysis revealed an evident increase of apy-1–hybridizing transcripts, supporting a possible involvement of apy-1 in ER stress response mechanisms (Figure 2A).
Figure 2.
Transcriptional regulation of apy-1 under stress conditions. (A) Northern blot analysis of total RNA from untreated and 5 μg/ml tunicamycin (Tun)- or 6% ethanol (EtOH)-, or 25°C (T)-treated N2 animals for 6 h. (B) Northern blot analysis of total RNA from untreated and 5 μg/ml tunicamycin (Tun)-treated animals of the indicated mutant strains, each lacking a different UPR activation pathway. Quantification of the radiolabeled signal on the blot is shown in bottom part of each panel. The hybridization signal for each strain in the untreated condition was set as 1. These data represent one of three independent experiments giving the same result; error bars, SD.
IRE-1 protein, and ATF-6 and PEK-1 in lower manner, are the most important folding sensors in the ER and their function is the up-regulation of many UPR genes, like uda-1, as a result of ER stress (Mori et al., 1993, Urano et al., 2002). We then analyzed apy-1 transcriptional regulation in ire-1, atf-6, and pek-1 mutant strains. Total RNA from mixed-stage worms, treated and not with 5 μg/ml tunicamycin, was extracted and analyzed by Northern blot. Results showed a 3–4-fold increase of apy-1 mRNA expression in the treated pek-1 strain; this up-regulation was instead abolished, under the same conditions, in ire-1 and atf-6 mutant backgrounds (Figure 2B). Similar results were also observed in the other ER stress conditions used before (not shown). These experiments directly demonstrate a role for ire-1 and atf-6 genes in transcriptional regulation of apy-1.
Activation of UPR Takes Place in apy-1(RNAi) Animals
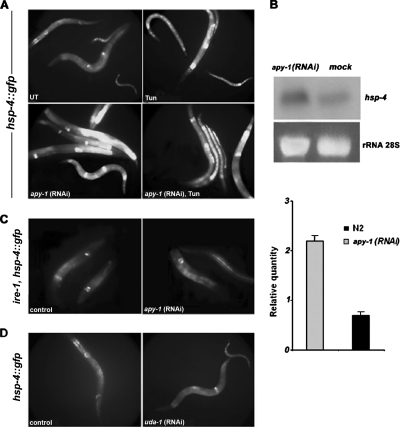
Because UDP, accumulated in ER as a by-product of reglucosylation reactions in protein folding control, might inhibit UDP-Glc:glycoprotein glucosyltransferase (GT; Parodi et al., 1983), cells probably have NDPases in the ER to alleviate product inhibition. The impairment of UDP removal would probably impinge upon folding control and originate ER stress conditions. To examine the apy-1 role in these circuits, we analyzed the expression of an ER stress reporter transgene, hsp-4, a C. elegans homologue of mammalian GRP-78/BiP. We used a transgenic strain (SJ4005) homozygous for a reporter gene (zcIs4), which consists of a fusion of the hsp-4 promoter to GFP (hsp-4::GFP). The hsp-4::gfp expression is induced by ER stress conditions, such as high temperature or tunicamycin treatment (Calfon et al., 2002). Inactivation of apy-1 function by RNAi strongly increased GFP expression (fourfold with respect to control), which reflected an hsp-4 up-regulation, indicating that loss of apy-1 may effectively cause ER stress (Figure 3A). This induction was slightly further enhanced after tunicamycin exposure. To confirm the up-regulation of hsp-4 expression, we performed a Northern blot analysis of hsp-4 mRNA from apy-1(RNAi) worms in the N2 background; this showed a net increase of hsp-4 transcription in comparison to the control preparation (Figure 3B). We then analyzed if this hsp-4 up-regulation was dependent on the UPR regulatory system by knocking down apy-1 in the SJ30 transgenic C. elegans strain, homozygous for hsp-4::GFP in a ire-1 mutant background (Calfon et al., 2002). In this case, induction of hsp-4::GFP by apy-1(RNAi) was not observed (Figure 3C), indicating that IRE-1 activity is required for the UPR induction resulting from apy-1(RNAi). UDA-1 and APY-1 were, so far, the only nucleotidases to be up-regulated at transcriptional level by treatment of the animals with tunicamycin. Therefore, we asked whether also uda-1(RNAi) would be able to increase expression of hsp-4::GFP similar to apy-1(RNAi); however, no UPR activation was observed in this case (Figure 3D).
Figure 3.
Induction of an ER stress reporter gene by apy-1 RNAi treatment. (A) Fluorescence micrographs of SJ4005 worms, containing an integrated copy of hsp-4::gfp, a green fluorescent protein transcriptional reporter driven by the hsp-4 promoter, untreated (UT), tunicamycin-treated (Tun; 5 μg/ml), or apy-1(RNAi) by feeding animals. Fluorescent images were obtained using the same exposure for all treatments. (B) Northern blot analysis of total RNA from N2 and apy-1(RNAi) worms. The blot was hybridized with hsp-4 probe that detects the endogenous gene. Quantification of the radiolabeled signal is shown in the bottom part of the panel. These data represent one of three independent experiments giving similar results; error bars, SD. (C) Fluorescence micrographs of SJ30 worms, containing in a ire-1 background an integrated copy of hsp-4::gfp, fed with bacteria carrying the empty vector (control) or fed with bacteria expressing apy-1(RNAi). (D) Fluorescence micrographs of SJ4005 worms fed with bacteria carrying the empty vector (control) or fed with bacteria expressing uda-1(RNAi). Fluorescent images were obtained using the same exposure for all treatments.
Because uda-1 and apy-1 transcripts resulted in up-regulation under ER stress, we investigated whether some nucleotidase activities in C. elegans increased under similar conditions. To this aim, total membrane fractions from N2 mixed-stage worms treated and untreated with 5 μg/ml tunicamycin were obtained and assayed for nucleotidase activity and substrate specificity. In the presence of calcium as a cofactor, membranes from worms treated with tunicamycin showed a 75% increase of UDPase activity with respect to control ones. The GDPase activity, although significant, was not enhanced by this treatment as well as the ADPase activity (Supplementary Figure S2A). To correlate the increased UDPase activity with ER responses originated by unfolded proteins, we performed the same experiment as before in the ire-1 mutant background, where the UPR pathway is not induced (Calfon et al., 2002). The UDPase activity of membranes from treated worms, in this case, increased only of ∼20% compared with the control (Supplementary Figure S2B).
The assay revealed an optimal pH between 7 and 8 when UDP was used as substrate and incubated with N2 membrane fractions (Supplementary Figure S2C). Moreover, a strictly Ca2+-UDPase activity was observed in the same fractions, because neither magnesium nor manganese were able to reveal any activity when utilized as cofactors (Supplementary Figure S3D). Identical results in terms of cation requirements were obtained with GDP, ADP, UTP, and ATP as substrates (not shown).
APY-1 Plays a Role during Development and Aging
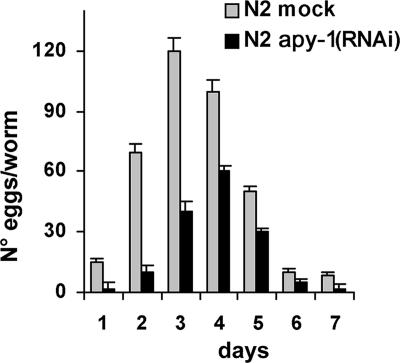
To investigate phenotypes linked to apy-1 loss-of-function, we observed RNAi worms to check for morphological and developmental defects and analyzed their brood size and growth rate of the progeny. The brood sizes of apy-1 worms were significantly decreased, on average a 50% reduction of progeny number compared with the wild type (Figure 4). In the daily progeny count apy-1 worms had significantly lower number of laid eggs compared with N2. In addition, most of the interfered animals showed slow growth, and the development to progressive larval stages was retarded with respect to control worms: in general, the RNAi worms took more than 20 h to reach adulthood (data not shown).
Figure 4.
Effects of apy-1(RNAi) inactivation on brood sizes. Average brood sizes per worm of N2 and apy-1 (RNAi by feeding) animals was reported. Worms were allowed to lay eggs at 16°C and all progeny was counted daily. The histogram represents four experiments; the values reported are averages of a minimum of 25 animals; error bars, SD.
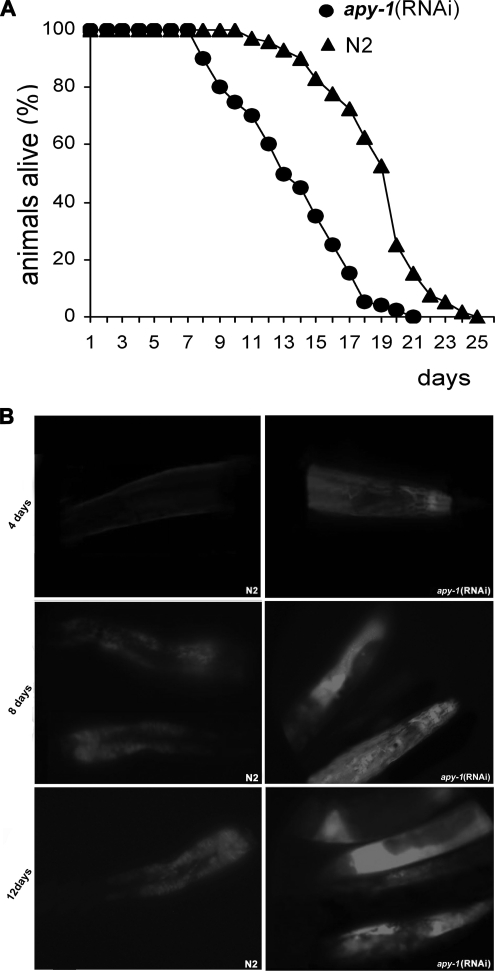
We have also analyzed the impact of APY-1 depletion on the life span by interfering only the L4 stage animals and then monitoring the survivors in order to avoid the developmental problems that could cause general sickness as observed in the F1 progeny. Indeed, apy-1(RNAi) individuals exhibited a reduced life span: half-life of 14 d, compared with 20 d for N2 worms (Figure 5A). This prompted us to monitor the accumulation of intestinal autofluorescence in adult animals. Intestinal autofluorescence, which is caused by lysosomal deposits of lipofuscin, accumulates over time in aging animal and is an established marker for aging (Garigan et al., 2002). In agreement with short life span, the apy-1(RNAi) animals accumulated intestinal autofluorescence more rapidly than the control worms (Figure 5B). In fact, at day 4 of treatment the RNAi animals started to accumulate intestinal fluorescence, whereas in the mock-treated animals, no fluorescence was detectable. At day 8 as well as at day 12 a strong difference in accumulated fluorescence was observed between the RNAi-treated and the mock-treated worms (Figure 5B).
Figure 5.
Effects of apy-1(RNAi) inactivation on lifespan and aging. (A) Lifespan analysis of apy-1(RNAi) and N2 mock-treated worms. Survival is plotted against days of adulthood, where 0 is the L4 larvae phase. Error bars, SD; n = 60 for each data point of single experiment. The results are the mean of three independent experiments. (B) Accumulation of lipofuscin autofluorescence with age. N2 mock-treated and apy-1(RNAi) worms at 4 and 8 d of adulthood were photographed under identical conditions.
Longevity in C. elegans is regulated by the insulin-like signaling pathway that includes the gene daf-2, encoding a protein most closely related to vertebrate insulin and insulin-like growth factor (IGF)-I receptors (Kenyon et al., 1993; Kimura et al., 1997). Mutations in daf-2 result in a two- to threefold increase in adult lifespan; in order to determine whether apy-1 interacts with the insulin pathway, daf-2 worms were interfered with apy-1 as was done for the N2 animals. The long living daf-2 animals showed, upon apy-1 interference, a reduction in their lifespan to the same extension that previously reported for the N2 background (S3A). The above data suggest that the role of APY-1 in maintaining a normal lifespan is independent of the DAF-2 activity.
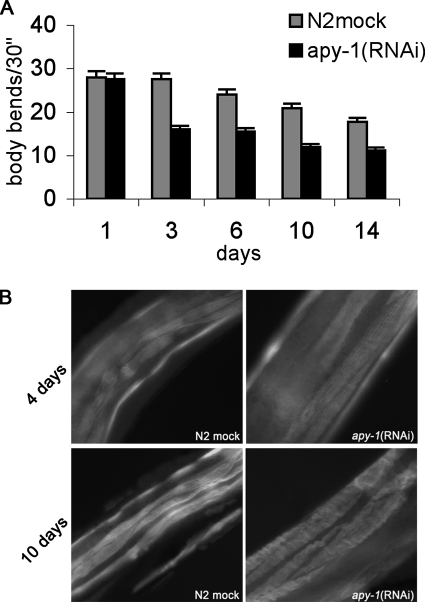
Because aging is usually also related to reduced motility, we then examined whether apy-1(RNAi) had any effects on motility by measuring the frequency of the body bends. The body-bending movements gradually decreased in mock- and RNAi-treated worms during aging (Figure 6A). However, the reduction in the body bends was more pronounced in apy-1(RNAi) than in the mock-treated animals. The frequency of body bends per 30 s was reduced by ∼60% in RNAi-treated worms compared with mock-treated worms along all the period analyzed. Similar results were also obtained in daf-2 strain (Supplementary Figure S3C).
Figure 6.
Muscle structure in apy-1(RNAi) animals. (A) Mean of body bends in apy-1(RNAi) animals and N2 mock-treated animals treated for the indicated period of time. Error bars, SD; n = 60 for each treatment. The histogram shows the mean of three different experiments. (B) Muscle structural integrity was examined by phalloidin staining of actin in body wall muscle at days 4 and 10 of adulthood in N2 mock- and RNAi-treated worms.
The reduced motility strongly suggests that the depletion of APY-1, during adulthood, reduces the rate of muscle contraction. On the other hand, aging in C. elegans is accompanied by sarcopenia, the progressive deterioration of muscle tissue (Huang et al., 2004). We then examined muscle structure during a period of adulthood. To this aim, we stained animals with phalloidin to visualize the actin-containing thin filaments of the body wall muscles. Phalloidin-stained muscles at adult days 4 of RNAi-treated worms showed patched or wrinkled sarcomeres, whereas the mock-treated animals exhibited straight and evenly stained filaments, as expected for intact muscle cells. These differences between the RNAi-treated and the control animals became even more evident at day 10 of treatment (Figure 6B).
To further characterize APY-1, we attempted to construct transgenic animals carrying apy-1::gfp by microinjection technique. A first round of microinjections at standard concentrations of the constructs failed to generate transgenic progeny, suggesting a toxic effect of apy-1 overexpression. Therefore, we reinjected the product at 15–30-fold dilution and obtained 80 F1 transformants over two independent assays. Only one of these animals gave rise to a stable transgenic line. The slight percentage of heritable transformation reinforces the idea of toxicity of the apy-1 gene product when expressed at higher levels than in native conditions. GFP expression pattern was analyzed in a portion of F1 roller animals and in the APY-1::GFP stable transgenic line. Fluorescence intensity was very faint, making difficult any conclusion about its localization. In animals showing a perceptible signal, it was localized mainly to the pharynx as a network pattern (data not shown). The experiments performed on the transgenic line showed a lifespan reduction compared with the wild-type counterpart (half-life of 10 d, compared with 18 d for N2 worms), further reinforcing the idea of the toxicity of APY-1 when over expressed, although we cannot rule out the possibility that the observed toxicity is a result of the APY-1::GFP fusion instead of a higher level of APY-1.
Altered Pharynx Morphogenesis Take Place in apy-1(RNAi) Animals
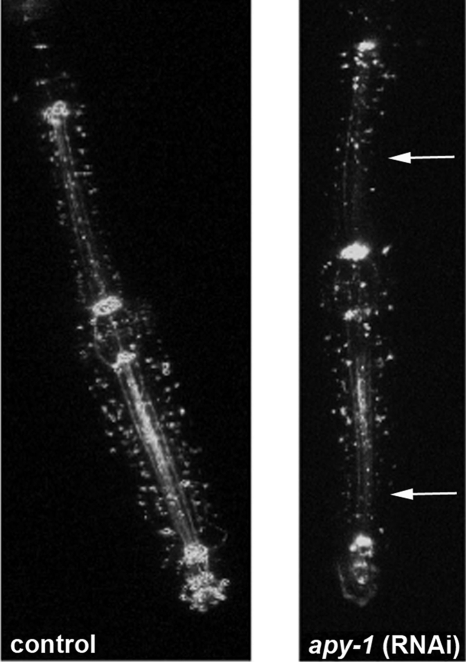
The C. elegans pharynx is regarded as a model system to study organ development; pharyngeal activity is one prominent characteristic of adult worms, and its functionality declines with age; to look for phenotypes in feeding behavior possibly correlated with depletion of APY-1, we visualized the pharyngeal region of the digestive tube using a transgenic strain carrying ajm-1::gfp, a fusion protein that localizes to epithelial cell junctions located at the most apical portion of their lateral membranes (Mohler et al.,1998). It was possible to observe an irregular and discontinuous distribution of fluorescence signals at the beginning and at the end of procorpus and at level of entire isthmus of apy-1(RNAi)–treated worms with respect to the more complete and organized appearance observed in the same regions of the control animals (Figure 7); this may suggest alterations in the morphology of the pharynx in animals depleted of APY-1.
Figure 7.
Pharynx analysis in apy-1(RNAi) animals. Fluorescence micrographs of pharyngeal region of the digestive tube from SU93 adult worm, containing an integrated copy of ajm-1::gfp, a marker for the epithelial apical junctions, treated and untreated with apy-1(RNAi) for 3 d. An irregular and reduced distribution of fluorescence signal was observed along the pharynx of apy-1(RNAi) worms (arrows).
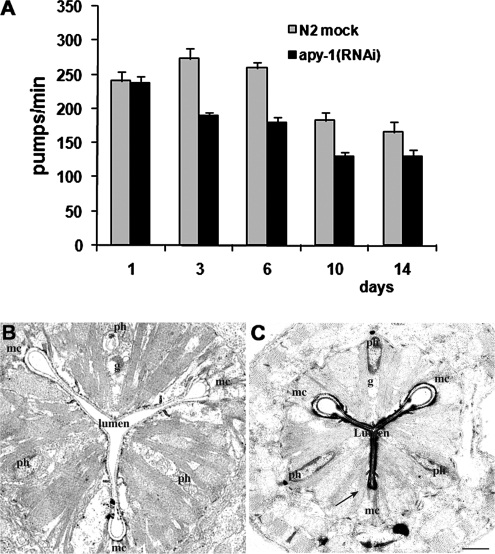
Furthermore, analysis of pharyngeal pumping revealed a reduction in pumping rate of apy-1(RNAi) worms when compared with control (Figure 8A). The mean pumping rate in apy-1(RNAi) animals was about 70% compared with mock-treated worms along all the period analyzed. We also analyzed the morphology of the pharynx of both the wild type and the mutant C. elegans at ultrastructural level. Pharyngeal morphology appeared normal in wild-type worms, with the typical three-radiate shape, surrounded by marginal and myoepithelial cells (Figure 8B). The lumen of the pharynx appeared normal (Figure 8B), and no intact E. coli cells accumulated in the pharyngeal lumen, suggesting an efficient pharyngeal function for feeding and pumping. On the other hand, cross-sections of mutant worms showed the typical shape of the pharynx with a striking morphological feature: the loss of a loop from the three-radiate shape of the pharynx (Figure 8C, arrow). Moreover, a very tight lumen was evident (Figure 8C, lumen); also in this case no intact E. coli cells were observed. These observations, together with the pumping results, suggest that mutant worms probably had a still adequate food transport through the pharynx, although a notably retarded pumping function was also present.
Figure 8.
Pharynx functionality in apy-1(RNAi) worms. (A) Mean pharyngeal pumping rate of apy-1(RNAi) animals and N2 mock-treated animals for the indicated period of time. Error bars, SD; n = 60 for each treatment. The histogram shows the mean of three different experiments. (C and B) Ultrastructural analysis of pharynx in apy-1(RNAi) animals and N2 mock-treated animals after 3 d of treatment in sample electronic photographs from transverse section. Mar1–3 represents three marginal cells; Ph1–3 represents three pharyngeal myoepithelial cells. The lumen is the internal part of the pharynx. Arrow points to the failing loop of the pharynx. Scale bar, 1 μm.
DISCUSSION
We have identified and named APY-1, a C. elegans homologue of the apyrase first found in the saliva of blood-sucking insects (Valenzuela et al., 1998; Charlab et al., 1999; Valenzuela et al., 2001); closely related enzymes have now been described in humans (Smith et al., 2002) and rats (Failer et al., 2002). The substrate specificity of APY-1 is very similar to that of the purified E-NDPase SCAN-1 of human and of the rat Ca2+-NDPase. It utilizes as a substrate mainly UDP and, to a minor extent, GDP. Only a small activity was detected with ADP, the preferred substrate of the apyrases from blood-sucking insects. Gene expression data (available on the World Wide Web at http://nematode.lab.nig.ac.jp/db2/index.php) indicate that the transcript for apy-1 (F08C6.6) can be detected in all developmental stages of the nematode. As mentioned above, NDPs are generated as by-products of glycosylation reactions and can inhibit glycosyltransferases if allowed to accumulate. NDP accumulation is normally prevented by the action of an NDPase that also generates the NMPs needed for the import of new nucleotide sugars (Hirschberg et al., 1998). This transport of nucleotide sugars into the Golgi lumen is necessary for subsequent addition of the corresponding sugars to proteins. NDPases are then though to play an essential role for efficient glycosylation process. Therefore, the “quality control” mechanisms, which ensure that only properly folded and assembled proteins exit the ER, rely on NDPase activity for the proper functioning of glycosyltransferases. Thus, we hypothesized that APY-1 could be required for an efficient organelle capacity to process unfolded proteins. The results presented in this article show that apy-1 is up-regulated under conditions that induce the UPR. Moreover such increase is dependent on ire-1 and atf-6 signaling resident in the ER. Consistently, the lack of APY-1 results in increased expression of the chaperone hsp-4, a signature of UPR activation, suggesting that worms with impaired apy-1 function are continuously under stress. Thus, the up-regulation of apy-1 in conditions that trigger the UPR response suggests that NDPases play an important role in protein quality control. Knocking down APY-1 correlated with some progeria-like phenotypes, as indicated by accumulation of lipofuscin, and, possibly, by reduction in body bends accompanied by alteration in the muscle cells. On the other hand, overexpression of APY-1 was toxic; a fine-tuning of APY-1 level appears therefore of physiological relevance. Numerous diseases are caused by defective UPR signaling (Schroder and Kaufman, 2006). There is evidence suggesting the existence of age-related deficit of the UPR signaling as a common pathogenic mechanism in several neurodegenerative disorders (Zhao and Ackerman, 2006), and several studies suggest that protein damage can be at least as important as DNA damage in the aging phenotype (Sierra, 2006). We might speculate that a lower efficiency in the glycosylation process, due to impairment of UDPase activity, could result in an increased fraction of misfolded or damaged proteins and premature aging phenotypes. Glycosylation changes have been observed, on the basis of DSA lectin binding, for progeria fibroblasts (Clark and Weiss, 1995), and other progeria cells display different, as yet uncharacterized, changes in glycosylation (Quentin et al., 1990). In this respect, elevated levels of the glycoprotein gp200 were consistently observed in all progeria fibroblast strains examined, and indeed gp200 was identified through glycan detection (Clark and Weiss, 1995). In any case, it should also be mentioned that it is possible that some of the premature aging phenotypes observed could be rather related to general sickness conditions of the individuals experiencing UPR stress.
Finally, underlying the complexity of the phenotype originated by apy-1 loss-of-function, we found that worms depleted of APY-1 showed structural and functional pharyngeal alterations. As a matter of fact, several mutants defective in proteoglycan synthesis exhibit pharyngeal defects; these include sqv-1 and sqv-8 mutants, defective in synthesis of chondroitin and heparin sulfate proteoglycans (Herman and Horvitz, 1999; Bulik et al., 2000), as well as pyr-1 animals defective in pyrimidine biosynthesis (Franks et al., 2006). These recurrent phenotypes highly suggest that impaired proteoglycan synthesis someway underlies the pharyngeal abnormalities seen in all of these mutants. Although protein glycosylation modulates a wide variety of intracellular events, the defect induced by the apy-1 loss-of-function has a particular effect on the pharynx development. A similar specific influence was observed with mutations of MIG-23, an NDPase that affects gonad morphogenesis through abnormal glycosylation of the MIG-17 ADAM protease (Nishiwaki et al., 2004). Thus it is possible that, beside the role in the UPR signaling, defects in apy-1 activity may affect a certain set of glycoproteins, resulting in cell type– or tissue-specific defects.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Talora and P. Berninsone for helpful comments, R. Legouis for technical suggestions, and F. Castelli for technical assistance. We are also grateful to Caenorhabditis Genetic Center for C. elegans strains. This work was partially supported by Ministero dell'Universita e della Ricerca Ateneo 2007 to C.P. and National Institutes of Health Grant GM 30365 to C.B.H. F.F. was supported, in part, by fellowship from the Pasteur Institute-Cenci Bolognetti Foundation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0547) on January 23, 2008.
REFERENCES
- Abeijon C., Yanagisawa K., Mandon E. C., Hausler A., Moremen K., Hirschberg C. B. Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccharomyces cerevisiae. J. Cell Biol. 1993;122:307–323. doi: 10.1083/jcb.122.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik D. A., Wei G., Toyoda H., Kinoshita-Toyoda A., Waldrip W. R., Esko J. D., Robbins P. W., Selleck S. B. sqv-3, -7, and -8, a set of genes affecting morphogenesis in Caenorhabditis elegans, encode enzymes required for glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA. 2000;97:10838–10843. doi: 10.1073/pnas.97.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Charlab R., Valenzuela J. G., Rowton E., Ribeiro J. M. Toward an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous sand fly Lutzomyia longipalpis. Proc. Natl. Acad. Sci. USA. 1999;96:155–160. doi: 10.1073/pnas.96.26.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. A., Weiss A. S. Hutchinson-Gilford progeria types defined by differential binding of lectin DSA. Biochem. Biophys. Acta. 1995;1270:142–148. doi: 10.1016/0925-4439(94)00081-z. [DOI] [PubMed] [Google Scholar]
- Costa M., Draper B. W., Priess J. R. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev. Biol. 1997;184:373–384. doi: 10.1006/dbio.1997.8530. [DOI] [PubMed] [Google Scholar]
- Devader C., Webb R. J., Thomas G. M., Dale L. Xenopus apyrase (xapy), a secreted nucleotidase that is expressed during early development. Gene. 2006;367:135–141. doi: 10.1016/j.gene.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Drosopoulos J. H., Broekman M. J., Islam N., Maliszewski C. R., Gayle R., 3rd, Marcus A. J. Site-directed mutagenesis of human endothelial cell ecto-ADPase/soluble CD39, requirement of glutamate 174 and serine 218 for enzyme activity and inhibition of platelet recruitment. Biochemistry. 2000;39:6936–6943. doi: 10.1021/bi992581e. [DOI] [PubMed] [Google Scholar]
- Failer B. U., Braun N., Zimmermann H. Cloning, expression, and functional characterization of a Ca2+-dependent endoplasmic reticulum nucleoside diphosphatase. J. Biol. Chem. 2002;277:36978–36986. doi: 10.1074/jbc.M201656200. [DOI] [PubMed] [Google Scholar]
- Franks D. M., Izumikawa T., Kitagawa H., Sugahara K., Okkema P. G. C. elegans pharyngeal morphogenesis requires both de novo synthesis of pyrimidines and synthesis of heparan sulfate proteoglycans. Dev. Biol. 2006;296:409–420. doi: 10.1016/j.ydbio.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Garigan D., Hsu A. L., Fraser A. G., Kamath R. S., Ahringer J., Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H. Electron microscopy and three-dimensional image reconstruction. Methods Cell Biol. 1985;48:395–436. doi: 10.1016/s0091-679x(08)61397-7. [DOI] [PubMed] [Google Scholar]
- Handa M., Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem. Biophys. Res. Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- Herman T., Horvitz H. R. Three proteins involved in Caenorhabditis elegans vulval invagination are similar to components of a glycosylation pathway. Proc. Natl. Acad. Sci. USA. 1999;96:974–979. doi: 10.1073/pnas.96.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg C. B., Robbins P. W., Abeijon C. Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 1998;67:49–69. doi: 10.1146/annurev.biochem.67.1.49. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Huang C., Xiong C., Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek E., Koziak K., Sevigny J., Siegel J. B., Anrather J., Beaudoin A. R., Bach F. H., Robson S. C. Identification and characterization of CD39/vascular ATP diphosphohydrolase. J. Biol. Chem. 1996;271:33116–331122. doi: 10.1074/jbc.271.51.33116. [DOI] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Koelle M. R., Horvitz H. R. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- Mateo J., Harden T. K., Boyer J. L. Functional expression of a cDNA encoding a human ecto-ATPase. Br. J. Pharmacol. 1999;128:396–402. doi: 10.1038/sj.bjp.0702805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C., Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mohler W. A., Simske J. S., Williams-Masson E. M., Hardin J. D., White J. G. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr. Biol. 1998;8:1087–1090. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- Mori K., Ma W., Gething M., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nishiwaki K., Kubota Y., Chigira Y., Roy S. K., Suzuki M., Schvarzstein M., Jigami Y., Hisamoto N., Matsumoto K. An NDPase links ADAM protease glycosylation with organ morphogenesis in C. elegans. Nat. Cell Biol. 2004;6:31–37. doi: 10.1038/ncb1079. [DOI] [PubMed] [Google Scholar]
- Parodi A. J., Mendelzon D. H., Lederkremer G. Z. Transient glucosylation of protein-bound Man9GlcNAc2, Man8GlcNAc2, and Man7GlcNAc2 in calf thyroid cells. A possible recognition signal in the processing of glycoproteins. J. Biol. Chem. 1983;258:8260–8265. [PubMed] [Google Scholar]
- Patil C. K., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Quentin E., Gladen A., Roden L., Kresse H. A genetic defect in the biosynthesis of dermatan sulfate proteoglycan: galactosyltransferase I deficiency in fibroblasts from a patient with a progeroid syndrome. Proc. Natl. Acad. Sci. USA. 1990;87:1342–1346. doi: 10.1073/pnas.87.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter H. Protein glycosylation lessons from Caenorhabditis elegans. Curr. Opin. Struct. Biol. 2004;14:607–616. doi: 10.1016/j.sbi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. Divergent roles of IRE1alpha and PERK in the unfolded protein response. Curr. Mol. Med. 2006;6:5–36. doi: 10.2174/156652406775574569. [DOI] [PubMed] [Google Scholar]
- Schulte am Esch J., 2nd, Sevigny J., Kaczmarek E., Siegel J. B., Imai M., Koziak K., Beaudoin A. R., Robson S. C. Structural elements and limited proteolysis of CD39 influence ATP diphosphohydrolase activity. Biochemistry. 1997;38:2248–2258. doi: 10.1021/bi982426k. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sierra F. Is (your cellular response to) stress killing you? J. Gerontol. A Biol. Sci. Med. Sci. 2006;6:557–561. doi: 10.1093/gerona/61.6.557. [DOI] [PubMed] [Google Scholar]
- Smith T. M., Hicks-Berger C. A., Kim S., Kirley T. L. Cloning, expression, and characterization of a soluble calcium-activated nucleotidase, a human enzyme belonging to a new family of extracellular nucleotidases. Arch. Biochem. Biophys. 2002;406:105–115. doi: 10.1016/s0003-9861(02)00420-4. [DOI] [PubMed] [Google Scholar]
- Smith T. M., Kirley T. L. Cloning, sequencing, and expression of a human brain ecto-apyrase related to both the ecto-ATPases and CD39 ecto-apyrases1. Biochem. Biophys. Acta. 1998;1386:65–78. doi: 10.1016/s0167-4838(98)00063-6. [DOI] [PubMed] [Google Scholar]
- Timmons L., Tabahara H., Mello C. C., Fire A. Z. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol. Biol. Cell. 2003;14:2972–2983. doi: 10.1091/mbc.E03-01-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta S. E., Helenius A. Glycoprotein reglucosylation and nucleotide sugar utilization in the secretory pathway: identification of a nucleoside diphosphatase in the endoplasmic reticulum. EMBO J. 1999;18:3282–3292. doi: 10.1093/emboj/18.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uccelletti D., O'Callaghan C., Bernisone P., Zemtseva I., Abeijon C., Hirschberg C. B. ire-1-dependent transcriptional up-regulation of a lumenal uridine diphosphatase from Caenorhabditis elegans. J. Biol. Chem. 2004;279:27390–27398. doi: 10.1074/jbc.M402624200. [DOI] [PubMed] [Google Scholar]
- Urano F., Calfon M., Yoneda T., Yun C., Kiraly M., Clark S. G., Ron D. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J. Cell Biol. 2002;158:639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela J. G., Charlab R., Galperin M. Y., Ribeiro J. M. Purification, cloning, and expression of an apyrase from the bed bug Cimex lectularius. A new type of nucleotide-binding enzyme. J. Biol. Chem. 1998;273:30583–30590. doi: 10.1074/jbc.273.46.30583. [DOI] [PubMed] [Google Scholar]
- Valenzuela J. G., Belkaid Y., Rowton E., Ribeiro J. M. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J. Exp. Biol. 2001;204:229–237. doi: 10.1242/jeb.204.2.229. [DOI] [PubMed] [Google Scholar]
- Wang T. F., Ou Y., Guidotti G. The transmembrane domains of ectoapyrase (CD39) affect its enzymatic activity and quaternary structure. J. Biol. Chem. 1998;273:24814–24821. doi: 10.1074/jbc.273.38.24814. [DOI] [PubMed] [Google Scholar]
- Xiao-Dong G., Kaigorodov V., Jigami Y. YND1, a homologue of GDA1, encodes a membrane-bound apyrase required for Golgi N- and O-Glycosylation in Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:21450–21456. doi: 10.1074/jbc.274.30.21450. [DOI] [PubMed] [Google Scholar]
- Yanagisawa K., Resnick D., Abeijon C., Robbins P. W., Hirschberg C. A guanosine diphosphatase enriched in Golgi vesicles of Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 1990;265:19351–19355. [PubMed] [Google Scholar]
- Zhang K., Kaufman R. J. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66:102–109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- Zhao L., Ackerman S. L. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Zhong X., Guidotti G. A yeast Golgi E-type ATPase with an unusual membrane topology. J. Biol. Chem. 1999;274:32704–32711. doi: 10.1074/jbc.274.46.32704. [DOI] [PubMed] [Google Scholar]
- Zimmermann H. Extracellular metabolism of ATP and other nucleotides. Naunyn Schmiedebergs Arch. Pharmacol. 2000;362:299–309. doi: 10.1007/s002100000309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.