Abstract
The generation of novel genes and proteins throughout evolution has been proposed to occur as a result of whole genome and gene duplications, exon shuffling, and retrotransposition events. The analysis of such genes might thus shed light into the functional complexity associated with highly evolved species. One such case is represented by TBC1D3, a primate-specific gene, harboring a TBC domain. Because TBC domains encode Rab-specific GAP activities, TBC-containing proteins are predicted to play a major role in endocytosis and intracellular traffic. Here, we show that the TBC1D3 gene originated late in evolution, likely through a duplication of the RNTRE locus, and underwent gene amplification during primate speciation. Despite possessing a TBC domain, TBC1D3 is apparently devoid of Rab-GAP activity. However, TBC1D3 regulates the optimal rate of epidermal growth factor–mediated macropinocytosis by participating in a novel pathway involving ARF6 and RAB5. In addition, TBC1D3 binds and colocalize to GGA3, an ARF6-effector, in an ARF6-dependent manner, and synergize with it in promoting macropinocytosis, suggesting that the two proteins act together in this process. Accordingly, GGA3 siRNA-mediated ablation impaired TBC1D3-induced macropinocytosis. We thus uncover a novel signaling pathway that appeared after primate speciation. Within this pathway, a TBC1D3:GGA3 complex contributes to optimal propagation of signals, ultimately facilitating the macropinocytic process.
INTRODUCTION
Gene duplication, exon shuffling, and retrotransposition events played crucial roles during vertebrate evolution (Long, 2001; McLysaght et al., 2002; Brosius, 2003). About 5% of the human genome is composed of duplicated segments that emerged during the past 35 million years of primate evolution, resulting in the generation of novel protein functions (Courseaux and Nahon, 2001; Taylor and Raes, 2004).
One of the cellular processes, whose regulation has became more and more complex and tightly regulated during evolution is endocytosis. Endocytosis serves to maintain cellular and organismal homeostasis by mediating the uptake of fluids, solutes, and signaling molecules and their receptors. Multiple mechanisms of endocytosis operate within a single cell (Conner and Schmid, 2003). Among them, two major categories are phagocytosis and pinocytosis, which mediate the uptake of particles or of fluid, respectively (Conner and Schmid, 2003).
Pinocytosis encompasses a variety of different membrane-entry routes and is invariably characterized by the relatively small size (50–150 nm) of the internalized particles. Large volumes of fluid are, instead, engulfed by extension, folding, and closure of plasma membranes (PM) through a process termed macropinocytosis (Swanson and Watts, 1995). Macropinocytosis displays many similarities to phagocytosis; the biochemical and cellular components of these endocytic mechanisms have been best characterized in hematopoietic cells, in processes such as phagocytosis by macrophages in response to Fc receptor stimulation, or macropinocytosis of the antigen-presenting dendritic cells (DCs; Swanson and Watts, 1995; Aderem and Underhill, 1999). In all these cases the dynamic assembly of actin filaments generates forces, which drive internalization (Cardelli, 2001; Amyere et al., 2002). In nonhematopoietic cells, the exact functional role, molecular components, and regulatory signaling pathways of macropinocytosis are only now beginning to be defined.
A robust body of evidence has established that dynamic actin structures, defined as dorsal or circular ruffles, are required for macropinocytosis (Buccione et al., 2004 and reference within). Consistent with this finding, is the fact that the de novo Arp2/3-dependent actin filament elongation, mediated by the WAVE family of proteins, is essential for the formation of dorsal membrane extensions needed to engulf fluids (Suetsugu et al., 2003; Innocenti et al., 2005). This pathway is under the control of the RhoGTPase, RAC, which mediates actin remodeling at the plasma membrane (PM) by controlling WASP family VE rprolin-homologous proteins (WAVEs) (Stradal et al., 2004). However, the sole activation of RAC is apparently insufficient to induce circular ruffles, underscoring the need for additional and coordinated signaling cascades (Lanzetti et al., 2004). In line with this, dynamin, a large GTPase best characterized for its ability to mediate vesicle scission during clathrin-dependent endocytosis, has been implicated in macropinocytosis through its binding to the actin regulatory protein, cortactin (McNiven et al., 2000). Finally, the concomitant occurrence of signals emanating from RAC, RAS-PI3K, and RAB5 was shown to be necessary for platelet-derived growth factor–mediated circular ruffles and macropinocytosis, providing evidence of cross-talk between signaling cascades controlled by different small GTPases (Lanzetti et al., 2004).
This concept seems to apply to other GTPases that govern actin membrane protrusions and trafficking at the PM, such as RAB34 (Sun et al., 2003) and, in particular, ARF6 (Donaldson, 2002). ARF6 is localized at the PM, where it participates with the regulation of membrane trafficking and actin remodeling (Schafer et al., 2000; Donaldson, 2002; D'Souza-Schorey and Chavrier, 2006). Notably, ARF6-containing, specialized endosomes have been identified (Donaldson, 2002), and, in some cases, these vesicles can fuse with RAB5-associated endosomes (Naslavsky et al., 2003), supporting the idea of cross-talk between different entry routes and various GTPases controlling the fate of internalized proteins. In this scenario, regulators of the biochemical activity and effectors of GTPases, particularly of RAB5 and ARF6, are likely to play a critical role. Among these regulators are the ARF effectors of the GGA family (Bonifacino, 2004), and the vast and poorly characterized family of TBC (Tre-2, Bub2, Cdc16) domain–containing proteins (Bernards, 2003).
GGAs are ubiquitously expressed adaptor proteins, involved in ARF-dependent sorting of cargos, such as the mannose-6-phosphate receptor, between the trans-Golgi network (TGN) and endosomes (Bonifacino, 2004). More recently GGA3, one of the family members, has also been implicated in plasma membrane receptor trafficking and down-regulation (Bonifacino, 2004; Puertollano and Bonifacino, 2004).
TBC domains are found in more than 80 mammalian proteins. In some cases, TBC-containing proteins act as GAPs (GTPase-activating proteins) for RABs, with the TBC domain encoding the catalytic activity (Cuif et al., 1999; Lanzetti et al., 2000; Haas et al., 2005; Miinea et al., 2005; Zhang et al., 2005). It is not clear, however, whether all TBC-containing proteins display similar activity. Within the superfamily of human TBC-containing proteins, three distinct proteins belong to a distinct subfamily: RNTRE (USP6NL; Matoskova et al., 1996), USP6 (also known as Tre17 or Tre2; Richardson and Zon, 1995), and TBC1D3. RNTRE regulates clathrin-dependent endocytosis and macropinocytosis by acting as a GAP on RAB5 (Lanzetti et al., 2000, 2004), and possibly on other RABs (Haas et al., 2005). Conversely, USP6, whose TBC domain is apparently devoid of GAP activity, is involved in the control of plasma membrane–endosomal trafficking in an ARF6-dependent manner (Martinu et al., 2004). Little is known, instead, about TBC1D3. Because the TBC1D3 gene originated during primate speciation (Paulding et al., 2003), we reasoned that studies of its function might provide a unique opportunity to understand how the machinery of membrane traffic and endocytosis contributes to evolution. Thus, the present studies were undertaken to understand the biological function of TBC1D3.
MATERIALS AND METHODS
Constructs and Plasmids
FLAG- and hemagglutinin (HA)-tagged TBC1D3 under the transcriptional control of the CMV promoter were generated by subcloning a fragment of the human TBC1D3 cDNA (kindly provided by Dr. M. Barbacid, Molecular Oncology Programme, Centro Nacional de Investigaciones Oncológicas (CNIO), E-28029 Madrid, Spain), obtained by PCR and encompassing positions 2-549 (containing the entire open reading frame [ORF]), in the pcDNA3.1-FLAG or pcDNA-1-HA vectors, respectively. Deletion mutants TBC1D3-92-319, TBC1D3-2-319, TBC1D3-92-549, and TBC1D3-319-549 were obtained by PCR and subcloned in the pcDNA3.1-FLAG vector. FLAG-TBC1D3-R147A and FLAG-TBC1D3-R147A-R154A mutants were generated by site-directed mutagenesis PCR (Stratagene, La Jolla, CA; Quick Change Mutagenesis Kit). All GST fusion proteins were obtained by recombinant PCR using the FLAG-TBC1D3 cDNA as a template and subcloned in the pGEX-KT vector (Amersham Pharmacia, Piscataway, NJ). Full-length human GGA3 was obtained by PCR, using the cDNA from RZPD Clone ID-IRAKp961D15167Q2 as a template (RZPD Deutsches Ressourcenzentrum fuer Genomforschung GmbH, Berlin, Germany) and subcloned into the pcDNA3.1FLAG vector. pcDNA3-FLAG-EFA6 was kindly provided by Dr. D. Talarico. (Department of Molecular Biology and Functional Genomics, DIBIT, Università Vita Salute San Raffaele, Via Olgettina 58, 20132, Milano, Italy) GEX-4T1-GGA3 was kindly provided by Dr. P. Chavrier (Membrane and Cytoskeleton Dynamics Group, UMR 144 CNRS/Institut Curie, Paris, France) and encodes human GGA3 VHS domain + N-terminal portion of the GAT domain (residues 1-226). pGEX-4T3-RAB1, RAB2, RAB3A, RAB7, RAB9, RAB11A, RAB17, RAB23, RAB25, RAB28, and RAB37 were generated by PCR. MYC-RAB5-S34N were generated as described before (Lanzetti et al., 2000), HA-ARF6-T27N, HA-ARF6-Q67L and GFP-PLCδ (pleckstrin homology [PH] domain) domain were from Dr. P. Chavrier. HA-RNTRE and glutathione S-transferase (GST)-RNTRE TBC domains used in this article were previously generated and described (Lanzetti et al., 2000). Details of the engineering strategies and sequences of the oligonucleotides used are available upon request. All fragments obtained by PCR were sequence verified.
Tissue Culture
HeLa cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine calf serum (FCS), 100 μg/ml streptomycin, 100 μg/ml penicillin, and 2 mM glutamine. Phoenix cells were grown in DMEM supplemented with 10% bovine North American serum, 100 μg/ml streptomycin, 100 μg/ml penicillin, and 2 mM glutamine. COS-7 cells were grown in DMEM supplemented with 10% bovine South American serum, 100 μg/ml streptomycin, 100 μg/ml penicillin, and 2 mM glutamine. Transfections were performed using either calcium phosphate, FUGENE (Invitrogen), or LipofectAMINE (Invitrogen) reagents, according to manufacturer's instructions. HeLa cells were transfected, for biochemical studies, using the LipofectAMINE reagent, and for immunofluorescence, using FUGENE 6. Phoenix cells were transfected using the calcium phosphate procedure. For actin analysis HeLa cells were seed on gelatin-coated coverslips and after 24 h, cells were processed for immunofluorescence (IF) as described below.
Pulldown and Coimmunoprecipitation Experiments
HeLa cells were lysed in a buffer containing 1% Triton X-100 (Pierce, Rockford, IL), 50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 1.5 mM MgCl2, and 5 mM EGTA, adding the Calbiochem protease inhibitor cocktail (ID-539134; La Jolla, CA) and phosphatase inhibitors (10 mM sodium orthovanadate and 20 mM sodium pyrophosphate). Bovine serum albumin was added to the lysates at 0.5% final concentration, and lysates were left rocking on ice for 15 min. For pulldown experiments, 40 μg of GST-fusion proteins attached to the glutathione-Sepharose beads were incubated in the presence of 1 mg of total cell lysate for 1 h at 4°C rocking. After three washes in lysis buffer, bound proteins were resolved by SDS-PAGE and visualized with the appropriate antibodies. For coimmunoprecipitation studies, cells were lysed and cell lysates were used immediately, without freeze/thawing. Immunoprecipitations and coimmunoprecipitation experiments were performed for 1 h in the presence of the appropriate antibody, and immune complexes were recovered by adsorption to protein G-Sepharose beads (Zymed, South San Francisco, CA), and samples were subjected to one additional round of immunopurification. After three washes in lysis buffer, bound proteins were resolved by SDS-PAGE and visualized with the appropriate antibodies. [35S]methionine-labeled protein of full-length and deletion mutants of TBC1D3, used in the experiment in Figure 7C, was synthesized by in vitro transcription–translation using a commercial kit (Promega, Madison, WI).
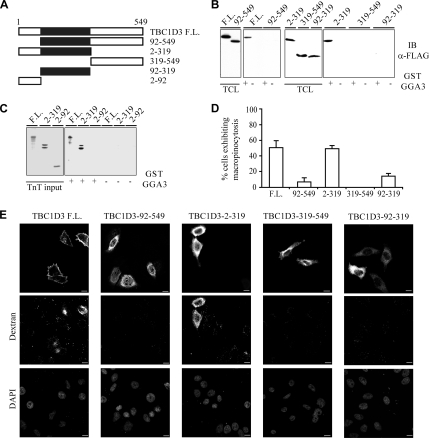
Figure 7.
The TBC1D3:GGA3 complex is required for TBC1D3-induced macropinocytosis. (A) A schematic representation of TBC1D3 and its deletion mutants. The TBC domain is shown in black. Amino acids boundaries are indicated. (B) Cell lysates obtained from 293T cells transfected with full-length FLAG-TBC1D3 or deletion mutants, as indicated at the top, were subjected to in vitro binding experiments with GST-GGA3 (+), or GST (−), as control. Total cell lysates (TCL) and bound proteins were analyzed by immunoblotting (IB) with the indicated Abs. Equal amounts of GST-fusion proteins were used. Shown are 3% of input and 100% of bound material. (C) [35S]methionine-labeled TBC1D3 (F.L.) or indicated deletion mutant were transcribed and translated in vitro and subjected to in vitro binding experiments with GST-GGA3 (+), or GST (−), as control. Total input proteins (TnT input) and bound proteins were analyzed by autoradiography. Shown are 3% of input and 100% of bound material. (D) HeLa cells transfected with full-length FLAG-TBC1D3 (F.L.) or FLAG-tagged deletion mutants, as indicated, were incubated with rhodamine-conjugated dextran. The percentage of transfected cells incorporating dextran was determined. Data are expressed as the mean ± SD of two independent experiments in which at least 100 cells were counted. (E) HeLa cells transfected with full-length FLAG-TBC1D3 (F.L.) or FLAG-tagged deletion mutants, as indicated, were fixed and stained with anti-FLAG Ab and processed for confocal microscopy to detect transfected cells (top panel), or incorporated rhodamine-conjugated dextran (dextran, middle panel). DAPI nuclear staining (DAPI, bottom panel) is also shown. Bars, 10 μm.
Antibodies
Rabbit polyclonal antibodies against human TBC1D3 were raised using as immunogen, the peptide 501–549 of human TBC1D3 protein, produced as a GST fusion protein by the IFOM-IEO-Campus Antibody Facility in collaboration with Eurogentec (Serain, Belgium). Before immunization, purified proteins were eluted from GSH beads (glutathione-Sepharose 4B purchased from Amersham Pharmacia), subjected to overnight dialysis in PBS buffer, and kept at −80°C. Rabbit polyclonal antibodies anti-HA tag (HA.11) and mouse monoclonal antibodies anti-c-MYC (9E10) were from Eurogentec. Mouse monoclonal anti-HA tag antibodies (16B12) were from BabCO (Richmond, CA). Mouse monoclonal anti-FLAG M2 were from Sigma (St. Louis, MO). Anti-ARF6 (3A-1) and anti-RAB5A (S-19) rabbit polyclonal antibodies and mouse monoclonal antibodies anti-GFP (B-2) were from Santa Cruz Biotechnology (Santa Cruz, CA).
Immunofluorescence Staining
Cells grown on coverslips were fixed freshly prepared 4% paraformaldehyde for 10 min at room temperature or 30 min on ice and permeabilized with Ca2/Mg2-free phosphate-buffered saline (PBS) containing 0.1% Triton X-100, 1% bovine serum albumin for 5 min at room temperature. Coverslips were then incubated for 1 h with the appropriate primary antibody, washed extensively, and then incubated with the secondary antibodies for 30 min. Coverslips were mounted in Fluoromount (Fisher Scientific, Pittsburg, PA) containing 1 mg/ml para-phenylenediamine. F-actin was detected by staining with rhodamine-conjugated phalloidin (Sigma) at a concentration of 6.7 U/ml. Indirect IF was analyzed under an AX-70 Provis (Olympus, Melville, NY) fluorescence microscope equipped with a b/w cooled CCD camera (c5985; Hamamatsu, Bridgewater, NJ), or with a Leica TCS SP2 AOBS confocal microscope (Deerfield, IL) equipped with 405-, 488-, 543-, and 633-nm laser lines.
GAP Assay
GAP assays using GST-TBC1D3 or GST-RNTRE TBC domains or immunoprecipitated TBC1D3 or RNTRE proteins were performed using 0.2 μM substrate [γ-32P]GTP-loaded Rab proteins and catalytic concentrations of GST-TBC1D3 (60 nM) and GST-RNTRE TBC domain (40 nM) or the immunoprecipitated full-length proteins as previously described (Lanzetti et al., 2000).
Dextran Internalization Assay
Cells were plated on glass coverslips, without gelatin coating, 1 d after cells were transfected with different constructs. Twenty-four hours after the transfection, cells were serum-starved for 16 h and then incubated with 1 mg/ml rhodamine-conjugated dextran, 70,000 MW (Molecular Probes, Eugene, OR; D-1818) for 30 min at 37°C, washed four times with ice-cold PBS, fixed in 4% paraformaldehyde (in PBS) for 10 min, washed four times with PBS, and processed for confocal analysis, as described above. Each assay was done in triplicate and at least 100 cells were counted in each experiment.
Dextran Internalization Assay for Fluorescence-activated Cell Sorting Analysis
HeLa cells were plated in six-well plates, without gelatin coating. For data presented in Figure 3 cells were transfected twice with siRNA TBC1D3-specific or scrambled control oligos. Twenty-four hours after the second transfection, cells were serum-starved for 16 h and then incubated with 1 mg/ml fluorescein-conjugated dextran, 70,000 MW (Molecular Probes; D1822) for 30 min at 37°C; at the same time cells were stimulated with different doses of epidermal growth factor (EGF; 1, 10, and 30 ng/ml). Cells were then harvested, washed four times with ice-cold PBS, and fixed in 1% formaldehyde for 15 min on ice. After two final washes, cells were analyzed on a FACSCan flow cytometer (Becton Dickinson, Franklin Lakes, NJ) using Cell Quest software. For data presented in Figure 8 cells were transfected twice with small interfering RNA (siRNA) GGA3-specific or scrambled control oligos, and 24 h after the second transfection, cells were transfected with FLAG-TBC1D3. Twenty-four hours after transfection cells were serum-starved for 16 h and then incubated with 1 mg/ml fluorescein-conjugated dextran, 70,000 MW (Molecular Probes; D1822) for 30 min at 37°C and processed for FACSCan analysis as described above.
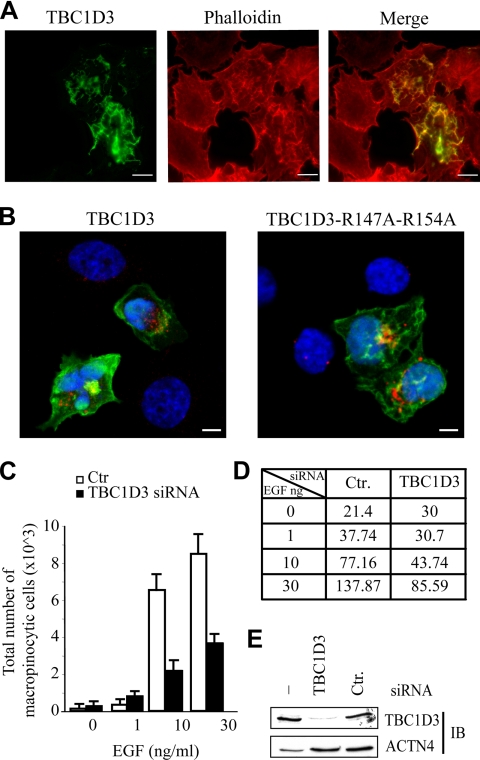
Figure 3.
TBC1D3 induces dorsal ruffling and is required for optimal EGF-mediated macropinocytosis. (A) HeLa cells were transfected with FLAG-tagged TBC1D3. Cells were fixed and stained with anti-FLAG antibody (green, TBC1D3) or rhodamine-phalloidin (red, phalloidin). Merged image is also shown. (B) Serum-starved HeLa cells, transfected with FLAG-tagged TBC1D3 wild type (TBC1D3) and a TBC1D3 double-point mutant (TBC1D3-R147A-R154A), were processed for rhodamine-conjugated dextran (red) uptake, fixed and stained with anti-FLAG Ab (green), and analyzed by confocal microscopy. Merged images are shown. DAPI (blue) indicates nuclear staining. Bars, 10 μm. (C) HeLa cells transfected as above with TBC1D3-specific (TBC1D3) siRNA or control scrambled oligos (Ctr), were serum-starved and incubated with 1 mg/ml fluorescein-conjugated dextran immediately followed by treatment with the indicated concentrations of EGF or mock treatment (0). Cells were processed for FACS analysis. Data are expressed as the average of total number of events scoring positive for fluorescein-dextran. Similar number of cells (∼50.000) was analyzed for each time point. At least three independent experiments were performed in triplicate. Similar data were obtained using two independent siRNA oligos (not shown). (D) Quantification of dextran uptake from C: data represent the mean intensity fluorescence, and are expressed in arbitrary unit. (E) HeLa cells were transfected (siRNA) with TBC1D3-specific (TBC1D3) siRNA or control scrambled oligos (Ctr). Cell lysates were analyzed by immunoblotting (IB) with the indicated Abs. Actinin-4 levels were used to verify equal loading.
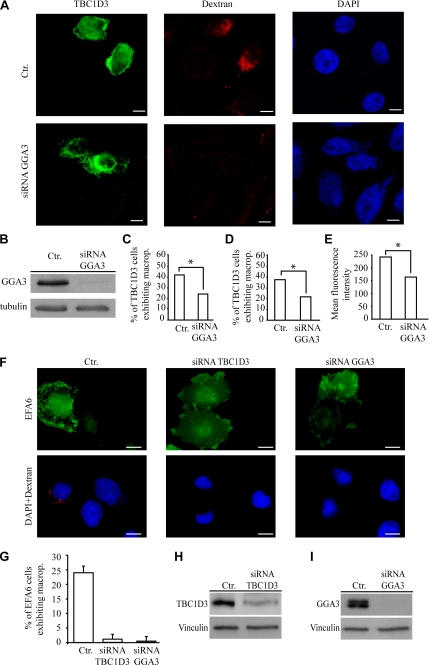
Figure 8.
GGA3 is required for TBC1D3-induced macropinocytosis. (A) HeLa cells were transfected with control scrambled oligos (top panels) or GGA3-specific siRNA (bottom panels). After 48 h cells were transfected with FLAG-TBC1D3 (left panels) and subsequently processed for rhodamine-conjugated dextran uptake (middle panels), and fixed and stained with the appropriate Abs; DAPI nuclear staining (right panels) is also shown. (B) Control lysates from scrambled (Ctr) or GGA3 siRNA (siRNA GGA3) HeLa cells were immunoblotted with the indicated Abs; tubulin levels were used to verify equal loading. (C) HeLa cells transfected as described above were serum-starved, incubated with 1 mg/ml fluorescein-conjugated dextran, fixed and stained with the appropriate Abs, and processed to detect TBC1D3 and rhodamine-conjugated dextran-positive cells by direct immunofluorescence or (D and E) were processed for FACS analysis. Data are expressed as the percentage of TBC1D3 and fluorescein-dextran–positive cells with respect to total number of TBC1D3-positive cells. Numbers represent the average of three independent experiments performed in triplicate (*p < 0.001) in which at least 100 cells in direct immunofluorescence experiments or ∼50.000 cells for FACSCan analysis were counted (D) or mean fluorescence intensity were calculated (E). Similar data were obtained using two independent GGA3 siRNA oligos (not shown). (F) HeLa cells were transfected with control scrambled oligos (left panels), TBC1D3-specific siRNA (middle panels) or GGA3-specific siRNA (right panels). After 48 h cells were transfected with FLAG-EFA6 (top panels) and subsequently processed for rhodamine-conjugated dextran uptake (bottom panels). (G) Data are expressed as the percentage of EFA6 and fluorescein-dextran–positive cells with respect to total number of EFA6-positive cells. Numbers represent the average of three independent experiments performed in triplicate in which at least 100 cells were counted. Control lysates (H) from scrambled (Ctr) or TBC1D3, and (I) from scrambled (Ctr.) or GGA3 siRNA HeLa cells. Lysates were immunoblotted with the indicated Abs; vinculin levels were used to verify equal loading.
Gene Silencing
TBC1D3 and GGA3 gene silencing in HeLa cells was obtained by transfecting siRNA oligos for TBC1D3 or GGA3 (Dharmacon, Boulder, CO) specific target sequence. HeLa cells were transfected according to suppliers' instructions, in six-well plates, with oligofectamine (Invitrogen). HeLa cells were subjected to double transfection with 200 nM siRNA oligos, and silencing was verified at various time points by immunoblot analysis with the specific antibodies against TBC1D3 or GGA3. As control, scrambled oligos were used. Each experiment was performed using two independent siRNA oligos. Oligonucleotide sequences are available upon request.
RESULTS
TBC1D3, the Paralog of Human RNTRE, Underwent Gene Duplication during Evolution
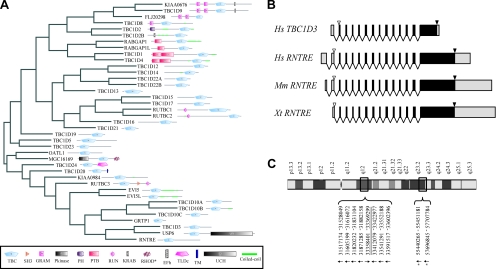
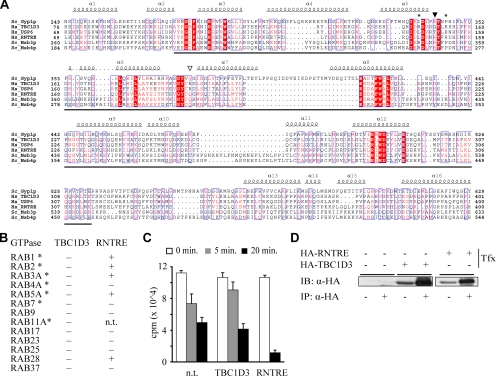
Among TBC-containing human proteins (Figure 1A), RNTRE is the most related to TBCID3 (34% identity, 49% similarity). Importantly, the gene structure of TBCID3 and RNTRE is extremely well conserved, even when compared with a distant RNTRE ortholog, such as the Xenopus tropicalis RNTRE (Figure 1B). This suggests that the recent origin of TBCID3, during primate speciation (Paulding et al., 2003), is due to a duplication of the RNTRE locus. Accordingly, all TBCID3-coding exons are identical in length to those of RNTRE, with the exception of the last one, which also contains the 3′ untranslated regions (UTRs; Figure 1B and Figure S1 in Supplementary Material). Gene structure conservation is a common feature among ortholog and paralog genes, and it has been proposed to represent a record of key events in evolution, even when protein sequence similarity is low (Batzoglou et al., 2000; Betts et al., 2001; Koonin, 2005). This applies to TBC1D3, which despite having originated only after the initial primate radiation (∼60 million years ago; Paulding et al., 2003), accumulated, in a remarkably short period of time, a considerable number of mutations, accounting for the relatively low degree of similarity between the TBC1D3 and RNTRE gene products (Figure S2 in Supplementary Material). This also suggests that TBC1D3 has been subject to positive evolutionary pressure, likely resulting in the acquisition of novel functions with respect to RNTRE.
Figure 1.
TBC1D3, the human paralog of RNTRE, underwent gene duplications during evolution. (A) Relatedness and domain composition of the human TBC-containing protein family. The dendrogram based on the sequence similarity of TBC-containing protein family members was generated based on the alignment of the entire amino acid sequence of each protein, obtained using the ClustalX program, and visualized with Hypertree software (Bingham and Sudarsanam, 2000). The domain composition for each protein, as detected by SMART (Letunic et al., 2006), is shown. Asterisk indicates that the rhodanese homology domain (RHOD) present in the MGC16169 protein is predicted to be catalytically inactive. (B) Schematic representation of the genomic structure of Homo sapiens TBC1D3 and of RNTRE from H. sapiens, Mus musculus, and X. tropicalis. Exons are drawn to scale; UTR regions are shown in gray; start and stop codons are indicated by empty and solid arrows, respectively. (C) Regions on chromosome 17, which display more than 97% identity with the TBC1D3 mRNA (NM_032258). The numbers refer to the base pair positions, as indicated in the human genome, May 2004 assembly (hg17). The underlined region indicates the official position assigned to the TBC1D3 gene (NCBI GeneID: 84218). Arrows indicate the orientation of the genes, and asterisks indicate loci from which the full ORF could not be reconstructed due to frameshifts or stop codons and can thus be considered to be pseudogenes.
A bioinformatics analysis revealed an additional peculiar feature of the TBC1D3 gene. We found at least nine additional regions displaying more than 97% identity to TBC1D3 mRNA present on chromosome 17 (Figure 1C and Figure S1 in Supplementary Material). The similarity among these genomic regions is not limited to the exons, but also extends to the introns, indicating that entire TBC1D3 locus underwent duplication. Seven of these TBC1D3-related regions (showing ∼99% of identity over >10 kb) contain ORFs potentially coding for full-length proteins (data not shown) and display a complex pattern of expression in several tissues (Hodzic et al., 2006). In addition specific patterns of expression of these paralogs were observed in different normal human tissues and altered in several prostate tumors in comparison to healthy prostate tissue (Hodzic et al., 2006). Conversely, the remaining two paralogs display in-frame stop codons and frameshifts caused by insertion/deletion of single nucleotides in the genomic sequence. In-depth bioinformatics analysis excluded that the redundancy of the TBC1D3 locus is due to an artifact resulting from a wrong assembly of the human genome (Figure S1 in Supplementary Material).
Duplication of large regions with high identity is not unusual throughout the human genome (Bailey et al., 2001). The duplication of the TBC1D3 locus, however, seems peculiar in that it occurred at least nine times, in regions that are not always contiguous on chromosome 17 and with a degree of sequence conservation that reflects its recent evolutionary origin. Thus, TBC1D3 encode for a TBC-domain–containing protein of particular interest because it is present only in primates and underwent remarkable gene duplication very recently in evolution giving raise to a novel, highly redundant gene that was susceptible to develop novel functions.
The TBC Domain of TBC1D3 Displays No Apparent GAP Activity
TBC1D3 possesses an N-terminal TBC domain, which displays closest similarity to the TBC domains of USP6 and RNTRE (Figure 2A). The TBC domain of the latter protein was shown to be a Rab-GAP, with in vitro specificity for RAB5 (Lanzetti et al., 2000) and RAB30 and RAB41 (Haas et al., 2005). Conversely, no GAP activity could be detected for the TBC domain of USP6 (Bizimungu et al., 2003). Consistent with this, the TBC domain of USP6 lacks conserved residues (Figure 2A) that are essential for catalytic activity (Pan et al., 2006; Bos et al., 2007). The same critical residues are also absent in TBC1D3, suggesting that it might not be a GAP. Despite this, TBC1D3 has been reported to possess a modest, but measurable RAB5-specific GAP activity (Pei et al., 2002). We analyzed the ability of TBC1D3 to stimulate the intrinsic GTPase activity of a collection of 13 different RABs (Figure 2B), using either native, immunoprecipitated, full-length TBC1D3 or its isolated and recombinantly produced TBC domain. In all cases, we could not detect any Rab-GAP activity (Figure 2, B–D). In particular, no activity on RAB5 was detected, at variance with a previous report (Pei et al., 2002). The reason for this discrepancy is not clear. We note, however, that in the mentioned study (Pei et al., 2002), a 10-fold molar excess of TBC1D3 (PRC17 in that study) with respect to RAB5 was used, whereas we used catalytic concentrations of the protein.
Figure 2.
The TBC domain of TBC1D3 does not possess any GAP activity. (A) Multiple alignments of TBC domains of human TBC1D3, USP6, and RNTRE with the TBC domain of the Yeast Gyp1p, Ms3p, and Msb4p proteins. Conserved and identical residues are in red and reversed red, respectively. The secondary structure of the TBC domain of Gyp1p is reported at the top of the alignment. The boundaries of the TBC domain were determined according to the SMART and PFAM programs and are underlined. The conserved arginine and glutamine residues essential for the GAP activity of TBC domains are indicated by a filled or empty arrowheads, respectively. Asterisks indicate the arginine residues mutated to alanine to generate the TBC1D3-R147A-R154A mutant shown in Figure 3. (B) The indicated Rab proteins (200 nM) were loaded with [γ-32P]GTP and then incubated for 6 min with purified TBC domains of GST-TBC1D3 or GST-RNTRE (60 and 40 nM, respectively) or with HA-immunoprecipitated full-length proteins (asterisks). The presence (+) or the absence (−) of GAP activity was assessed by filter binding assay as described in Lanzetti et al. (2000). n.t., not tested. (C) Time-dependent kinetics of the GAP activity of immunoprecipitated HA-TBC1D3 or HA-RNTRE on [γ-32P]GTP loaded RAB5. Bound [γ-32P]GTP was measured. Nontransfected COS-7 cells (n.t.) were used as control and revealed the high rate of spontaneous hydrolysis of RAB5. Data are expressed as the mean ± SD of three independent experiments. (D) Immunoprecipitation results with anti-HA antibodies referring to the experiment described in C. Cell lysates from COS-7 cells transfected with HA-tagged TBC1D3 and RNTRE, as indicated (Tfx), were subjected to immunoprecipitation (IP: α-HA +) with Ab against HA. Total cell lysates and immunoprecipitated proteins were analyzed by immunoblotting (IB) with the Ab against HA. Shown are 2.5% of input and 100% of the bound material.
We cannot exclude that TBC1D3 is a GAP for Rabs not tested in this study; however, various considerations argue against this possibility. First, as already mentioned, TBC1D3 lacks residues that are critical for the GAP activity of TBC domains. Second, mutations to reintroduce these critical residues (G151R and R188Q) did not confer GAP activity to TBC1D3 (data not shown). Third, RNTRE, used as a control in our assays, displayed detectable activity on at least five different Rabs (Figure 2, B–D). Although it cannot be said whether this in vitro promiscuity reflects similar promiscuity in vivo, our data allow us to conclude that, under our assay conditions, the activity of a bona fide Rab-GAP can be readily detected. Finally, biological evidence described below also argues against a role for TBC1D3 as a GAP.
TBC1D3 Induces Dorsal Ruffling and Is Required for Optimal EGF-mediated Macropinocytosis
To gain insight into the biological function of TBC1D3, we initially performed overexpression experiments. Ectopic expression of TBC1D3 led to alterations of the actin cytoskeleton at the dorsal surface of HeLa cells (Figure 3A) and of a variety other cell types (not shown), with the formation of ruffle-like structures where TBC1D3 and F-actin colocalized (Figure 3A, merge). These protrusions are typically associated with the process of macropinocytosis, which can easily be monitored after the incorporation of the classical macropinocytic tracer 70-kDa rhodamine-labeled dextran (Dharmawardhane et al., 2000; Lanzetti et al., 2004). After 30 min of incubation little or no dextran internalization was observed in untransfected, serum-starved control cells. Conversely, about ∼50% of TBC1D3-overexpressing cells, displayed internalized, dextran-filled vesicles (Figure 3B and Table 1).
Table 1.
Both RAB5 and ARF6 activity are required for TBC1D3-induced macropinocytosis
| Transfection | % cells exhibiting macropinocytosis |
|---|---|
| TBC1D3 | 51 ± 15 |
| TBC1D3 R147A-R154A | 42 ± 16 |
| TBC1D3 + ARF6-T27N | 5 ± 6 |
| TBC1D3 + RAB5-S34N | 15 ± 5 |
| RAB5 WT + ARF6-T27N | 98 ± 2 |
| ARF6-Q67L + RAB5-S34N | 12 ± 3 |
| ARF6-Q67L | 30 ± 3 |
| EFA6 | 26 ± 1 |
| ARF6-T27N | 0 |
| RAB5-S34N | 2 ± 1 |
| RAB5-WT | 98 ± 2 |
Quantification of dextran uptake. The percentage of transfected cells incorporating dextran was determined. Data are expressed as the mean ± SD of two independent experiments where at least 100 cells were counted.
The availability of a bioassay for TBC1D3, allowed further testing of the possibility that TBC1D3 is a GAP for a yet undetermined Rab in vivo. A “dual finger” mechanism has been recently proposed for GAP catalysis, in which an arginine (Arg) residue, the “arginine finger,” and a glutamine (Gln) residue, the “glutamine finger,” of the GAP moiety are essential to stabilize the transition state of the GTP hydrolysis reaction (Pan et al., 2006; Bos et al., 2007). In TBC domains, these residues (corresponding to aa 150 and 187 of RNTRE, respectively) are highly conserved and have been shown to be essential for GAP activity (Lanzetti et al., 2000; Pan et al., 2006). In TBC1D3, these residues are not conserved (Figure 2A). However, two arginine residues at positions 147 and 154 might substitute for the “arginine finger” function (Figure 2A), exploiting an alternative mechanism of action that might render the “glutamine finger” dispensable for the catalytic process. Thus, we engineered a TBC1D3 mutant, harboring a double Arg→Ala substitution in these two positions. This mutant was capable of inducing macropinocytosis with an efficiency similar to that of WT TBC1D3 (Figure 3B and Table 1), further suggesting that a putative Rab-GAP function of TBC1D3 is not responsible for this effect.
From the above data, we concluded that overexpression of TBC1D3 is per se sufficient to elicit macropinosome formation in absence of stimulation, indicating that it may be part of a signaling cascade controlling this event. To assess whether TBC1D3 also is also needed for macropinocytosis under physiological conditions, we performed RNA interference of TBC1D3 in HeLa cells (Figure 3C) and measured the extent of macropinocytosis after stimulation with EGF, a known inducer of dextran incorporation (West et al., 1989). These experiments were performed using a FACS (fluorescent-activated cell sorter)-based analysis, to quantify the extent of dextran incorporation. EGF-stimulated, TBC1D3-knockdown HeLa cells showed a significant decrease in dextran internalization, compared with control cells both with respect to the total number of cell exhibiting macropinocytosis (Figure 3C) and to dextran uptake, as assessed by calculating the mean fluorescence intensity, by FACS analysis, of dextran-positive cells (Figure 3D). Notably and in agreement with previous published data (West et al., 1989), the levels of macropinocytosis in unstimulated cells were minimal and unaffected by ablation of TBC1D3. Together, these results suggest that TBC1D3 is physiologically required for optimal macropinocytosis in response to EGF stimulation. The observation that the TBC1D3 gene emerged during primate speciation, whereas macropinocytosis is conserved throughout the mammalian kingdom (Cardelli, 2001), supports the notion that, in higher eukaryotes TBC1D3 may have evolved to provide a novel function of optimization of the macropinocytic process.
TBC1D3 Mediates Macropinocytosis by Acting in a Pathway That Requires RAB5 and ARF6
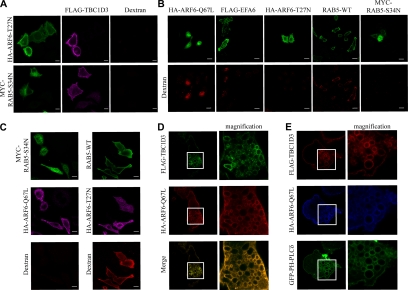
ARF6 and RAB5 are GTPases known to control macropinocytosis (Roberts et al., 2000; Donaldson and Honda, 2005). The mechanisms underlying their action and their functional links in this process are, however, only partially defined. To identify the signaling pathways in which TBC1D3 might be involved, a molecular genetics approach was utilized, using dominant negative and activated mutants of these GTPases. Both dominant negative ARF6 (ARF6-T27N) and RAB5 (RAB5-S34N) efficiently inhibited TBC1D3-mediated internalization of rhodamine-dextran (Figure 4A and Table 1), indicating that TBC1D3 requires the activity of both ARF6 and RAB5.
Figure 4.
TBC1D3 mediates macropinocytosis in a RAB5- and ARF6-pathway and localizes into ARF6-positive endosomes. HeLa cells were (A) cotransfected with FLAG-TBC1D3 and either HA-ARF6-T27N or MYC-RAB5-S34N; (B) transfected with HA-tagged ARF6 mutants (HA-ARF6-Q67L or HA-ARF6-T27N), or FLAG-tagged EFA6, or RAB5-WT or MYC-RAB5-S34N; or (C) cotransfected either with WT RAB5 and HA-ARF6-T27N (right panels), or with MYC-RAB5-S34N and HA-ARF6-Q67L (left panels). Cells were processed for rhodamine-conjugated dextran (red) uptake, fixed and stained with the appropriate Abs, as indicated at top of each image, and analyzed by confocal microscopy. Bars, 10 μm. The rhodamine-dextran signals may appear quite dissimilar in the different transfectants, possibly due to the stage at which ARF6 or RAB5 act in and perturb the macropinocytic process. (D) HeLa cells cotransfected with FLAG-tagged TBC1D3 (top) and HA-tagged ARF6-Q67L (middle) were fixed and stained with the appropriate Abs. A merged image (bottom) and magnifications of selected areas (right panels) to highlight colocalization are also shown. (E) HeLa cells cotransfected with FLAG-tagged TBC1D3 (red), HA-tagged ARF6-Q67L (blue), and GFP-PH-PLCδ (green) were fixed and stained with the appropriate Abs, as indicated at the left of each image, and analyzed by confocal microscopy. Magnifications of selected areas (right panels) to highlight colocalization are also shown.
To gain further insight into the signaling events shaped by these interactions, the relationship between ARF6 and RAB5 in macropinocytosis was explored. The expression of a constitutive active mutant of ARF6 (ARF6-Q67L), or of EFA6, an exchange factor for ARF6 (Franco et al., 1999), induced dextran internalization, whereas a dominant negative ARF6, as expected, did not (Figure 4B and Table 1). Similarly, RAB5, but not a dominant negative RAB5, potently induced macropinocytosis (Figure 4B and Table 1). Additionally, RAB5-induced macropinocytosis was unaffected by a dominant negative ARF6, whereas a dominant negative RAB5 inhibited ARF6-mediated macropinocytosis (Figure 4C and Table 1). Thus, ARF6 acts upstream of RAB5 in mediating dextran internalization, at least under the conditions of ectopic expression used here.
On expression of ARF6-Q67L, TBC1D3 was recruited to and accumulated on enlarged vesicles (Figure 4D), which have been previously characterized as ARF6-positive endosomes (Caplan et al., 2002), suggesting that ARF6-dependent relocalization of TBC1D3 might be part of its mechanism of action. To confirm this, we investigated the colocalization of TBC1D3 with another marker of this compartment, phosphatidylinositol 4,5-bisphosphate (PIP2), which can be detected in living cells by the GFP-tagged PH domain of phospholipase Cδ (Katan and Allen, 1999). On expression of ARF6-Q67L, TBC1D3 and GFP-PH-PLC-δ colocalized in vesicle-like structures (Figure 4E), providing evidence that TBC1D3 not only stimulates actin reorganization and macropinocytosis, but also cotraffics with ARF6 in specialized endosomal vesicles. Notably, ARF6 mediates the internalization of MHCI and of the IL-2 receptor α-subunit, TAC (Donaldson, 2005). These cargos are subsequently delivered to RAB5-associated endosomes, supporting the existence of a hierarchical link between ARF6 and RAB5 and connecting endosome-dependent internalization to trafficking events (Naslavsky et al., 2003). Thus, collectively, these data support a possible mechanism of signaling whereby TBC1D3 acts in an ARF6-dependent pathway, requiring RAB5 activity, leading to macropinocytosis.
TBC1D3 Interacts and Colocalize with GGA3 at the Plasma Membrane and in ARFQ67L-induced Enlarged Endosomes
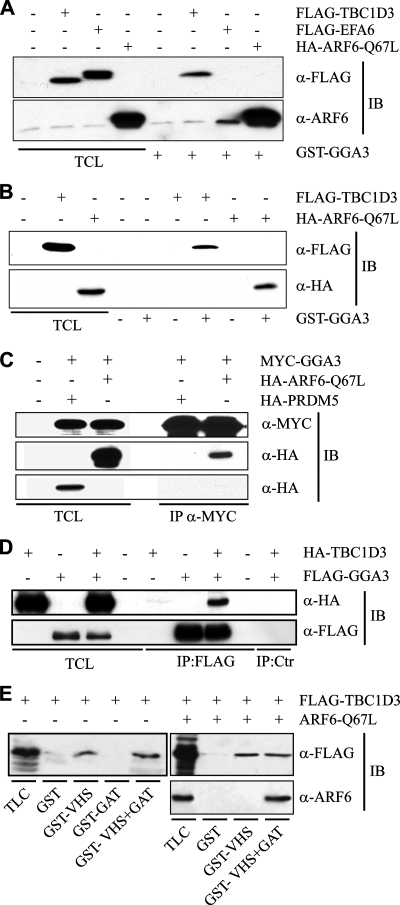
One obvious, possible mechanism of action for TBC1D3 would be to induce ARF6 activation, similarly to what has been reported for USP6 (Martinu et al., 2004). Thus, we analyzed ARF6-GTP levels in cells overexpressing TBC1D3, or EFA6 or ARF6-Q67L as controls. We used an assay based on the ability of a γ-ear–containing fragment of GGA3, an ARF-binding protein, to bind to ARF6 in a GTP-dependent manner (Dell'Angelica et al., 2000; Martinu et al., 2004). No significant activation of ARF6 in TBC1D3-overexpressing cells could be detected, under conditions in which expression of EFA6 induced a significant increase in ARF6-GTP levels (Figure 5A). Surprisingly, however, the GGA3 bait interacted with TBC1D3. The strength of this interaction was not dissimilar to that between the γ-ear of GGA3 and ARF6-GTP (ARF6-Q67L; Figure 5B), at least under the conditions of our pulldown assay. Furthermore, coimmunoprecipitation experiments indicated that the association between ARF6 and GGA3 and between TBC1D3 and GGA3 also occurs in vivo, supporting the physiological relevance of this interaction (Figure 5, C and D). Next, we tested whether TBC1D3 and ARF6 bind GGA3 on difference surfaces. GGA3, like all other GGAs, binds to ARFs through its GAT domain (Takatsu et al., 2002). Conversely, the association of GGA3 to TBC1D3 is mediated by its VHS domain and is thus independent from the GAT domain (Figure 5E). These results also suggest that the possibility that the GGA3 may simultaneously bind to ARF6 and TBC1D3. Accordingly, ectopically coexpressed ARF6 and TBC1D3 associated with immobilized GST-GGA3 construct encompassing both the VHS and the GAT domain, whereas only TBC1D3, but not ARF6 bound to the immobilized GST-VHS domain (Figure 5E), suggesting that TBC1D3, ARF6-GTP, and GGA3 may form a trimeric complex.
Figure 5.
TBC1D3 and ARF6 bind to distinct regions of GGA3. (A) Cell lysates obtained from serum-starved HeLa cells transfected with either FLAG-TBC1D3, FLAG-EFA6, HA-ARF6-Q67L or were mock-transfected (Ctr), as indicated at the top, were subjected to in vitro binding experiments with GST-GGA3(1-226). Lysates (TCL) and specifically bound proteins were analyzed by immunoblot (IB) with the indicated Abs. Shown are 2.5% of lysates and 100% of the bound material. (B) Cell lysates obtained from HeLa cells transfected with either FLAG-TBC1D3, or HA-ARF6-Q67L, or with the empty vector (Crt), as indicated on the top, were subjected to in vitro binding experiments with GST-GGA3 (+) or GST (−), as negative control. Total cell lysates (TCL), and specifically bound TBC1D3 and ARF6 were analyzed by immunoblot (IB) with the indicated Abs. Shown are 10% of inputs and 100% the bound material. (C) Cell lysates from HeLa cells cotransfected with MYC-tagged GGA3 and either HA-tagged ARF6-Q67L or the HA-tagged PRDM5 as control unrelated protein were subjected to coimmunoprecipitation (IP) with Ab against MYC. Total cell lysates (TCL) and coimmunoprecipitated proteins were analyzed by immunoblotting (IB) with the indicated Abs. Shown are 1% of input and 100% of the bound material. (D) Cell lysates from HeLa cells transfected with HA-tagged TBC1D3 and FLAG-tagged GGA3 were subjected to coimmunoprecipitation (IP) with Ab against FLAG or unrelated anti-GFP Ab (Ctr). Total cell lysates (TCL) and coimmunoprecipitated proteins were analyzed by immunoblotting (IB) with the indicated Abs. Shown are 1% of input and 100% of the bound material. (E) Cell lysates obtained from HeLa cells transfected with either FLAG-TBC1D3 alone (left panel) or in combination with ARF6-Q67L (right panels) were subjected to in vitro binding experiments with the indicated GST-fusion fragments of GGA3. Lysates (TLC) and specifically bound proteins (IVB) were analyzed by immunoblot with the appropriate antibodies. Shown are 2.5% of lysates and 100% of bound material.
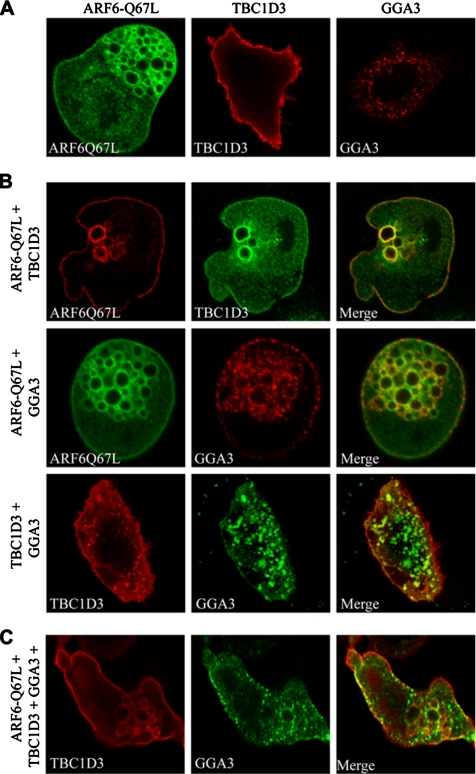
To better characterize these interactions and get some clues as to which cellular compartments TBC1D3, GGA3, and ARF6 exert their functions, we compare their cellular localization when expressed alone or in combination. Individually expressed TBC1D3 and GGA3 were mainly localized along the plasma membrane and in a vesicle-like pattern, respectively (Figure 6A), whereas ARF6-Q67L induced and localized on enlarged endo-membranes (Figure 6A), as previously described (Brown et al., 2001). On ARF6-Q67L coexpression, a significant fraction (>50%, Figure S3, top panel, in Supplementary Material) of TBC1D3 was detectable in ARF6-Q67L-induced enlarged endosomes (Figure 6B, top panel). More importantly, also GGA3 was recruited to plasma membrane and accumulated and extensively colocalized with ARF6-positive enlarged endosomes (Figure 6B, middle panel, and Figure S3, in Supplementary Material), suggesting that GGA3 can cotraffic with ARF6. We next tested whether GGA3 localization was perturbed by TBC1D3. In cells concomitantly expressing TBC1D3 and GGA3, the latter protein was partially recruited to the PM and to vesicle-like structures at the cell periphery, where, in both cases, colocalized with TBC1D3 (Figure 6B, bottom panel, and Figure S3, in Supplementary Material), thus supporting the notion that the two proteins not only coimmunoprecipitate, but can be found together in defined subcellular compartments. This latter notion was further reinforced after expression of activated ARF6. Under these conditions, GGA3 and TBC1D3 displayed significant and extensive colocalization (more than 80% of total protein colocalized; Figure S3, in Supplementary Material) along the plasma membrane, and at the level of ARF6-induced enlarged endosomes (Figure 6C). In cell expressing GGA3, TBC1D3, and ARF6Q67L or ARF6Q67L and TBC1D3, endosomes appear somewhat larger than in cells expressing only ARF6Q67L, suggesting that TBC1D3 may have an effect on endosome morphology. However, no statistically significant endosome size difference could be revealed when a larger number of TBC1D3/GGA3 and control cells were analyzed. Thus, collectively these findings suggest that the ARF6 induces relocalization of TBC1D3 and GGA3 to cellular compartments, where they are likely to promote and facilitate the macropinocytic process.
Figure 6.
GGA3 is recruited and colocalizes with ARF6 and TBC1D3 at the plasma membrane and into ARF6-positive endosomes. HeLa cells were (A) transfected with ARFQ67L, HA-TBC1D3, and FLAG-GGA3 alone as indicated on top of each image; or (B) cotransfected with ARFQ67L and HA-TBC1D3 (top panel), ARFQ67L and FLAG-GGA3 (middle panel), or HA-TBC1D3 and FLAG-GGA3 (bottom panel). Cells were fixed and stained with the appropriate Abs, as indicated, and analyzed by confocal microscopy. (C) HeLa Cells cotransfected with ARFQ67L, HA-TBC1D3, and FLAG-GGA3 were fixed and stained with the appropriate Abs, as indicated, and analyzed by confocal microscopy. The extents of colocalization are provided in Figure S3 in Supplementary Materials.
GGA3 Is Required for TBC1D3-induced Macropinocytosis
Experiments provided so far suggest that TBC1D3 may act in an ARF6-GGA3–dependent pathway, regulating and optimizing a signaling cascade that leads to macropinocytosis. To supply further functional evidences in this direction, we initially determined the surface of interaction of TBC1D3 with GGA3, using various fragments of TBC1D3 (Figure 7A). The minimal region required for binding to GGA3 comprised the TBC domain plus the N-terminal portion of TBC1D3 (aa 2-319). Notably, a deletion mutant lacking the first 91 amino acids, but retaining the TBC domain (aa 92-549), was no longer able to associate with GGA3, whereas the TBC domain plus the N-terminal portion of TBC1D3 (aa 2-319) retains this ability (Figure 7B). Attempts to express the isolated 1-92 N-terminal region of TBC1D3 upon transfection of 293T cells failed. We thus produced this region exploiting the in vitro transcription/translation system (TnT) and tested its ability to bind to immobilized GST-GGA3. An extended TBC domain of TBC1D3, including also the N-terminal region (aa 2-319), or the full-length protein bound to GGA3, was instead unable to interact with the isolated N-terminal region (aa 2-92; Figure 7C). These results also argue that interaction between TBC1D3 and GGA3 is likely direct, because the use of an in vitro TnT system would in principle exclude the contribution of other cellular proteins to the interaction They further suggest that the association of TBC1D3 with GGA3 either requires a novel binding surfaces encompassing the N-terminal region and part of the TBC domain or that an extended TBC domain is required for this interaction. This latter notion may be supported by observation that in the TBC domain of Gyp1p (Figure 2A) two additional N-terminal α-helices are present, which however are poorly conserved in TBC1D3 and are not part of the core of the domain, as defined by SMART and PFAM.
Next, we tested the relevance of the interaction of GGA3 with TBC1D3 in the process of macropinocytosis. To this end, various TBC1D3 constructs (Figure 7A), were ectopically expressed in HeLa cells, and their ability to induce dextran uptake was determined (Figure 7, D and E). Only the fragment encompassing the N-terminal portion and the TBC domain (aa 2-319) was able to elicit dextran uptake (Figure 7, D and E). All the constructs unable to associate with GGA3 (Figure 7, A and B) were also unable to stimulate macropinocytosis (Figure 7, D and E), thus providing correlative molecular evidence in support of the relevance of the TBC1D3:GGA3 interaction. This was further reinforced by examining the ability of TBC1D3 to promote macropinocytosis after siRNA-mediated ablation of GGA3. In cells were GGA3 expression was successfully reduced by siRNA specific oligos, TBC1D3-mediated dextran uptake was significantly impaired by more than 50% (Figure 8, A–D). Notably, similar levels of inhibition by GGA3 siRNA of TBC1D3-induced macropinocytosis was obtained in totally independent experiments measuring the extent of dextran uptake either by direct immunofluorescence (Figure 8C) or FACS analysis (Figure 8, D and E). To further demonstrate that both TBC1D3 and GGA3 are directly required for ARF6-mediated macropinocytosis, we individually depleted these proteins by siRNA and examined the extent of dextran incorporation induced by EFA6, the physiological activator of ARF6 (Franco et al., 1999). Remarkably, individual removal of either TBC1D3 or GGA3 significantly impaired EFA6-induced dextran uptake (Figure 8, F–I).
Next, we reasoned that if TBC1D3 and GGA3 act in concert, they should be concomitantly required to promote optimal macropinocytosis. Indeed, expression of TBC1D3 and GGA3 (Figure 9, bottom panel) led to about twofold increase in the extent of macropinocytosis, compared with TBC1D3-only–expressing cells (Figure 9, top panel). Importantly, the overexpression of GGA3 alone (Figure 9, middle panel) did not significantly induced dextran uptake in HeLa cells, supporting the idea that the effects of TBC1D3 and GGA3 are not merely additive, but rather cooperative/synergistic. Thus, collectively our data provide a cogent set of evidence, indicating that TBC1D3 by forming a complex with GGA3 promote macropinocytosis, most likely by acting as an effector unit in the pathways involving ARF6 and RAB5, thus revealing some of the components of a novel signaling cascades important in the regulation of specific endocytic routes.
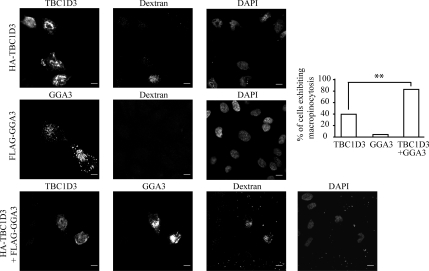
Figure 9.
TBC1D3 and GGA3 cooperate in inducing dextran uptake. HeLa cells transfected with full-length HA-TBC1D3 or FLAG-GGA3 alone or in combination, as indicated on the left, were tested for dextran incorporation. Cells were fixed and stained with the appropriate Abs and processed for confocal microscopy to detect transfected cells (left panels) or incorporated rhodamine-conjugated dextran (dextran). DAPI nuclear staining (DAPI, right panels) is also shown. Bars, 10 μm. Quantification of dextran uptake is provided as the percentage of cells incorporating dextran with respect to the total number of cells transfected with TBC1D3, GGA3, or GGA3 + TBC1D3. Data are expressed as the mean ± SD of two independent experiments in which at least 100 cells were counted (**p < 0.01).
DISCUSSION
In this study, we characterized the biological role of TBC1D3, a primate-specific gene that underwent multiple gene duplications during primate evolution. The origin of this gene can be tracked back to a duplication event that occurred very recently in evolutionary time. Despite this recent origin, TBC1D3 underwent a significant number of mutations with respect to its closest homologue, RNTRE, acquiring the features of an adaptor molecule involved, together with GGA3, in a pathway that regulates, through ARF6, the macropinocytic process. Our data establish that TBC1D3 is necessary for optimal macropinocytosis in human cells. Furthermore, molecular genetics evidence is compatible with the participation of TBC1D3 in a pathway leading to macropinocytosis, which also requires ARF6 and RAB5. In this pathway, TBC1D3 acts in concert with GGA3.
Several questions remain to be answered. First, where does TBC1D3 act? In the simplest scenario it might act at the PM where it might participate in actin remodeling and in the formation of dorsal ruffles associated with macropinocytosis. The facts that overexpression of TBC1D3 alone can induce both dorsal ruffling and macropinocytosis and that TBC1D3 colocalizes with F-actin in dynamic actin-rich structures at the PM, together with ARF6, support this possibility. In this straightforward setting, ARF6 and RAB5 might be functionally connected to TBC1D3 at the PM, because both proteins have been reported to function in that location in association with dynamic actin events leading to macropinocytosis (Schafer et al., 2000; Lanzetti et al., 2004).
More complex scenarios can, however, be envisioned. Endocytic events seem not only to be a consequence of actin remodeling at the PM, but also to be required for it. For instance, a complex interplay of endocytic routes is necessary to ensure proper localization of RAC and RHO in the formation of membrane protrusions (reviewed in Polo and Di Fiore, 2006). In addition, TBC1D3 might work, at least in part, in a vesicular/endosomal compartment, as witnessed by its localization in ARF6-induced endosomes and by its in vivo interaction with GGA3 in a vesicular compartment. Thus, TBC1D3 might also participate in endocytic/rerouting events required to properly localize the molecular machinery of ruffle formation and macropinocytosis.
A second important question relates to the biochemical function of TBC1D3. Our molecular genetics analysis indicated a hierarchical relationship TBC1D3-ARF6-RAB5. If the pathway were linear, then TBC1D3 might work as an activator of ARF6. However, we failed to detect any increase in ARF6-GTP in TBC1D3-overexpressing cells. Hierarchy does not necessarily mean, however, a linear relationship. This might be especially true in an endocytic/rerouting scenario (see above). In this framework, evidences provided in this work might help shed light on the function of TBC1D3. We demonstrate the existence of a TBC1D3:GGA3 and GGA3:ARF6-GTP complexes in vivo, both at the plasma membrane and ARF6-induced endosomes, and implicated them in the control of macropinocytosis; more importantly both TBC1D3 and GGA3 are required for ARF6-induced macropinocytosis. This body of evidence favors the possibility that TBC1D3 is involved in an effector function of ARF6, in which a putative GGA3:TBC1D3 dimeric effector is recruited to ARF6-GTP through GGA3 and acts downstream through TBC1D3 or through the GGA3:TBC1D3 complex. An alternative possibility is that TBC1D3 stabilizes an ARF6-GTP:GGA3 complex. In this case TBC1D3 might be required for macropinocytosis only in the presence of rate-limiting concentration of components of the complex (e.g., low ARF-GTP): an “optimizing” role is in line with the fact that macropinocytosis also occurs in species in which the TBC1D3 gene is not present.
A final question concerns the molecular links between ARF6 and RAB5, in the novel TBC1D3-controlled pathway described here. A recent set of observations indicated that GGAs can directly associate with the RAB5 activator complex Rabaptin-5:Rabex-5 in the trafficking between the trans-Golgi network and endosomes (reviewed in Kawasaki et al., 2005). Whether this interaction leads to activation of the GEF activity of the Rabaptin-5:Rabex-5 complex or regulates its proper localization or whether TBC1D3 participates to these events remains to be established.
An additional intriguing issue to point out arises from the observation that TBC1D3 is present in multiple copies on chromosome 17, in a region that also contains the CCL3 and CCL4 genes. CCL3 and CCL4 are chemokines, which are the natural ligands of the receptor utilized by the HIV virus to infect T-cells. Importantly, the copy number of these two genes varies in the human population as a consequence of segmental duplication events, correlating with HIV susceptibility (Gonzalez et al., 2005). A similar fluctuation in the population of TBC1D3 copy number may also occur, possibly accounting for variable individual rates of macropinocytosis. Notably, macropinocytosis can be utilized by viruses for infectious entry and/or postentry events (reviewed in Pelkmans, 2005). It will therefore be interesting to investigate whether TBC1D3 copy number impacts on viral infection. In more general terms, genomic regions embedded within segmental duplications are frequently enriched in genes associated with immunity and defense, likely contributing to the enhanced ability of humans to adapt to their environment (Bailey et al., 2002). This might provide one possible explanation for the positive selection and amplification of the TBC1D3 gene, which may have evolved to promote an efficient immune response via enhanced macropinocytosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Barbacid for providing the TBC1D3 cDNA, Mario Faretta for technical help, and Pascale Romano for critically reading and editing the manuscript. This work was supported by the following grants: Associazione Italiana Ricerca sul Cancro (G.S., L.L., and P.P.D.F.); Human Science Frontier Program (G.S.); the Italian Ministry of Health (G.S. and P.P.D.F.); European Community, Grant FP6 (G.S., L.L., and P.P.D.F.); Ministero della Salute, Ministero dell'Universita e della Ricerca, Fondazione Monzino (P.P.D.F.); Association for International Cancer Research (L.L.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0594) on January 16, 2008.
REFERENCES
- Aderem A., Underhill D. M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- Amyere M., Mettlen M., Van Der Smissen P., Platek A., Payrastre B., Veithen A., Courtoy P. J. Origin, originality, functions, subversions and molecular signalling of macropinocytosis. Int. J. Med. Microbiol. 2002;291:487–494. doi: 10.1078/1438-4221-00157. [DOI] [PubMed] [Google Scholar]
- Bailey J. A., Gu Z., Clark R. A., Reinert K., Samonte R. V., Schwartz S., Adams M. D., Myers E. W., Li P. W., Eichler E. E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey J. A., Yavor A. M., Massa H. F., Trask B. J., Eichler E. E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzoglou S., Pachter L., Mesirov J. P., Berger B., Lander E. S. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- Betts M. J., Guigo R., Agarwal P., Russell R. B. Exon structure conservation despite low sequence similarity: a relic of dramatic events in evolution? EMBO J. 2001;20:5354–5360. doi: 10.1093/emboj/20.19.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham J., Sudarsanam S. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 2000;16:660–661. doi: 10.1093/bioinformatics/16.7.660. [DOI] [PubMed] [Google Scholar]
- Bizimungu C., De Neve N., Burny A., Bach S., Bontemps F., Portetelle D., Vandenbol M. Expression in a RabGAP yeast mutant of two human homologues, one of which is an oncogene. Biochem. Biophys. Res. Commun. 2003;310:498–504. doi: 10.1016/j.bbrc.2003.09.051. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Bos J. L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- Brown F. D., Rozelle A. L., Yin H. L., Balla T., Donaldson J. G. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccione R., Orth J. D., McNiven M. A. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 2004;5:647–657. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- Caplan S., Naslavsky N., Hartnell L. M., Lodge R., Polishchuk R. S., Donaldson J. G., Bonifacino J. S. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardelli J. Phagocytosis and macropinocytosis in Dictyostelium: phosphoinositide-based processes, biochemically distinct. Traffic. 2001;2:311–320. doi: 10.1034/j.1600-0854.2001.002005311.x. [DOI] [PubMed] [Google Scholar]
- Conner S. D., Schmid S. L. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Courseaux A., Nahon J. L. Birth of two chimeric genes in the Hominidae lineage. Science. 2001;291:1293–1297. doi: 10.1126/science.1057284. [DOI] [PubMed] [Google Scholar]
- Cuif M. H., Possmayer F., Zander H., Bordes N., Jollivet F., Couedel-Courteille A., Janoueix-Lerosey I., Langsley G., Bornens M., Goud B. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 1999;18:1772–1782. doi: 10.1093/emboj/18.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., Bonifacino J. S. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhane S., Schurmann A., Sells M. A., Chernoff J., Schmid S. L., Bokoch G. M. Regulation of macropinocytosis by p21-activated kinase-1. Mol. Biol. Cell. 2000;11:3341–3352. doi: 10.1091/mbc.11.10.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G. Arf6 and its role in cytoskeletal modulation. Methods Mol. Biol. 2002;189:191–198. doi: 10.1385/1-59259-281-3:191. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G. Arfs, phosphoinositides and membrane traffic. Biochem. Soc. Trans. 2005;33:1276–1278. doi: 10.1042/BST0331276. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G., Honda A. Localization and function of Arf family GTPases. Biochem. Soc. Trans. 2005;33:639–642. doi: 10.1042/BST0330639. [DOI] [PubMed] [Google Scholar]
- Franco M., Peters P. J., Boretto J., van Donselaar E., Neri A., D'Souza-Schorey C., Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Haas A. K., Fuchs E., Kopajtich R., Barr F. A. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- Hodzic D., Kong C., Wainszelbaum M. J., Charron A. J., Su X., Stahl P. D. TBC1D3, a hominoid oncoprotein, is encoded by a cluster of paralogues located on chromosome 17q12. Genomics. 2006;88:731–736. doi: 10.1016/j.ygeno.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Innocenti M., et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 2005;7:969–976. doi: 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- Katan M., Allen V. L. Modular PH and C2 domains in membrane attachment and other functions. FEBS Lett. 1999;452:36–40. doi: 10.1016/s0014-5793(99)00531-1. [DOI] [PubMed] [Google Scholar]
- Kawasaki M., Nakayama K., Wakatsuki S. Membrane recruitment of effector proteins by Arf and Rab GTPases. Curr. Opin. Struct. Biol. 2005;15:681–689. doi: 10.1016/j.sbi.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Koonin E. V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Lanzetti L., Palamidessi A., Areces L., Scita G., Di Fiore P. P. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 2004;429:309–314. doi: 10.1038/nature02542. [DOI] [PubMed] [Google Scholar]
- Lanzetti L., Rybin V., Malabarba M. G., Christoforidis S., Scita G., Zerial M., Di Fiore P. P. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- Letunic I., Copley R. R., Pils B., Pinkert S., Schultz J., Bork P. SMART5, domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. Evolution of novel genes. Curr. Opin. Genet. Dev. 2001;11:673–680. doi: 10.1016/s0959-437x(00)00252-5. [DOI] [PubMed] [Google Scholar]
- Martinu L., Masuda-Robens J. M., Robertson S. E., Santy L. C., Casanova J. E., Chou M. M. The TBC (Tre-2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane-endosomal trafficking through activation of Arf6. Mol. Cell. Biol. 2004;24:9752–9762. doi: 10.1128/MCB.24.22.9752-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoskova B., Wong W. T., Seki N., Nagase T., Nomura N., Robbins K. C., Di Fiore P. P. RN-tre identifies a family of tre-related proteins displaying a novel potential protein binding domain. Oncogene. 1996;12:2563–2571. [PubMed] [Google Scholar]
- McLysaght A., Hokamp K., Wolfe K. H. Extensive genomic duplication during early chordate evolution. Nat. Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- McNiven M. A., Kim L., Krueger E. W., Orth J. D., Cao H., Wong T. W. Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J. Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miinea C. P., Sano H., Kane S., Sano E., Fukuda M., Peranen J., Lane W. S., Lienhard G. E. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem. J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N., Weigert R., Donaldson J. G. Convergence of non-clathrin- and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Mol. Biol. Cell. 2003;14:417–431. doi: 10.1091/mbc.02-04-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Eathiraj S., Munson M., Lambright D. G. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- Paulding C. A., Ruvolo M., Haber D. A. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc. Natl. Acad. Sci. USA. 2003;100:2507–2511. doi: 10.1073/pnas.0437015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L., Peng Y., Yang Y., Ling X. B., Van Eyndhoven W. G., Nguyen K. C., Rubin M., Hoey T., Powers S., Li J. PRC17, a novel oncogene encoding a Rab GTPase-activating protein, is amplified in prostate cancer. Cancer Res. 2002;62:5420–5424. [PubMed] [Google Scholar]
- Pelkmans L. Viruses as probes for systems analysis of cellular signalling, cytoskeleton reorganization and endocytosis. Curr. Opin. Microbiol. 2005;8:331–337. doi: 10.1016/j.mib.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Polo S., Di Fiore P. P. Endocytosis conducts the cell signaling orchestra. Cell. 2006;124:897–900. doi: 10.1016/j.cell.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Bonifacino J. S. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- Richardson P. M., Zon L. I. Molecular cloning of a cDNA with a novel domain present in the tre-2 oncogene and the yeast cell cycle regulators BUB2 and cdc16. Oncogene. 1995;11:1139–1148. [PubMed] [Google Scholar]
- Roberts R. L., Barbieri M. A., Ullrich J., Stahl P. D. Dynamics of rab5 activation in endocytosis and phagocytosis. J. Leukoc. Biol. 2000;68:627–632. [PubMed] [Google Scholar]
- Schafer D. A., D'Souza-Schorey C., Cooper J. A. Actin assembly at membranes controlled by ARF6. Traffic. 2000;1:892–903. doi: 10.1034/j.1600-0854.2000.011108.x. [DOI] [PubMed] [Google Scholar]
- Stradal T. E., Rottner K., Disanza A., Confalonieri S., Innocenti M., Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Suetsugu S., Yamazaki D., Kurisu S., Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- Sun P., Yamamoto H., Suetsugu S., Miki H., Takenawa T., Endo T. Small GTPase Rah/Rab34 is associated with membrane ruffles and macropinosomes and promotes macropinosome formation. J. Biol. Chem. 2003;278:4063–4071. doi: 10.1074/jbc.M208699200. [DOI] [PubMed] [Google Scholar]
- Swanson J. A., Watts C. Macropinocytosis. Trends Cell Biol. 1995;5:424–428. doi: 10.1016/s0962-8924(00)89101-1. [DOI] [PubMed] [Google Scholar]
- Takatsu H., Yoshino K., Toda K., Nakayama K. GGA proteins associate with Golgi membranes through interaction between their GGAH domains and ADP-ribosylation factors. Biochem. J. 2002;365:369–378. doi: 10.1042/BJ20020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S., Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- West M. A., Bretscher M. S., Watts C. Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J. Cell Biol. 1989;109:2731–2739. doi: 10.1083/jcb.109.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. M., Walsh B., Mitchell C. A., Rowe T. TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem. Biophys. Res. Commun. 2005;335:154–161. doi: 10.1016/j.bbrc.2005.07.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.