Abstract
A role of Arp2/3 complex in lamellipodia is well established, whereas its roles in filopodia formation remain obscure. We addressed this question in neuronal cells, in which motility is heavily based on filopodia, and we found that Arp2/3 complex is involved in generation of both lamellipodia and filopodia in growth cones, and in neuritogenesis, the processes thought to occur largely in Arp2/3 complex-independent manner. Depletion of Arp2/3 complex in primary neurons and neuroblastoma cells by small interfering RNA significantly decreased the F-actin contents and inhibited lamellipodial protrusion and retrograde flow in growth cones, but also initiation and dynamics of filopodia. Using electron microscopy, immunochemistry, and gene expression, we demonstrated the presence of the Arp2/3 complex-dependent dendritic network of actin filaments in growth cones, and we showed that individual actin filaments in filopodia originated at Arp2/3 complex-dependent branch points in lamellipodia, thus providing a mechanistic explanation of Arp2/3 complex functions during filopodia formation. Additionally, Arp2/3 complex depletion led to formation of multiple neurites, erratic pattern of neurite extension, and excessive formation of stress fibers and focal adhesions. Consistent with this phenotype, RhoA activity was increased in Arp2/3 complex-depleted cells, indicating that besides nucleating actin filaments, Arp2/3 complex may influence cell motility by altering Rho GTPase signaling.
INTRODUCTION
Actin polymerization generates force to drive leading edge protrusion in motile cells. Two major protrusive organelles, lamellipodia and filopodia, differ by their morphology, function, cell type distribution, structure, and dynamics of the underlying actin cytoskeleton. Filopodia are characteristic for cells displaying exploratory behavior, especially for neuronal cells. They contain a bundle of long actin filaments undergoing continuous barbed end elongation at their tips (Small et al., 1978; Mallavarapu and Mitchison, 1999). In contrast, lamellipodia dominate in fast moving cells, such as fish keratocytes, but they are not abundant in growth cones, where they are frequently called veils. Lamellipodia contain a network of branching filaments that undergo constant de novo nucleation (Svitkina et al., 1997; Svitkina and Borisy, 1999; Borisy and Svitkina, 2000). Accordingly, the important molecular players for filopodial protrusion include proteins protecting barbed ends from capping, such as Ena/vasodilator-stimulated phosphoprotein (VASP) proteins (Bear et al., 2002; Lebrand et al., 2004; Barzik et al., 2005) and formins (Peng et al., 2003; Pellegrin and Mellor, 2005; Schirenbeck et al., 2005), and a bundling protein fascin (Svitkina et al., 2003; Adams, 2004; Vignjevic et al., 2006), whereas lamellipodial protrusion requires Arp2/3 complex-dependent nucleation (Rogers et al., 2003; Biyasheva et al., 2004; Steffen et al., 2006) followed by rapid capping (Mejillano et al., 2004).
Despite the significant recent progress in understanding the molecular mechanisms of actin-based protrusion, the identity of actin filament nucleators responsible for formation of filopodia remains unknown. We previously proposed that Arp2/3 complex-dependent nucleation provides a filamentous resource to build filopodial bundles (Svitkina et al., 2003). Other data showing Arp2/3 complex at the bases of filopodia (Lorenz et al., 2004; Diez et al., 2005) are also consistent with this scenario. Furthermore, lamellipodial protrusion could be shifted to filopodial protrusion by inhibiting capping protein (Mejillano et al., 2004), suggesting that lamellipodia and filopodia are dynamically interrelated and their filaments may be generated by common nucleator(s). An alternative model proposes that formins are solely responsible for nucleation of filopodial filaments, whereas Arp2/3 complex is specialized for lamellipodia and is not necessary for filopodia formation (Millard et al., 2004; Nicholson-Dykstra et al., 2005; Faix and Rottner, 2006). This idea has been inspired by the discovery of nucleating activity of formins (Pruyne et al., 2002; Zigmond, 2004) and their involvement in filopodia formation (Peng et al., 2003; Pellegrin and Mellor, 2005; Schirenbeck et al., 2005), and supported by reports that Arp2/3 complex is functionally dispensable for filopodia formation (Strasser et al., 2004; Steffen et al., 2006). Disagreement between these sets of data prompted us to apply a previously unexplored combination of functional, kinetic, and structural approaches to thoroughly investigate a role of Arp2/3 complex in filopodia formation using neuronal cells as a model system. We report here that Arp2/3 complex functionally contributes to generation of filopodia and also seems to be involved in regulation of RhoA activity.
MATERIALS AND METHODS
Reagents
Plasmids and Constructs.
Human enhanced yellow fluorescent protein (EYFP)-Arp3 and mouse pDsRed2-cortactin were gifts of Shin-ichiro Kojima (Northwestern University, Chicago, IL); enhanced green fluorescent protein (EGFP)-VASP was a gift of Frank Gertler (Massachusetts Institute of Technology, Cambridge, MA); the constitutively active green fluorescent protein (GFP)-Rac1V12 was a gift of Kozo Kaibuchi (Nagoya University, Japan), and EYFP-actin was from Clontech (Mountain View, CA). Transient transfection of cells with DNA constructs was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Antibodies.
The following rabbit polyclonal antibodies were used: p16-Arc subunit of Arp2/3 complex (Vignjevic et al., 2003), nonmuscle myosin II from bovine spleen (Verkhovsky et al., 1987); spectrin (Chemicon International, Temecula, CA); Arp3 (Santa Cruz Biotechnology, Santa Cruz, CA), and p34-Arc subunit of Arp2/3 complex (Upstate Biotechnology, Charlottesville, VA). The following mouse monoclonal antibodies were used: cortactin (clone 411F; Upstate Biotechnology), fascin (clone 55K2; Chemicon International), and vinculin (Sigma-Aldrich, St. Louis, MO). Secondary antibodies were from Invitrogen or Jackson ImmunoResearch Laboratories (West Grove, PA). AlexaFluor 488-, 594-, and 647-labeled phalloidins were from Invitrogen. All other reagents were from Sigma-Aldrich unless indicated otherwise.
Cell Culture
B35 rat neuroblastoma cells (Otey et al., 2003) and NG108-15 rat/mouse hybrid neuroblastoma cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 2 mM glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). For microscopy, cells were plated onto coverslips or glass-bottomed dishes coated sequentially with 1 mg/ml poly-l-lysine and 1.25 μg/ml laminin. To induce differentiation, cells were first cultured in a regular culture medium for 24 h after plating and then transferred to a differentiation medium consisting of DMEM with 0.1 mM dbcAMP and N2 supplement (Invitrogen).
Xenopus neural crest cells were prepared as described previously (Chang et al., 1999) and plated on glass coverslips precoated with 0.1 to 1.0 μg/cm2 concanavalin A. The cultures were kept at 20°C in a culture medium consisting of (vol/vol) 50% Leibovitz L-15 medium (Invitrogen), 49% Ringer's solution (115 mM NaCl, 2 mM CaCl2, 2.5 mM KCl, and 10 mM HEPES, pH 7.6), 1% fetal bovine serum (Invitrogen), and 50 ng/ml neurotrophin-3. Experiments were performed 24–48 h after cell culture preparation (Zakharenko and Popov, 2000).
Rat hippocampal neurons isolated as described previously (Wilcox et al., 1994) and grown on poly-l-lysine-coated (1 mg/ml) coverslips in serum-free Neurobasal media (Invitrogen) with B27 supplement (Invitrogen) and without antibiotics were kindly provided by Marc Dichter or Rita Balice-Gordon (University of Pennsylvania). Growth cones were analyzed after cultivation for 3–5 d in vitro.
Immunofluorescence
For detection of F-actin in cells, staining with fluorescently labeled phalloidin (0.033 μM) was performed after fixation with 4% paraformaldehyde and permeabilization with 0.1% Triton X-100 in phosphate-buffered saline (PBS). Immunostaining was usually performed after cell extraction for 5 min with 1% Triton X-100 in PEM buffer (100 mM PIPES-KOH, pH 6.9, 1 mM MgCl2, and 1 mM EGTA) optionally containing 4% polyethelene glycol (PEG) (mol. wt. 35,000) and 2 μM phalloidin, followed by fixation with 4% paraformaldehyde or 0.2% glutaraldehyde. Glutaraldehyde-fixed samples were quenched with 2 mg/ml NaBH4. For p34-Arc immunostaining, extracted unfixed cells were incubated with primary antibody in PEM buffer containing 5 μM unlabeled phalloidin for 15 min, and then they were washed with PEM buffer, fixed with 4% paraformaldehyde, and stained with secondary antibody. For VASP staining, cells were extracted with a mixture containing 0.5% paraformaldehyde and 1% Triton X-100 in PEM buffer, and then they were fixed with 4% paraformaldehyde and costained with phalloidin. Fascin staining was performed after methanol fixation without extraction. Extracted and paraformaldehyde-fixed samples were used for myosin II staining. For costaining of F-actin, labeled phalloidin was added to the secondary antibody solution.
Microscopy
Light microscopy was performed using an inverted microscope (Eclipse TE2000; Nikon, Tokyo, Japan) equipped with Planapo 100 × 1.3 or 20 × 0.75 objectives and Cascade 512B charge-coupled device camera (Roper Scientific, Trenton, NJ) driven by MetaMorph imaging software (Molecular Devices, Sunnyvale, CA). For live cell imaging, cells were plated on glass-bottomed dishes coated with appropriate substrates and kept on the microscope stage at 35°C under a layer of mineral oil. No oil was applied to dishes imaged for correlative microscopy. For overnight movies, cells were transferred into bicarbonate-free DMEM.
Samples for platinum replica electron microscopy (EM) and for correlative light and electron microscopy were processed essentially as described previously (Svitkina and Borisy, 2006). Hippocampal neurons for EM were extracted in a mixture of 0.25% glutaraldehyde and 0.5% Triton X-100 in PEM buffer for ∼5 min and postfixed with 2% glutaraldehyde. Other cells were extracted as for immunofluorescence but without addition of PEG. Immunoelectron microscopy with p16-Arc and p34-Arc antibodies was performed as described previously (Svitkina and Borisy, 2006). Primary p34-Arc antibody was applied to unfixed cells as for immunofluorescence, followed by fixation with 2% glutaraldehyde. Samples were analyzed using JEOL 1200EX (JEOL, Tokyo, Japan) transmission EM operated at 120 kV. For three-dimensional (3D) images, stereo pairs were taken at +/−6–20° angles by rotating a grid holder. Anaglyph images were produced by placing one image of the stereo pair into the red channel and another into the green and blue channels of the same RGB image by using Adobe Photoshop software (Adobe Systems, Mountain View, CA). Red/cyan or red/green anaglyph glasses (left eye is red) should be used to view 3D images.
RNA Interference (RNAi)
Small interfering RNA (siRNA) duplexes against following target sequences were used: 5′-GAAAGGCGTGGATGACCTA-3′ (rat Arp3) and 5′-AAGGAACTTCAGGCACACGGA-3′ (rat p34-Arc subunit of Arp2/3 complex, a.k.a. ARPC2) were obtained from Dharmacon RNA Technologies (Lafayette, CO); control siRNA was obtained from QIAGEN (Valencia, CA). siRNAs were labeled with cyanine (Cy)3 or fluorescein isothiocyanate (FITC) by using a Silencer siRNA labeling kit (Ambion, Austin, TX) and used at 20 nM concentration. Transfection with siRNA was performed using Lipofectamine 2000 (Invitrogen) 24 h after plating for neuroblastoma cells and after 1–3 d in vitro for hippocampal neurons. Effects of siRNA were analyzed 2–3 d after transfection. Efficiency of siRNA transfection was ∼95% of the cell population for B35 cells and ∼30–50% for hippocampal neurons. For immunoblotting, B35 cell lysates were prepared 2 or 3 d after siRNA transfection. For rescue experiments, B35 cells were cotransfected with Arp3 siRNA and human pEYFP-Arp3, and they were analyzed 2 d posttransfection as undifferentiated cells, or they were switched to differentiation conditions 24 h after transfection and analyzed 1–2 d later. For the overnight differentiation assay, cell cultures transfected with Cy3-p34-Arc or FITC-control siRNAs were mixed and plated into the same dish. Control and inhibited cells were distinguished by the color of the siRNA label.
Western Blot
Cells were washed with PBS at 4°C and lysed either in a buffer containing 10 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol, and a protease inhibitor tablet (Roche Diagnostics) or in 1% SDS solution in water. Protein concentration in lysates was determined using bicinchoninic acid reagent (Pierce Chemical, Rockford, IL). SDS-polyacrylamide gel electrophoresis (10% polyacrylamide), and immunoblotting were performed according to standard protocols. Spectrin was used as a loading control. Immunoblots were developed using enhanced chemifluorescence Western blotting kit (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Determination of RhoA Activity
B35 cells growing in six-well plates were transfected with control or p34-Arc siRNA, and they were allowed to grow to ∼60% confluence. After 24 h of transfection, cells were switched to differentiating conditions, and cell lysates were prepared 1 h later. After equalization of total protein concentrations, lysates were assayed using G-LISA Active RhoA Assay kit (Cytoskeleton, Denver, CO) according to the manufacturer's protocol. In this assay, active RhoA from lysates binds to Rhotekin RBD domain immobilized in the 96-well plate and is subsequently detected by immunostaining with a primary RhoA antibody and an HRP-conjugated secondary antibody. The intensity of HRP reaction product is then determined by colorimetry at 490-nm wavelength, and results are expressed in units of optical density.
Image Analysis and Statistics
All morphometric measurements were done using MetaMorph Imaging software (Molecular Devices) unless indicated otherwise, and they were repeated for at least two independent experiments. At least 10 cells were analyzed to get a value for each parameter. Data were analyzed using Excel (Microsoft, Redmond, WA) or SigmaPlot (Systat Software, Point Richmond, CA) software packages. Significance was determined using a two-tailed t test.
To determine changes in the F-actin contents in growth cones of B35 neuroblastoma, cells were extracted as described above, and then they were fixed with 4% paraformaldehyde, stained with fluorescently labeled phalloidin, and imaged using the same acquisition settings. An average intensity of phalloidin fluorescence after background subtraction was measured for regions of interest containing a 2-μm-wide area adjacent to the growth cone's leading edge. To determine changes in Arp2/3 complex contents in hippocampal neurons after p34-Arc siRNA treatment, cells were extracted and stained with p34-Arc antibody and phalloidin as described above. An average intensity of phalloidin and p34-Arc staining after background subtraction was determined for entire growth cone area.
For quantification purposes, lamellipodia in growth cones were defined as regions of dense actin network adjacent to the leading edge and positioned between neighboring filopodial bundles. The width of lamellipodia was determined using Adobe Photoshop software by measuring the distance from the leading edge of the growth cone to the beginning of a significantly sparser actin network along a line orthogonal to the leading edge. Actin filament branch frequency (number of branches per unit area), and distribution of Arp2/3 complex immunogold labeling (number of gold particles per unit area) in lamellipodial regions of growth cones was determined using high-resolution EM images. Stereo images were used for quantification of branches. Areas between neighboring filopodial bundles spanning a zone from the leading edge to up to 3 μm inward were used for the analysis. Branches or gold particles in 0.2-μm-wide and 1-μm-long slices were scored, expressed as density per square micrometer, and plotted against the distance from the edge. Only clearly visible gold particles and Y-shaped actin filament configurations were scored.
Parameters of lamellipodia and filopodia dynamics were determined using phase-contrast or fluorescence (for YFP-actin–expressing cells) time-lapse sequences acquired with 2- to 3-s intervals between frames at 100× magnification. Both approaches gave quantitatively similar results for each data set. Rates of lamellipodia protrusion and retrograde flow were measured using kymographs generated along straight lines drawn perpendicular to the leading edge. Slopes produced by advancing leading edges or by retrogradely moving lamellipodial features were used to calculate the rates. For quantification of filopodia dynamics, a distance from a fixed point within a filopodium to the filopodial tip was measured every 10 s while the filopodium existed and movie lasted. A filopodium was considered pausing if its length change was <2 pixels for at least two time intervals. Data are expressed as a fraction of time which filopodia spent protruding, retracting or pausing.
The filopodia frequency was defined as the number of filopodia per unit length of growth cone perimeter, and it was determined using still phase-contrast images. Only filopodia extending beyond the cell edge were counted. The length of the growth cone perimeter was determined using MetaMorph line function. The rate of filopodia initiation was determined by counting newly formed filopodia during the period of observation (∼4–5 min) per 10 μm of the growth cone perimeter per minute.
Neurite extension during differentiation of B35 neuroblastoma cells was analyzed using cells plated on coverslips or glass-bottomed dishes 1–2 d after transfection with control or inhibitory siRNA and induced to differentiate 1 d later. Total number and length of neurites was determined using fixed samples on coverslips. A number of neurites in hippocampal neurons was determined after 2–3 d in vitro and 24 h after transfection with control or inhibitory siRNA by using fixed and phalloidin-stained samples. For dynamics of neurite extension, phase-contrast time-lapse sequences were acquired from the beginning of differentiation for 10–12 h with 5-min time intervals using 20× objective. The length of each neurite was plotted against time.
RESULTS
Initiation of Filopodia in Neuronal Cells
Significant domination of filopodia over lamellipodia in neuronal growth cones suggests that filopodia in these cells may be formed not from lamellipodia, but from any site on the cell surface. We used time-lapse microscopy to determine a pathway of filopodia initiation in four cell types of neuronal origin, two neuroblastoma cell lines (rat B35 and rat/mouse hybrid NG108) and two primary neuronal cultures (rat hippocampal and Xenopus neural crest neurons).
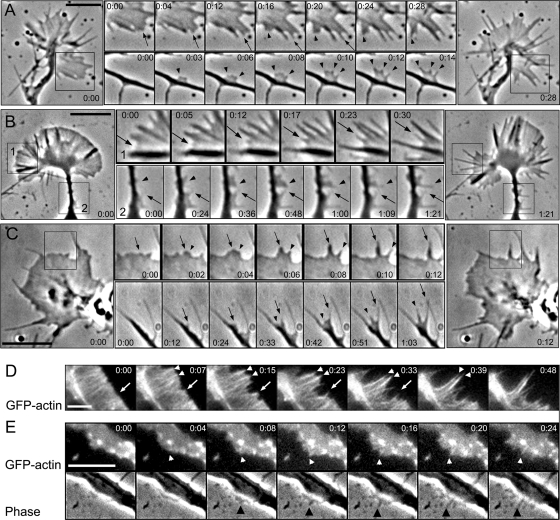
Analysis of phase contrast (Figure 1, A–C; and Supplemental Video 1) and fluorescence (Figure 1, D and E) sequences demonstrated that the majority of nascent filopodia arose from a lamellipodium-like protrusion in all tested neuronal cells. In growth cones, the common sites of filopodia formation were interfilopodial veils. The filopodia initiation at these sites began as a small phase-dense bulge (Figure 1, A–C) similar to previously described “nubs” in chicken dorsal root ganglia neurons (Steketee et al., 2001) and to filopodial Λ-precursors in B16F1 melanoma cells (Svitkina et al., 2003). Filopodia also emerged from neuronal shafts, usually close to growth cones. Although these sites seemed to lack lamellipodia, careful analysis of such events revealed that initiation of lateral filopodia also began as a lamellipodial structure, which then quickly produced filopodia through intermediate formation of small bulges (Figure 1, A–C; and Supplemental Video 2). Dynamics of YFP-actin in B35 neuroblastoma cells also showed filopodia initiation from lamellipodia-like protrusions (Figure 1, D and E). Both protruding filopodia and internal filopodial bundles (frequently called microspikes or ribs) were formed by gradual condensation of actin fluorescence with intermediate formation of cone-shaped actin densities (Figure 1D). Similarly, local actin densities served as precursors for formation of lateral filopodia on neuronal shafts (Figure 1E), consistent with published data (Loudon et al., 2006). Together, we have analyzed at least 10 cells for each cell type, and ∼90% of filopodia in these cells were formed from lamellipodia. The remaining ∼10% were ambiguous cases, in which we could not clearly determine how filopodia were formed. We had only one case across all four cell types where a filopodium formed from a phase-dense bulge that did not have a flattened lamellipodia-like shape (Figure 1C, arrow in the lower sequence).
Figure 1.
Filopodia initiation in neuronal cells. (A–C) Phase-contrast time-lapse sequences of rat hippocampal neuron (A), differentiated B35 neuroblastoma cell (B), and Xenopus neuron (C). Large panels at left and right show the beginning and the end of the sequence, respectively. Montages in the middle show individual time frames for the boxed regions in the corresponding large panels (B, all; A and C, top) or for other time-lapse sequences (A and C, bottom). Arrows and arrowheads mark individual filopodia formed during the sequence. Filopodia initiation occurs from lamellipodia-like regions and is usually preceded by formation of a small bulge at the leading edge of a growth cone (A–C, top), or at the tip of a secondary neurite (C, bottom), or within a lateral protrusion (A and B, bottom). (D and E) Dynamics of YFP-actin in a growth cone (D) and a lateral protrusion (E) of B35 cells. A corresponding phase contrast sequence is shown for D. Cone-shaped condensation of actin fluorescence precedes filopodia formation (arrows and arrowheads). Time in minutes:seconds. Bars, 5 μm.
Thus, filopodia in neuronal cells were initiated in association with lamellipodia, similar to B16F1 melanoma cells (Svitkina et al., 2003), despite a notable paucity of lamellipodia in neuronal cells.
Structural Organization of Neuronal Growth Cones
Lamellipodia-like “veils,” which fill the narrow spaces between filopodia in growth cones look very similar to conventional lamellipodia; however, branched filaments and Arp2/3 complex were not detected in growth cones of rat hippocampal neurons (Strasser et al., 2004), but they were found in chicken dorsal root ganglion neurons (Mongiu et al., 2007). To control for potential variability between different neuronal cells regarding organization and molecular composition of growth cones, we analyzed the distribution of lamellipodial and filopodial markers and the structural organization of growth cones and lateral protrusions in four different types of neuronal cells.
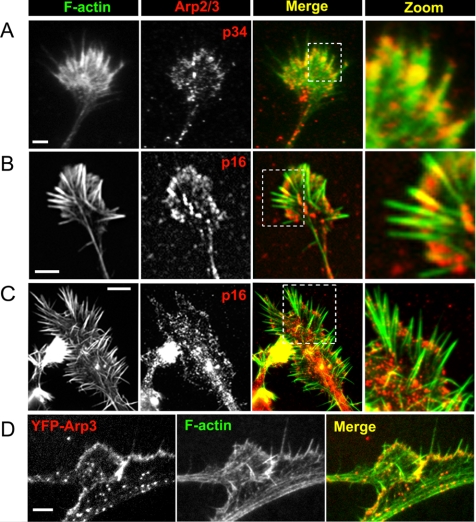
Immunostaining of hippocampal neurons and neuroblastoma cells revealed enrichment of Arp2/3 complex at the periphery of growth cones (Figure 2, A–C), and in lateral protrusions extending from neuronal shafts (data not shown). Arp2/3 complex antibodies also stained the cell body, neurites, and central domains of growth cones, as reported previously (Strasser et al., 2004), but the intensity and pattern of staining in these locations varied depending on cell type, antibody, and sample preparation protocol. Expression of YFP-Arp3 (Figure 2D) in B35 cells confirmed the enrichment of this protein at the growth cone periphery. Another lamellipodial marker, cortactin (Weaver et al., 2003), was also enriched at the growth cone periphery (Supplemental Figure S1B). Time-lapse observations revealed that during filopodia initiation cortactin was present in the pre-existing interfilopodial veils and at the bases of emerging bulges, which served as filopodial precursors, but they became excluded from emerged filopodia (Supplemental Figure S1C). Similar to the previous reports (Cohan et al., 2001; Lebrand et al., 2004), the filopodial markers, fascin and VASP, were associated with filopodial bundles along the length (fascin) or at the tips (VASP); VASP also localized to the edge of veils (Supplemental Figure S1).
Figure 2.
Arp2/3 complex in neuronal cells. (A–C) Localization of Arp2/3 complex (red) by immunostaining with p34-Arc antibody (A) or p16-Arc antibody (B and C) in hippocampal neurons after 4 d in culture (A), or differentiated B35 (B) or NG-108 neuroblastoma cells (C). (D) Expression of YFP-Arp3 in differentiated B35 cells on a background of depletion of endogenous Arp3. Actin (green) is stained by phalloidin (A–D). Boxed regions for A–C are enlarged at right. Arp2/3 complex is present at the periphery of growth cones and variably in the central domains of growth cones and in neuronal shafts. Bars, 2 μm (A, B, and D) or 5 μm (C).
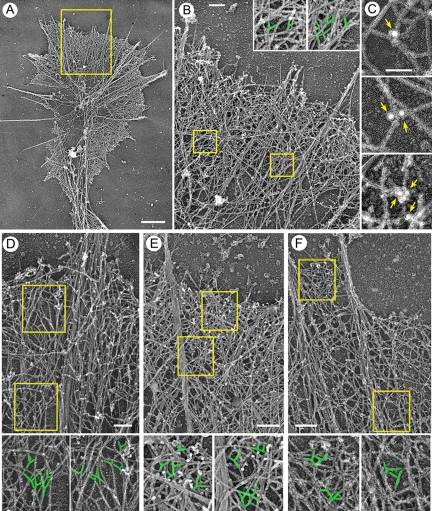
Platinum replica EM was used to determine the structural organization of neuronal protrusions (Figure 3). As expected (Bridgman and Dailey, 1989), long bundled actin filaments comprising the core of a filopodium were abundant in growth cones in all four studied cell types. Bundles either protruded beyond the growth cone margins as genuine filopodia, or they remained within a growth cone, as microspikes or ribs (Figure 3A). Some filopodial bundles were relatively thick and long, and they extended rearward far into the central domain of the growth cone. Others were relatively thin and short and terminated within the peripheral domain, where they splayed apart into thinner bundles and individual filaments.
Figure 3.
Structural organization of actin network in neuronal growth cones. EM of differentiated B35 cells (A–C), differentiated NG-108 cells (D), Xenopus neurons (E), or hippocampal neurons (F). (A) Overview of a growth cone. (B and D–F) Periphery of growth cones contains branched actin filaments. Boxed regions are enlarged in insets (B) or below main panels (D–F); branched filaments are highlighted in green. (C) Immunogold (18 nm) staining of growth cones with p34-Arc antibody. Arrows point to gold particles located at branch points. Bars, 1 μm (A), 0.2 μm (B and D–F), or 0.1 μm (C).
An important question was whether interfilopodial veils contain branched filaments, a signature of Arp2/3 complex-depended nucleation. We used stereo EM to better discriminate branches from filament overlaps (Supplemental Figure S2). This analysis showed that the peripheral actin network in growth cones of all four types of neuronal cells contained numerous branched filaments (Figure 3, B and D–F), especially between major filopodial bundles. To confirm that these branches are formed by Arp2/3 complex, we performed immunoEM with p34-Arc antibody (Supplemental Figure S3E), and we showed localization of gold particles at the branch points (Figure 3C). Filopodial (bundled) and lamellipodial (branched) filaments did not form two distinct sets. On contrary, we observed some filaments originating at a branch point that merged into the bundle and other filaments that branched off a filament in the bundle. EM analysis of lateral protrusions revealed that bulge-like structures contained a dendritic network of actin filaments (Supplemental Figure S2, C and D), whereas lateral filopodia contained actin filament bundles. Gradual transitions between two types of organizations were frequently observed.
These results show that growth cone veils of all tested neuronal cell types contain dendritic networks and the lamellipodial markers, Arp2/3 complex and cortactin, just like conventional lamellipodia, and that individual actin filaments in this network can merge into or arise from an actin bundle.
Age-dependent Lamellipodia-to-Filopodia Progression in Neuronal Protrusions
We performed EM of growth cones with known life histories to correlate the kinetics of filopodia initiation with the structural organization of the underlying actin cytoskeleton. Because the actin network predominantly assembles at the leading edge (Wang, 1985; Watanabe and Mitchison, 2002) and then moves backward relative to the advancing edge without significant reorganization except for some loss of filaments due to depolymerization (Theriot and Mitchison, 1991; Mallavarapu and Mitchison, 1999), the structure of the network allows one to deduce the recent history of the network assembly (Katoh et al., 1999; Svitkina et al., 2003). This analysis demonstrated that protruding veils of differentiated B35 cells contained dense actin networks rich in branched filaments (Supplemental Figure S3, A–D), which confirms the lamellipodial character of these protrusions. A similar observation was also recently reported by Mongiu et al. (2007) for chicken dorsal root ganglia neurons.
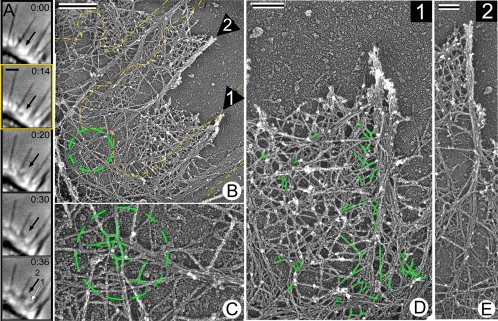
EM analysis of filopodia with known histories showed that the structural organization of filopodia correlated with their age (Figure 4). Nascent filopodia, whether they were formed in the growth cone or at the neuronal shafts, were associated with the branched lamellipodial network at their roots. Two examples of recently formed filopodia are shown in Figure 4, A–E. Branched filaments were especially numerous at the roots of the youngest filopodia, such as filopodium 1 in Figure 4, which was not yet recognizable in the last live image (Figure 4A), but was visible by EM (Figure 4, B and D). Apparently, it formed during a short interval between acquisition of the last live image and cell extraction. In the slightly older filopodium 2 (Figure 4, A–E), which was formed a few seconds before extraction (Figure 4A), the actin network in the root was relatively sparse, apparently due to actin depolymerization, but branched filaments could still be detected there (Figure 4, B, C, and E). The EM structure can be correlated not only with the last frame of the movie, but also with the previous frames. When the life history of filopodium 2 was projected onto respective EM images, the phase contrast bulge, a filopodium precursor that occurred at time 0:14, corresponded to the filopodium root with branched filaments (Figure 4B), suggesting that the branched network served as a precursor for the filopodium formation. Importantly, some filaments in the nascent filopodial bundles could be traced back to their origin at a branch point (Figure 4C). In older filopodia, a fraction of filaments terminated with apparently free (not engaged in branch formation) “pointed” (as predicted from their orientation relative to the cell edge) ends (data not shown). The origin of many other filaments was obscured.
Figure 4.
Nascent filopodia contain branched filaments in their roots. Correlative EM of B35 cells. (A) Time-lapse sequence of a growth cone segment shows formation of nascent filopodia (arrows). Black arrow marks formation of a small bulge (0:14), which then forms a filopodium (0:20); the filopodium then becomes partially engulfed by an advancing lamellipodium (0:30–0:36). White arrow marks a small protrusion adjacent to a mature filopodium (0:36). (B) EM of the region shown in A overlaid with a cell contour (yellow line) from 0:14 frame, when the bulge first occurred. Numbered arrows point to corresponding regions in A (0:36); they contain filopodial bundles. Encircled region is enlarged in C. (C–E) High magnifications of nascent filopodia 1 and 2 show branched filaments in their roots (C and D, highlighted in green). Note, that branched filaments in the root of filopodium 2 (C) correspond to the bulge region (B), from which the filopodium emerged. Some actin filaments originated as branches enter the filopodial bundles. Time in minutes:seconds. Bars, 1 μm (A), 0.5 μm (B), or 0.2 μm (D and E).
These results show that nascent filopodia are intimately associated with the lamellipodial network and contain filaments originating at the branch points, while unbranched filaments are more apparent in older filopodia.
Arp2/3 Complex Depletion by siRNA Impairs Lamellipodia and Filopodia in Growth Cones
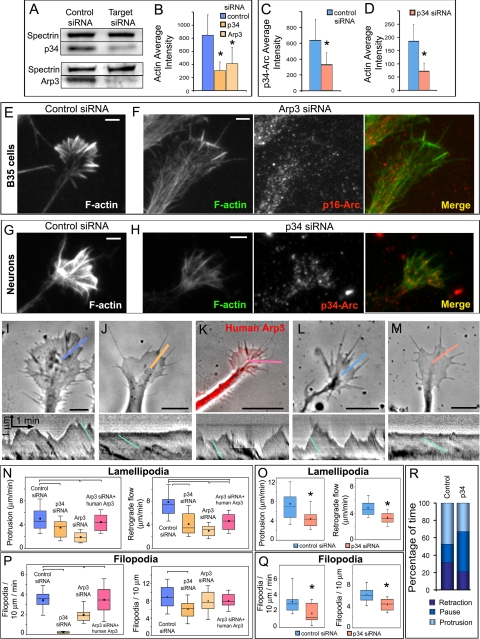
The functional contribution of Arp2/3 complex to growth cone dynamics and filopodia formation in B35 neuroblastoma cells and primary rat hippocampal neurons was investigated using RNAi (Figure 5). Transfection with fluorescently labeled siRNA directed against p34-Arc or Arp3 subunits of the Arp2/3 complex suppressed expression of these proteins after 2–3 d of transfection by ∼50%, as estimated by Western blotting for B35 cells (Figure 5A) and by immunostaining for hippocampal neurons (Figure 5C). Immunofluorescence images showed that Arp2/3 complex depletion was especially evident at the periphery of growth cones in siRNA-treated cells (Figure 5, F and H).
Figure 5.
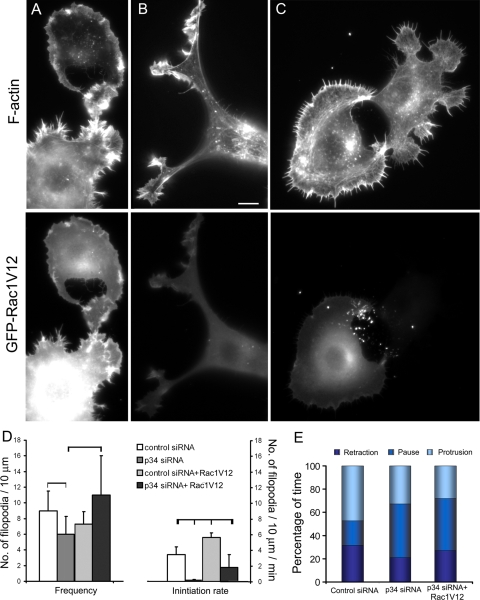
Arp2/3 complex knockdown inhibits lamellipodia and filopodia in B35 neuroblastoma cells and rat hippocampal neurons. (A) Western blotting of undifferentiated B35 cells transfected with control, p34-Arc or Arp3 siRNA. Spectrin is used as loading control. Expression levels of Arp2/3 complex subunits are decreased by ∼50%. (B) Quantification of F-actin in growth cones of differentiated B35 cells transfected with control (n = 25 growth cones), p34-Arc (n = 33) or Arp3 (n = 23) siRNA and stained with fluorescently labeled phalloidin (p < 0.001). (C and D) Quantification of p34-Arc immunofluorescence (C) or F-actin (D) in growth cones of hippocampal neurons transfected with control (n = 29 growth cones) or p34-Arc (n = 15) siRNA (p < 0.001). (E–H) Staining of F-actin by phalloidin and Arp2/3 complex by p16-Arc (F) or p34-Arc (H) antibody in B35 cells (E and F) or hippocampal neurons (G and H) transfected with indicated siRNA. Both actin and Arp2/3 complex components are decreased at the growth cone periphery of knockdown cells (compare phalloidin staining in E and F and in G and H). Images of control and knockdown cells were acquired and processed identically. (I–M) Growth cone dynamics in control (I and L), p34-Arc siRNA-treated (J and M), and rescued (cotransfected with Arp3 siRNA and human YFP-Arp3, K) differentiated B35 cells (I–K) and hippocampal neurons (L and M). Lower panels show kymographs generated along color lines in top panels. Lamellipodia in control (I and L) and rescued (K) growth cones undergo repetitive cycles of protrusion and retraction, whereas siRNA-treated cells (J and M) have relatively quiescent leading edges. Rates of retrograde flow are decreased in knockdown cells (J and M) compared with control (I and L) and rescued cells (K). Examples of slopes used for quantification are shown in kymographs by light blue lines. (N and O) Rates of lamellipodia protrusion (left plots) and retrograde flow (right plots) in differentiated B35 cells (N) and hippocampal neurons (O) transfected as indicated. (P and Q) Filopodia frequency (left plots) and rates of filopodia initiation (right plots) in differentiated B35 cells (P) and hippocampal neurons (Q) transfected as indicated. Plots in N–Q are generated based on analysis of phase contrast movies. Top and bottom of a box indicate 75th and 25th quartiles; whiskers indicate 10th and 90th percentiles; dot is the mean; line is the median. Differences between data sets connected by brackets (N, P) are statistically significant (p < 0.001). Asterisks (O and Q) indicate statistical significance compared with control (p < 0.001). n = 21–49 events for N and O and 11–39 growth cones for P and Q). (R) Filopodia dynamics in control and p34-Arc siRNA-treated differentiated B35 cells. Bar segments represent percentage of time spent by filopodia in protrusion, pause, or retraction phases, as indicated (n = 57–119 filopodia). Bars, 2 μm (E–H) or 5 μm (I–M).
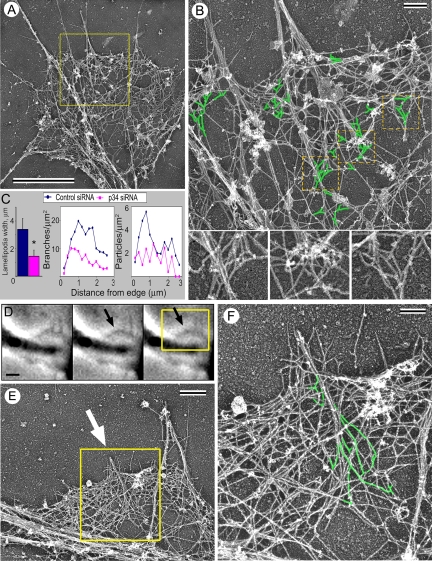
Arp2/3 complex knockdown caused an approximately twofold decrease in the F-actin content in growth cones of both cell types (Figure 5, B, D, and E–H). Decreased amount of actin filaments in growth cones of Arp2/3 complex-depleted B35 cells, especially in their internal regions, was also clearly visible by electron microscopy (Figure 6A, compare with Figure 3A).
Figure 6.
EM of Arp2/3 complex knockdown growth cones of B35 cells. (A) Growth cone of p34-Arc siRNA-treated differentiated cell shows sparse actin network in lamellipodia and few remaining filopodial bundles (compare with Figure 3). (B) Enlarged boxed region from A shows filopodia and lamellipodia in detail. Branched filaments (green) are seen in the sparse lamellipodial network. Many filopodial filaments originate from branch points. Boxed regions in B are further enlarged at bottom without highlighting. (C) Inhibition of lamellipodia in growth cones of differentiated p34-Arc siRNA-treated cells, as revealed by quantification of the lamellipodium width (left), density of actin filament branches (middle), or density of gold particles after immunostaining with p16-Arc antibody (right) (n = 6 cells for each). Branch and immunogold densities are plotted against the distance from the leading edge. (D–F) Correlative EM of nascent filopodia in p34-Arc siRNA-treated differentiated B35 cells. (D) Time-lapse sequence of a growth cone shows formation of a small bulge at the end of the sequence (arrow). (E) EM of the region corresponding to the boxed area in E. Arrow marks a filopodial bundle formed at the position of the bulge. (F) Enlarged boxed region in E. Branched filaments (green) are seen in the root of this thin nascent filopodium. Time in minutes:seconds. Bars, 2 μm (A and D), 0.5 μm (B and E), or 0.2 μm (F).
Arp2/3 depletion severely inhibited lamellipodia in both differentiated and nondifferentiated B35 cells and in hippocampal neurons. Thus, the rates of lamellipodia protrusion and retrograde flow were significantly decreased compared with cells treated with control siRNA (Figure 5, I–O; Supplemental Figure S3; and Supplemental Videos 3 and 4). Coexpression of human YFP-Arp3 resistant to rat-specific siRNA rescued the inhibition of the lamellipodial dynamics in Arp3 siRNA-treated B35 cells (Figure 5, K and N; Supplemental Figure S3; and Supplemental Video 3), demonstrating the specificity of the phenotype. Electron microscopy of Arp2/3 complex-depleted cells demonstrated that lamellipodia were narrower and contained a sparser actin network with fewer branches and decreased immunogold labeling by p16-Arc antibody (Figure 6, A–C).
Importantly, depletion of the Arp2/3 complex inhibited growth cone filopodia in differentiated B35 cells and hippocampal neurons (Figure 5, I–M), as judged from the significantly reduced rates of filopodia initiation and decreased filopodia frequency along the growth cone perimeter (Figure 5, P and Q). Two siRNAs (p34-Arc and Arp3) produced comparable effects in B35 cells, but Arp3 siRNA seemed less potent and significantly inhibited the initiation rate, whereas a decrease in the steady-state filopodia frequency was not statistically significant. Inhibition of filopodia initiation by Arp3 siRNA could be rescued by coexpression of human YFP-Arp3. Additionally, p34-Arc siRNA treatment decreased the dynamic behavior of filopodia in B35 cells, namely, filopodia spent more time pausing and less time protruding or retracting (Figure 5R), whereas the velocity of filopodia protrusion during protrusive phases did not change. Filopodia in Arp2/3 complex-depleted B35 cells, like in control cells, contained VASP at their tips and fascin along the length of the bundles, but the intensity of staining was lower, apparently reflecting the decreased actin contents (Supplemental Figure S1).
Because of incomplete inhibition of filopodia in siRNA-treated cells and partial Arp2/3 complex depletion, we investigated how the remaining filopodia were formed. A very sparse actin network in growth cones of Arp2/3 complex-depleted B35 cells allowed us to follow many filopodial actin filaments back to their origin (Figure 6). Remarkably, many filopodial filaments originated as a branch on another filament (Figure 6B), similar to normal filopodia (Figure 4, B–E). Branched actin filaments entering filopodial bundles were found in all analyzed filopodia (10 filopodia in 4 cells), suggesting that filopodial bundles in Arp2/3 complex-depleted cells might be formed preferentially at sites of remaining activity of the Arp2/3 complex. Consistent with this idea, correlative EM of cells with known histories showed that nascent filopodia in Arp2/3 complex-depleted cells were associated with the dendritic network and their filopodial filaments originated at branch points (Figure 6, D–F).
Together, these results indicate that Arp2/3 complex is functionally involved in generation of both lamellipodia and filopodia, because Arp2/3 complex depletion inhibits lamellipodial protrusion, as well as filopodia initiation and dynamics. Incomplete inhibition of filopodia is likely related to partial knockdown of Arp2/3 complex, because the remaining filopodia were formed in association with the remaining dendritic network.
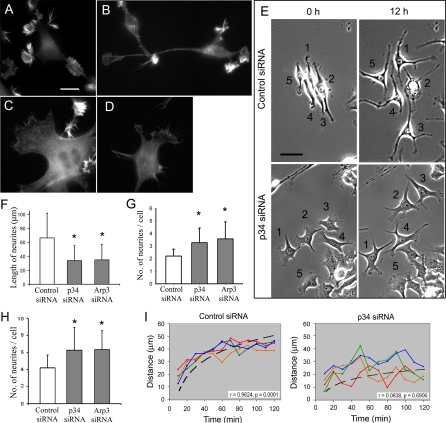
Arp2/3 Complex Is Important for Neuronal Differentiation and Motility
We next investigated whether Arp2/3 complex depletion changes neuritogenesis. We primarily used B35 neuroblastoma cells, because their differentiation can be induced at a desired time, when Arp2/3 complex is sufficiently depleted. Undifferentiated B35 cells had certain features of differentiated neurons, such as abundant filopodial bundles in protrusions and neurite-like processes. However, these “neurites” were short and usually completely covered by lateral protrusions (Figure 7A). Induction of differentiation in B35 cells led to extension of one to three dominant neurites, gradual loss of protrusive activity at the sides of neurites, and rounding of the cell body (Figure 7B).
Figure 7.
Neuritogenesis is impaired in Arp2/3 complex-depleted B35 cells and hippocampal neurons. (A–D) Phalloidin staining of control (A and B) and p34-Arc-treated (C and D) undifferentiated (A and C) and differentiated (B and D) B35 cells. (E) Mixed population of B35 cells transfected with Cy3-p34-Arc or FITC-control siRNA were imaged immediately (left column) or 12 h (right column) after initiation of differentiation. Numbers mark the same cells at both time points. In the top row, all numbered cells contain control siRNA except cell 2, which contains p34-Arc siRNA, but dies during the movie. In the lower row, all numbered cells contain p34-Arc siRNA. (F and G) Length (F) and number (G) of neurites 48 h after initiation of differentiation in B35 cells treated by control or inhibitory siRNA, as indicated. Asterisks indicate statistical significance compared with control (p < 0.001). n = 152–242 neurites (F) and 63–74 cells (G). (H) Number of neurites in hippocampal neurons 48 h after transfection with control or inhibitory siRNA, as indicated. Asterisks indicate statistical significance compared with control (p < 0.001; n = 56–60). (I) Time course of neurite extension after initiation of differentiation (t = 0) in B35 cells treated with control (left) or p34-Arc (right) siRNA. Each line represents an individual neurite. Dashed lines are curve fitting to a logarithmic function of neurites shown in red in each plot (r, r for plotted variables; p, significance). Bars, 10 μm (A–D) or 5 μm (E).
Arp2/3 complex depletion changed the overall morphology of undifferentiated B35 cells and impaired neuritogenesis upon induction of differentiation. Undifferentiated Arp2/3 complex-depleted cells had fibroblast-like morphology with large lamellas fringed by residual lamellipodia and filopodia (Figure 7C). In differentiating conditions, cells formed neurites, but instead of one to three thin and long conventional neurites they extended multiple relatively short and broad processes (Figure 7, F and G), whereas their bodies remained large and flat instead of rounding up (Figure 7, D and E). Instead of small well-defined growth cones, neurites of Arp2/3 complex-depleted B35 cells were usually associated with relatively large lamella, which might or might not be separated from the cell body by a quiescent neurite shaft. In the most severe cases, cells almost completely failed to form neurites and they remained associated with an extensive lamella (e.g., cell 5 in Figure 7E, bottom). Similar to B35 cells, depletion of Arp2/3 complex by p34-Arc or Arp3 siRNA in hippocampal neurons resulted in increased numbers of neurites per cell (Figure 7H), although we could not detect a difference in the neurite length in primary neurons, possibly because of significant extension of neurites that might occur before the protein was knocked down.
The time course of neurite extension in B35 cells after Arp2/3 complex depletion showed abnormal cycles of protrusion and retraction, whereas control neurites displayed relatively smooth growth (Figure 7I and Supplemental Video 5). During extension phases, however, the rate of neurite elongation was not significantly different in control and Arp2/3 complex-depleted cells.
Depletion of the Arp2/3 Complex Induces RhoA Activation in Cells
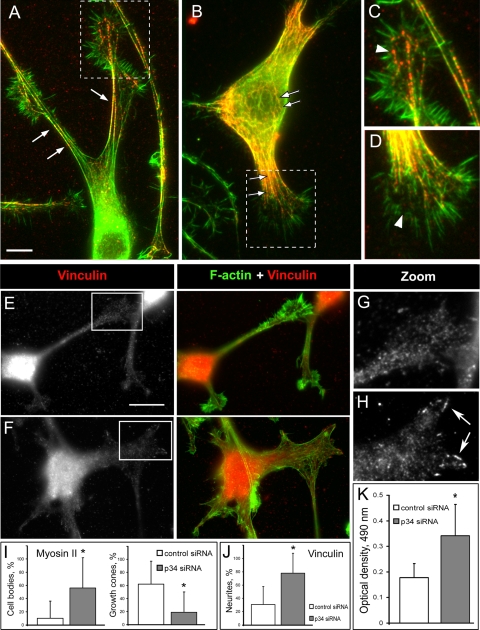
Defective neuritogenesis in Arp2/3 complex-depleted cells, in particular, an erratic style of neurite extension, is hard to explain by the decreased dynamics of the growth cones. Rather, it suggests additional changes, likely in adhesion and/or contraction. Stress fibers and focal adhesions perform major contractile and adhesive functions in many cell types. Therefore, we investigated the organization of stress fibers in control and Arp2/3 complex-depleted B35 cells.
Control differentiated B35 cells had a few slender F-actin bundles along the edges of neurites. Some shorter bundles were also present in the neurite interior and, in minor amounts, in the cell body (Figures 7B and 8A). Immunostaining with myosin II antibody revealed a characteristic punctate pattern along actin bundles (Figure 8, A and C), similar to that in fibroblast stress fibers (Verkhovsky et al., 1995). In addition, bright myosin II puncta were scattered in the central domains of growth cones, where they occasionally colocalized with filopodial roots (Figure 8C), as also reported previously (Bridgman and Dailey, 1989; Medeiros et al., 2006). Relatively dim myosin II structures were also found in the cell body (Figure 8A). In fibroblasts, stress fibers are usually associated with prominent focal adhesions, which can be visualized by a focal adhesion marker, vinculin. Vinculin immunostaining in differentiated B35 cells showed a mostly weak punctate distribution throughout the cell with only occasional enrichment at the tip of the bundles (Figure 8, E and G).
Figure 8.
Arp2/3 complex depletion induces stress fibers and activates RhoA. (A–D) Myosin II immunostaining (red) in control (A and C) and Arp3 siRNA-treated (B and D) cells. Actin is labeled by phalloidin (green). Boxed regions in A and B are enlarged in C and D, respectively. Note increased formation of actin bundles in neurites and cell body (arrows) and decreased amount of myosin II puncta in growth cones (arrowheads) of knockdown cells. (E–H) Vinculin immunostaining (red) in control (E and G) and p34-Arc siRNA-treated (F and H) cells. Actin is labeled by phalloidin (green). Boxed regions in E and F are enlarged in G and H, respectively. Note increased formation of focal adhesions in p34-Arc siRNA-treated cells (arrows). (I) Quantification of myosin II immunostaining showing percentage of cells with high contents of myosin II-containing bundles in the cell bodies (left) and percentage of growth cones with peripheral myosin II puncta (right) in B35 cells expressing control and p34-Arc siRNA. (J) Quantification of vinculin immunostaining showing percentage of neurites with focal contacts in B35 cells expressing control and p34-Arc siRNA. (K) RhoA activity in control and p34-Arc siRNA-treated cells. Asterisk indicates statistical significance (p < 0.05; n = 4). Bars, 5 μm (A and B) or 10 μm (E and F).
Arp2/3 complex-depleted B35 cells, which were induced to differentiate for 1–2 d, displayed more actin filament bundles in neurites and the cell body compared with control cells (Figure 8, B and D). Long bundles were present not only at the edges of neurites but also in their interior. Cell bodies were filled with numerous nonoriented short bundles (Figure 8B). Accordingly, myosin II staining was more pronounced in the cell body of knockdown cells compared with control cells (Figure 8I, left). Conversely, the number and brightness of myosin II puncta in the central domains of growth cones were significantly diminished or almost completely disappeared after Arp2/3 complex depletion except for puncta associated with the actin bundles extending from the neurite (Figure 8D). Quantification showed that myosin II puncta were significantly diminished in the central domain of Arp2/3 complex-depleted growth cones (Figure 8I, right). Expression of YFP-Arp3 reversed the excessive formation of myosin II-containing structures in neurites (Supplemental Figure S1E). The vinculin staining revealed a significant increase of focal adhesions in neurites of Arp2/3 complex-depleted cells (Figure 8, E–H and J).
The observed changes in the neurite behavior and organization of actin bundles after Arp2/3 complex depletion were reminiscent of a phenotype induced by activation of a small GTPase RhoA (Govek et al., 2005). Therefore, we tested whether activity of RhoA changed after Arp2/3 complex depletion. Lysates of differentiating control and siRNA-treated cells were assayed using G-LISA RhoA kit (see Materials and Methods). The results showed a significant increase in RhoA activity in p34-Arc siRNA-treated cells (Figure 8K).
Effects of Rac1V12 Expression on Arp2/3 Complex Knockdown Phenotype
Activities of RhoA and Rac1 in cells frequently display negative relationship with each other (Govek et al., 2005). Therefore, Arp2/3 complex depletion might inhibit filopodia indirectly through inhibition of Rac1 associated with up-regulation of RhoA. To control for this possibility, we investigated a phenotype of p34-Arc-depleted B35 cells after expression of a constitutively active Rac1, GFP-Rac1V12.
Control B35 cells transfected with Rac1V12 and induced to differentiate had variable cell morphology, but all were severely impaired in neuritogenesis (Figure 9, A and B). The majority of cells failed to produce neurites, and their cell bodies were surrounded by protrusions, either by a smooth lamellipodium with no signs of filopodia, or by a mixture of lamellipodia and filopodia (Figure 9A). Some weakly expressing cells displayed rudimentary neuritogenesis with multiple short processes tipped with lamellipodia and filopodia (Figure 9B). Quantification of filopodia in the subset of cells expressing filopodia (cells without filopodia were excluded from quantification) showed that the filopodia frequency did not change, but the rate of filopodia initiation increased by ∼70% after Rac1V12 expression (Figure 9D).
Figure 9.
Expression of GFP-Rac1V12 in B35 cells. (A–C) Phalloidin staining (top row) of differentiated control (A and B) and p34-Arc siRNA-treated (C) cells expressing GFP-Rac1V12 (bottom row). GFP-Rac1V12 and phalloidin image sets are processed identically, so the brightness of fluorescence reflects the level of Rac1V12 expression or F-actin contents, respectively. Representative cell morphologies are shown. (D) Filopodia frequency (left plot) and rates of filopodia initiation (right plot) in cells transfected with indicated constructs. Values connected by brackets indicate statistical significance (p < 0.01; n = 8–19 cells for each). (E) Filopodia dynamics in cells transfected as indicated. Bar segments represent percentage of time spent by filopodia in protrusion, pause, or retraction phases, as indicated (n = 48–119 filopodia). Bar, 5 μm (A–C).
Cotransfection of p34-Arc siRNA and Rac1V12 also largely prevented B35 cells from forming neurites in differentiating conditions, especially in highly Rac1V12-expressing cells, whereas low-expressing cells formed some rudimentary processes (Figure 9C). Rac1V12 partially rescued filopodia formation in p34-Arc–depleted cells. Thus, the filopodia frequency at the leading edge became indistinguishable from that in control cells with or without Rac1V12 (Figure 9D, left). However, the rate of filopodia initiation, although increased ∼10-fold after Rac1V12 expression in p34-Arc–depleted cells, remained approximately twofold lower than in control cells with or without Rac1V12. These data suggest that the filopodia deficiency in Arp2/3 complex-depleted cells may in part be mediated by altered activities of Rho GTPases, and in part by a deficiency in actin nucleation.
DISCUSSION
Arp2/3 Complex in Filopodia Formation
In this study, we found that Arp2/3 complex is present in neuronal growth cones and functionally contributes to protrusion of lamellipodia and initiation of filopodia.
Existing discrepancies regarding the presence of Arp2/3 complex in neuronal growth cones (Strasser et al., 2004; Mongiu et al., 2007) prompted us first to investigate the Arp2/3 complex requirement for lamellipodial dynamics. Using EM, immunochemistry, and gene expression, we showed the presence of branched actin filaments and Arp2/3 complex at the growth cone periphery of four different neuronal cells, consistent with findings in chicken dorsal root ganglion neurons and PC-12 pheochromocytoma cells (Mongiu et al., 2007) and the presence of the members of the Arp2/3 complex-activating WAVE complex, WAVE1 and Sra1, in growth cones of hippocampal neurons (Kawano et al., 2005). Furthermore, using RNAi we demonstrated that Arp2/3 complex is important for protrusion of growth cone veils, similar to lamellipodia in other cell types (Kunda et al., 2003; Rogers et al., 2003; Steffen et al., 2006; see however, Di Nardo et al., 2005). Discrepancies between our data and those of Strasser et al. (2004), who reported Arp2/3 complex-independent growth cone motility in hippocampal neurons, may potentially derive from an alternative functional approach. Thus, Strasser et al. (2004) expressed the CA fragment of neural Wiskott-Aldrich syndrome (N-WASP), which is expected to sequester Arp2/3 complex, but the expression level might be insufficient, as they used only moderately expressing cells; or the CA construct might be inefficient in this experimental system. In contrast, using a well established and controllable siRNA technique, we observed a clear and reproducible phenotype even after partial depletion of the Arp2/3 complex. The specificity of the phenotype was extensively controlled by using different cell types, different target sequences, nontargeting control siRNA, and rescue by a nontargeted gene.
Importantly, depletion of Arp2/3 complex inhibited filopodia, indicating the functional contribution of Arp2/3 complex to their generation. These observations, however, conflict with the reported absence of the filopodial phenotype in non-neuronal cells after inhibition of Arp2/3 complex (Steffen et al., 2006). Because these authors evaluated filopodia in the presence of the constitutively active Cdc42 after depletion of the WAVE complex, it is possible that N-WASP, another major Arp2/3 complex activator and a Cdc42 effector, produced an alternative source of Arp2/3 complex-nucleated filaments for filopodia initiation.
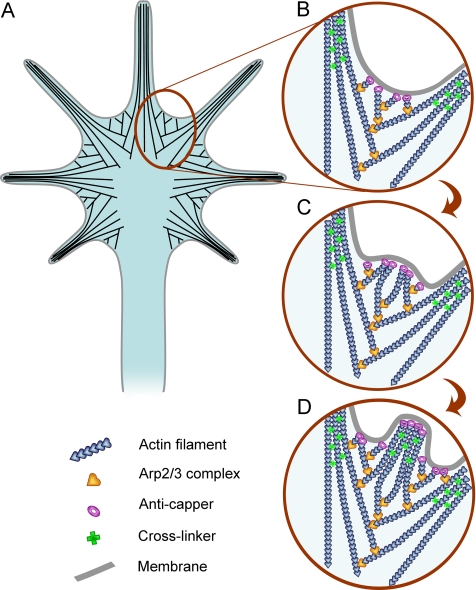
Our study also provides a mechanistic explanation of Arp2/3 complex functions during filopodia formation (Figure 10). Thus, correlative light and electron microscopy revealed that filopodia initiation in growth cones is associated with the dendritic network that is especially prominent in interfilopodial veils (Figure 10, A and B). During protrusion, this network is reorganized into bundles of long filaments (Figure 10, B–D). Activities of anti-capping proteins, such as Ena/VASP proteins (Bear et al., 2002; Lebrand et al., 2004) and formins (Zigmond et al., 2003), and cross-linkers, such as fascin (Vignjevic et al., 2006), are likely responsible for this reorganization. Overall, this process is consistent with the convergent elongation mechanism, we previously proposed for filopodia formation in B16F1 melanoma cells (Svitkina et al., 2003). Lateral filopodia, which were reported to arise from actin patches of unknown structure (Loudon et al., 2006), are formed by a similar mechanism. The constant presence of branched filaments at filopodial roots and their fusion with the filopodial bundles suggests that Arp2/3 complex-mediated nucleation supplies filaments for making filopodia. Although it is possible that some of these branches are formed not by Arp2/3 complex, but by another cross-linking protein, a parallel decrease in branched filaments and Arp2/3 complex labeling after siRNA treatment suggests that the majority of structurally observed branches were made by Arp2/3 complex. Branched filaments also constantly occur in the roots of filopodia remaining after Arp2/3 complex depletion, suggesting that remaining Arp2/3 complex forms preferred sites for filopodia initiation. If filopodia formation was mediated exclusively by formins, as proposed previously (Faix and Rottner, 2006), we would be able to see at least some filopodia not associated with branched filaments after Arp2/3 complex depletion. It should be noted, however, that until complete inhibition of the Arp2/3 complex is achieved, it is not possible to determine whether the dendritic network creates highly favorable conditions for filopodia formation or it is absolutely necessary for it.
Figure 10.
Actin filament organization (A) and the mechanism of filopodia formation (B–D) in neuronal growth cones. (A) Actin filaments (black lines) at the periphery of growth cones are organized into bundles of long filaments in filopodia and branching networks in lamellipodia; these filaments do not form distinct sets, but lamellipodial filaments may branch off of the filopodial filaments and/or merge into filopodial bundles. (B) New filopodia usually form from interfilopodial networks that are rich in Arp2/3 complex (yellow hearts) and branching actin filaments (blue hearts). (C) Arp2/3 complex-nucleated filaments elongate persistently with an assistance of anti-capping molecules (purple rings) and converge to form a bulge-like protrusion at the site of an incipient filopodium. (D) Continued elongation of barbed ends with a concomitant bundling by a cross-linker (green crosses) results in filopodia formation.
The functional contribution of Arp2/3 complex-dependent nucleation to filopodia formation does not contradict a role of formins in this process. On contrary, we found that some filopodial filaments display apparently free “pointed” ends as might be expected for formin-nucleated filaments. Although we cannot exclude the possibility that these linear filaments are produced by severing and/or debranching of Arp2/3 complex-nucleated filaments or simply by rupture during sample preparation, it seems likely based on other studies (Peng et al., 2003; Pellegrin and Mellor, 2005; Schirenbeck et al., 2005; Yang et al., 2007) that at least some of these filaments are nucleated by a formin.
The reorganization of the dendritic network into filopodia seemed more efficient in neuronal cells, compared with B16 F1 cells (Svitkina et al., 2003). We previously proposed a two-step mode of signaling leading to filopodia formation, in which a first step is responsible for dendritic nucleation and a second one for reorganization of the dendritic network into bundles (Biyasheva et al., 2004). Our current data suggest that the relative activities of two proposed signaling modules may be shifted toward the second one in neuronal cells. This hypothesis may also explain the domination of filopodia over lamellipodia in neurons.
Arp2/3 Complex in Neuronal Signaling
Functions of Arp2/3 complex in growth cone dynamics are predicted to influence neuritogenesis, as we indeed observed. Inhibition of axonal outgrowth was also observed after knockdown of Sra-1 or WAVE1 members of WAVE complex in hippocampal neurons (Kawano et al., 2005). However, the interpretation of these results is not straightforward, because we also detected increased RhoA activity after Arp2/3 complex depletion, which might explain the altered pattern of neurite extension analogous to that induced by constitutively active RhoA (Gallo, 2006). Although the mechanisms of Arp2/3 complex interaction with RhoA signaling remain unknown, this finding fits well to an emerging theme that actin filament nucleators may also function in signaling through Rho GTPases and form feedback loops. Thus, Arp2/3 complex was shown to bind and activate the Rac GEF Tiam1 (Ten Klooster et al., 2006), and several formins act upstream of Rho GTPases (Habas et al., 2001; Koka et al., 2003; Kitzing et al., 2007).
Involvement of Arp2/3 complex in signaling through Rho GTPases may indicate that its role in filopodia formation is indirect and mediated, for example, by decreased Rac activity. We addressed this concern by expressing Rac1V12 in siRNA-treated cells, and we found that it rescued filopodia formation, but only partially, suggesting that Arp2/3 complex may contribute to filopodia formation not only by nucleating actin filaments in lamellipodia but also by altering activities of Rho GTPases. How active Rac1 stimulates filopodia in Arp2/3 complex-depleted cells is still an open question. One possibility is that Rac1 fully activates the remaining Arp2/3 complex through WAVE complex. Alternatively, Rac1 may induce actin polymerization through other known or unknown actin nucleators. At present, we cannot distinguish between these nonexclusive possibilities. Rac1V12 expression also increased the rate of filopodia initiation in control cells, consistent with an idea of lamellipodia-dependent filopodia formation.
In conclusion, we showed here that Arp2/3 complex is a key component of actin-based protrusion in neuronal growth cones and that it is involved in generation of lamellipodia, and, importantly, filopodia. Besides its roles in constructing the actin machinery for protrusion, Arp2/3 complex seems to play a role in signaling through Rho GTPases and thus influences neuritogenesis during neuronal differentiation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. R. Balice-Gordon, M. Dichter, F. Gertler, H. Higgs, K. Kaibuchi, D. Kessler, S. Kojima, and C. Otey for generous gifts of reagents and cells; P. Sterling for permission to use a JEOL 1200EX electron microscope; S. Popov for help with Xenopus neural crest cell culture; and S. Zigmond for critical reading of the manuscript. This work was supported by National Institutes of Health grant GM-70898 (to T.S.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/10.1091/mbc.E07-09-0964) on February 6, 2008.
REFERENCES
- Adams J. C. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Barzik M., Kotova T. I., Higgs H. N., Hazelwood L., Hanein D., Gertler F. B., Schafer D. A. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J. E., et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Biyasheva A., Svitkina T., Kunda P., Baum B., Borisy G. Cascade pathway of filopodia formation downstream of SCAR. J. Cell Sci. 2004;117:837–848. doi: 10.1242/jcs.00921. [DOI] [PubMed] [Google Scholar]
- Borisy G. G., Svitkina T. M. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 2000;12:104–112. doi: 10.1016/s0955-0674(99)00063-0. [DOI] [PubMed] [Google Scholar]
- Bridgman P. C., Dailey M. E. The organization of myosin and actin in rapid frozen nerve growth cones. J. Cell Biol. 1989;108:95–109. doi: 10.1083/jcb.108.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Svitkina T. M., Borisy G. G., Popov S. V. Speckle microscopic evaluation of microtubule transport in growing nerve processes. Nat. Cell Biol. 1999;1:399–403. doi: 10.1038/15629. [DOI] [PubMed] [Google Scholar]
- Cohan C. S., Welnhofer E. A., Zhao L., Matsumura F., Yamashiro S. Role of the actin bundling protein fascin in growth cone morphogenesis: localization in filopodia and lamellipodia. Cell Motil. Cytoskeleton. 2001;48:109–120. doi: 10.1002/1097-0169(200102)48:2<109::AID-CM1002>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Di Nardo A., Cicchetti G., Falet H., Hartwig J. H., Stossel T. P., Kwiatkowski D. J. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc. Natl. Acad. Sci. USA. 2005;102:16263–16268. doi: 10.1073/pnas.0508228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez S., Gerisch G., Anderson K., Muller-Taubenberger A., Bretschneider T. Subsecond reorganization of the actin network in cell motility and chemotaxis. Proc. Natl. Acad. Sci. USA. 2005;102:7601–7606. doi: 10.1073/pnas.0408546102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J., Rottner K. The making of filopodia. Curr. Opin. Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Gallo G. RhoA-kinase coordinates F-actin organization and myosin II activity during semaphorin-3A-induced axon retraction. J. Cell Sci. 2006;119:3413–3423. doi: 10.1242/jcs.03084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek E. E., Newey S. E., Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Habas R., Kato Y., He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- Katoh K., Hammar K., Smith P. J., Oldenbourg R. Birefringence imaging directly reveals architectural dynamics of filamentous actin in living growth cones. Mol. Biol. Cell. 1999;10:197–210. doi: 10.1091/mbc.10.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y., Yoshimura T., Tsuboi D., Kawabata S., Kaneko-Kawano T., Shirataki H., Takenawa T., Kaibuchi K. CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol. Cell Biol. 2005;25:9920–9935. doi: 10.1128/MCB.25.22.9920-9935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzing T. M., Sahadevan A. S., Brandt D. T., Knieling H., Hannemann S., Fackler O. T., Grosshans J., Grosse R. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21:1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka S., Neudauer C. L., Li X., Lewis R. E., McCarthy J. B., Westendorf J. J. The formin-homology-domain-containing protein FHOD1 enhances cell migration. J. Cell Sci. 2003;116:1745–1755. doi: 10.1242/jcs.00386. [DOI] [PubMed] [Google Scholar]
- Kunda P., Craig G., Dominguez V., Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Lebrand C., Dent E. W., Strasser G. A., Lanier L. M., Krause M., Svitkina T. M., Borisy G. G., Gertler F. B. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- Lorenz M., Yamaguchi H., Wang Y., Singer R. H., Condeelis J. Imaging sites of N-WASP activity in lamellipodia and invadopodia of carcinoma cells. Curr. Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Loudon R. P., Silver L. D., Yee H. F., Jr, Gallo G. RhoA-kinase and myosin II are required for the maintenance of growth cone polarity and guidance by nerve growth factor. J. Neurobiol. 2006;66:847–867. doi: 10.1002/neu.20258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A., Mitchison T. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 1999;146:1097–1106. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros N. A., Burnette D. T., Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- Mejillano M. R., Kojima S., Applewhite D. A., Gertler F. B., Svitkina T. M., Borisy G. G. Lamellipodial versus filopodial mode of the actin nanomachinery; pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Millard T. H., Sharp S. J., Machesky L. M. Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem. J. 2004;380:1–17. doi: 10.1042/BJ20040176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiu A. K., Weitzke E. L., Chaga O. Y., Borisy G. G. Kinetic-structural analysis of neuronal growth cone veil motility. J. Cell Sci. 2007;120:1113–1125. doi: 10.1242/jcs.03384. [DOI] [PubMed] [Google Scholar]
- Nicholson-Dykstra S., Higgs H. N., Harris E. S. Actin dynamics: growth from dendritic branches. Curr. Biol. 2005;15:R346–R357. doi: 10.1016/j.cub.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Otey C. A., Boukhelifa M., Maness P. B35 neuroblastoma cells: an easily transfected, cultured cell model of central nervous system neurons. Methods Cell Biol. 2003;71:287–304. doi: 10.1016/s0091-679x(03)01013-6. [DOI] [PubMed] [Google Scholar]
- Pellegrin S., Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Peng J., Wallar B. J., Flanders A., Swiatek P. J., Alberts A. S. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 2003;13:534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A., Bretschneider T., Arasada R., Schleicher M., Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Small J. V., Isenberg G., Celis J. E. Polarity of actin at the leading edge of cultured cells. Nature. 1978;272:638–639. doi: 10.1038/272638a0. [DOI] [PubMed] [Google Scholar]
- Steffen A., Faix J., Resch G. P., Linkner J., Wehland J., Small J. V., Rottner K., Stradal T. E. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol. Biol. Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steketee M., Balazovich K., Tosney K. W. Filopodial initiation and a novel filament-organizing center, the focal ring. Mol. Biol. Cell. 2001;12:2378–2395. doi: 10.1091/mbc.12.8.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser G. A., Rahim N. A., VanderWaal K. E., Gertler F. B., Lanier L. M. Arp2/3 is a negative regulator of growth cone translocation. Neuron. 2004;43:81–94. doi: 10.1016/j.neuron.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Svitkina T. M., Borisy G. G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. M., Borisy G. G. Correlative light and electron microscopy studies of cytoskeletal dynamics. In: Celis J., editor. Cell Biology: A Laboratory Handbook; 3rd Edition. vol. 3. San Diego: Academic Press; 2006. pp. 277–285. [Google Scholar]
- Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M., Borisy G. G. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. M., Verkhovsky A. B., McQuade K. M., Borisy G. G. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Klooster J. P., Evers E. E., Janssen L., Machesky L. M., Michiels F., Hordijk P., Collard J. G. Interaction between Tiam1 and the Arp2/3 complex links activation of Rac to actin polymerization. Biochem. J. 2006;397:39–45. doi: 10.1042/BJ20051957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot J. A., Mitchison T. J. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- Verkhovsky A. B., Surgucheva I. G., Svitkina T. M., Tint I. S., Gelfand V. I. Organization of stress fibers in cultured fibroblasts after extraction of actin with bovine brain gelsolin-like protein. Exp. Cell Res. 1987;173:244–255. doi: 10.1016/0014-4827(87)90349-1. [DOI] [PubMed] [Google Scholar]
- Verkhovsky A. B., Svitkina T. M., Borisy G. G. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T., Borisy G. G. Role of fascin in filopodial protrusion. J. Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D., Yarar D., Welch M. D., Peloquin J., Svitkina T., Borisy G. G. Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. L. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J. Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Mitchison T. J. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Weaver A. M., Young M. E., Lee W. L., Cooper J. A. Integration of signals to the Arp2/3 complex. Curr. Opin. Cell Biol. 2003;15:23–30. doi: 10.1016/s0955-0674(02)00015-7. [DOI] [PubMed] [Google Scholar]
- Wilcox K. S., Buchhalter J., Dichter M. A. Properties of inhibitory and excitatory synapses between hippocampal neurons in very low density cultures. Synapse. 1994;18:128–151. doi: 10.1002/syn.890180206. [DOI] [PubMed] [Google Scholar]
- Yang C., Czech L., Gerboth S., Kojima S., Scita G., Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharenko S., Popov S. Plasma membrane recycling and flow in growing neurites. Neuroscience. 2000;97:185–194. doi: 10.1016/s0306-4522(00)00022-1. [DOI] [PubMed] [Google Scholar]
- Zigmond S. H. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Zigmond S. H., Evangelista M., Boone C., Yang C., Dar A. C., Sicheri F., Forkey J., Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.