Abstract
Mutations in unc-96 or -98 cause reduced motility and a characteristic defect in muscle structure: by polarized light microscopy birefringent needles are found at the ends of muscle cells. Anti-paramyosin stains the needles in unc-96 and -98 mutant muscle. However there is no difference in the overall level of paramyosin in wild-type, unc-96, and -98 animals. Anti-UNC-98 and anti-paramyosin colocalize in the paramyosin accumulations of missense alleles of unc-15 (encodes paramyosin). Anti-UNC-96 and anti-UNC-98 have diffuse localization within muscles of unc-15 null mutants. By immunoblot, in the absence of paramyosin, UNC-98 is diminished, whereas in paramyosin missense mutants, UNC-98 is increased. unc-98 and -15 or unc-96 and -15 interact genetically either as double heterozygotes or as double homozygotes. By yeast two-hybrid assay and ELISAs using purified proteins, UNC-98 interacts with paramyosin residues 31-693, whereas UNC-96 interacts with a separate region of paramyosin, residues 699-798. The importance of surface charge of this 99 residue region for UNC-96 binding was shown. Paramyosin lacking the C-terminal UNC-96 binding region fails to localize throughout A-bands. We propose a model in which UNC-98 and -96 may act as chaperones to promote the incorporation of paramyosin into thick filaments.
INTRODUCTION
The fundamental repeating unit of myofibrils is the sarcomere, a highly ordered assemblage of many proteins that performs the work of muscle contraction. Despite ever increasing knowledge of the components of sarcomeres and moderate understanding of their assembly, much less is known about their maintenance. To address questions in muscle biology, Caenorhabditis elegans is utilized as a model organism (Waterston, 1988; Moerman and Fire, 1997; Moerman and Williams, 2006). In addition to its attributes as an excellent system in which to carry out a mutational analysis in a whole organism, this nematode offers several advantages for studying muscle. These include its optical transparency, which allows evaluation of muscle structure by polarized light, and localization of GFP tagged proteins. In addition, its usual mode of self-fertilization allows propagation of muscle mutants that would be unable to mate.
The major muscle of C. elegans lies in the body wall and is required for its locomotion. In adults, there are 95 spindle-shaped cells that are divided among four quadrants, which lie just beneath a basement membrane, hypodermis, and cuticle. This striated muscle is organized similarly to vertebrate muscle in that within each sarcomere, thin filaments are attached to Z-disk-like structures (dense bodies) and overlap with thick filaments, which are organized around M-lines. However, rather than filling the entire cell as is the case for vertebrate striated muscle, the sarcomeres are restricted to a narrow region of ∼1.5 μm along one side of the cell. Moreover, all the M-lines and Z-disks are attached to the muscle cell membrane, making these structures good models for studying muscle costameres and focal adhesions of nonmuscle cells (Moerman and Fire, 1997; Moerman and Williams, 2006).
In nematode adult body wall muscle, thick filaments are ∼10 μm long and are composed primarily of myosin heavy chain A (MHC A), myosin heavy chain B (MHC B), and paramyosin, encoded by the genes myo-3, unc-54, and unc-15, respectively (Epstein et al., 1974; Miller et al., 1983; Kagawa et al., 1989). Within each thick filament these components are differentially localized: MHC A is in the middle of the thick filament and MHC B is in the polar regions (Miller et al., 1983). Paramyosin is an invertebrate-specific protein that is primarily an α-helical coiled-coil rod and is ∼40% identical in amino acid sequence to the rod domains of myosin heavy chains. The myosins and a portion of paramyosin are organized around a tubular core consisting of paramyosin and filagenins in a specific geometry (Deitiker and Epstein, 1993; Epstein et al., 1995; Muller et al., 2001).
Recently, we have reported that two proteins, UNC-98 and -96, originally identified through genetic analysis (Zengel and Epstein, 1980), localize to M-lines. UNC-98 is a 310-residue protein containing four C2H2 Zn fingers (Mercer et al., 2003). UNC-96 is a 418-residue protein that has no recognizable domains (Mercer et al., 2006). Each of these proteins is involved in linking integrin-associated complexes to thick filaments at the M-line: Associated with the cytoplasmic tail of integrins is a complex of four conserved proteins, including UNC-97(PINCH) (Moerman and Williams, 2006; Norman et al., 2007). Two independent sets of protein–protein interactions function to link UNC-97 to a myosin heavy chain, MHC A. First, UNC-97 interacts with the Zn fingers of UNC-98, and the N-terminal portion of UNC-98 interacts with the C-terminus of MHC A (Miller et al., 2006). Second, UNC-97 associates with LIM-8 and -9, which in turn interact with UNC-96, which binds to MHC A (Qadota et al., 2007).
Mutations in either unc-98 or -96 result in a characteristic defect in muscle structure: by polarized light microscopy, the myofibrils are less organized and there are birefringent needle-like structures at the ends of muscle cells (Zengel and Epstein, 1980; Mercer et al., 2003, 2006). In this article we demonstrate that the needles in unc-98 mutants contain accumulations of paramyosin and that UNC-98 interacts both genetically and biochemically with paramyosin. Previously, we obtained similar results with UNC-96 (Mercer et al., 2006). Furthermore, we show that separate regions of paramyosin interact with UNC-98 and -96: UNC-98 to paramyosin residues 31-693 and UNC-96 to paramyosin residues 699-798. The levels of UNC-98 depend on the level or mutant state of paramyosin. Paramyosin lacking the C-terminal UNC-96 binding region fails to localize to A-bands. We suggest a model in which UNC-98 and -96 may act as chaperones to promote incorporation of paramyosin into thick filaments.
MATERIALS AND METHODS
Nematode Strains and Genetics
The following C. elegans strains were used in this work: wild-type N2, unc-98(su130), unc-98(sf19), unc-96(su151), unc-96(r291), unc-96(sf18), dpy-7(e88) unc-98(su130), dpy-7(e88), unc-15 (e1215), unc-15(e73), and unc-15(e1214). To generate the unc-15(e1215)/+; dpy-7(e88) unc-98(su130)/+ double heterozygotes, unc-15(e1215)/+ males were crossed with dpy-7(e88) unc-98(su130) homozygous hermaphrodites. One-half of the resulting nondumpy progeny are double heterozygotes. The dumpy progeny from the same cross were allowed to self-cross, producing one-quarter unc-15(e1215); dpy-7(e88) unc-98(su130) homozygotes. unc-15(e1215); dpy-7(e88) was also generated.
Antibody Staining and Immunofluorescence Microscopy
N2, unc-98, and unc-15 animals were stained using procedures described in Mercer et al. (2003) modified from Benian et al. (1996). Mouse monoclonal antibodies used for immunofluorescence localization include: anti-paramyosin antibody 5-23 at 1:200 (vol/vol) dilution (Miller et al., 1983), anti-myosin heavy chain A antibody 5–6 at 1:400 dilution (Miller et al., 1983), anti-myosin heavy chain B antibody 5–8 at 1:400 dilution (Miller et al., 1983), and anti-α-actinin antibody MH35 at 1:200 (Francis and Waterston, 1985). Additionally, a polyclonal anti-UNC-98 antibody EU131 affinity-purified against the full-length UNC-98 protein was used at 1:200 dilution (Mercer et al., 2003). To visualize antibody localization, fluorescein isothiocyanate, tetramethylrhodamine B isothiocyanate, and Cy-3–conjugated secondary antibodies, purchased from Jackson ImmunoResearch Laboratories (West Grove, PA), were used at 1:400 dilution. Images depicting single antibody localization within body-wall muscle were captured using a Zeiss Axioskop microscope (Carl Zeiss, Jena, Germany) and were captured using a Zeiss D4 Databack 35-mm camera and Fuji Sensei 100 Film (Tokyo, Japan). Images depicting dual antibody localization within body-wall muscle were captured with a scientific-grade, cooled charge-coupled device (Cool-Snap HQ with ORCA-ER chip) on a multi-wavelength, wide-field, three-dimensional microscopy system (Intelligent Imaging Innovations, Denver, CO). Samples were imaged in successive 0.2-μm focal planes, and out-of-focus light was removed using either the nearest neighbor or the constrained iterative deconvolution algorithm (Weiner et al., 1999). Images were processed using Adobe Photoshop software (San Jose, CA).
Western Blots
Extracts from wild-type and unc-96, -98, and -15 mutant animals were prepared using the method described by Hannak et al. (2002). The protein concentrations of the extracts were determined as described in Minamide and Bamburg (1990). Extracts were separated on 10 or 12% SDS-PAGE gels and transblotted onto nitrocellulose membranes. To determine the amount of protein extract to run within the linear range of detection of film, trial experiments were performed in which the quantity of extract ranged from 1 to 50 μg for each antibody. The Western blots were exposed to anti-paramyosin monoclonal 5-23 at 1:1000 (vol/vol), anti-actin monoclonal C4 at 1:1000 (vol/vol; Chemicon International, Temecula, CA), affinity-purified rabbit antibodies to the N-terminal region of UNC-98 (Miller et al., 2006) at 1:200 (vol/vol), or affinity-purified rabbit antibodies to UNC-96 (Mercer et al., 2006) at 1:200 (vol/vol). Enhanced chemiluminescence (ECL; Amersham, Indianapolis, IN) was used to detect antibody reactions.
Actomyosin Preparation
Actomyosin was prepared from wild-type (N2), unc-96(sf18), and unc-98(sf19) animals as described in Epstein et al. (1974). Ten micrograms of each actomyosin preparation were separated on a 10% SDS-PAGE gel. The proteins were visualized by Coomassie staining.
Polarized Light and Stereoscopic Microscopy
The organization of the body-wall muscle in unc-15/+ heterozygotes, dpy-7 unc-98/+ heterozygotes, and unc-15/+; dpy-7 unc-98/+ double heterozygotes was examined by polarized light microscopy as described in Waterston et al. (1980). Stereoscopic images of unc-15, dpy-7 unc-98, unc-15; dpy-7 unc-98, and unc-15; dpy-7 homozygotes were captured using a Zeiss SV-11 microscope and a Nikon Coolpix 4500 camera (Melville, NY).
Production of Bacterially Expressed UNC-98 and -96
cDNA encoding the entire 310 amino acids of UNC-98 was generated using a random-primed cDNA library (a gift from R. Barstead, Oklahoma Medical Research Foundation, Oklahoma City, OK). The full-length UNC-98 cDNA was amplified using 5′ primer 98HIS5′ (gtacggatccatggatgacgacatcttcaaagaggc) and 3′ primer 98HIS3′ (gatgaagcttgaatcgcggagtcacgtatccgcttg). The amplified cDNA was inserted into the pET-24a expression vector (Novagen, Madison, WI) using restriction sites 5′ BamHI and 3′ HindIII. After sequencing the DNA insert to verify that it was error free, the plasmid was transformed into Escherichia coli BL21-CodonPlus (DE3)-RIL competent cells (Stratagene, La Jolla, CA). Construction of a similar vector for expression of the N-terminal 112 amino acids of UNC-98 was described in Miller et al. (2006). The UNC-98 proteins tagged with six histidine residues at their C-terminus were induced and purified using His-Bind Nickel Columns (Novagen). The Novagen His-Bind binding and wash buffers used for purification of the full-length UNC-98 His-tagged protein were modified to include 0.1% NP-40 and 750 mM NaCl. A 30-kDa molecular-weight cutoff Centricon centrifugal filter device (Millipore, Bedford, MA) was used to remove low-molecular-weight impurities from the full-length UNC-98 protein. The proteins were dialyzed in 50 mM Tris, pH 7.5, buffer. The His-tagged UNC-96 was produced as described in Mercer et al. (2006).
Paramyosin Enzyme-linked Immunosorbent Assays
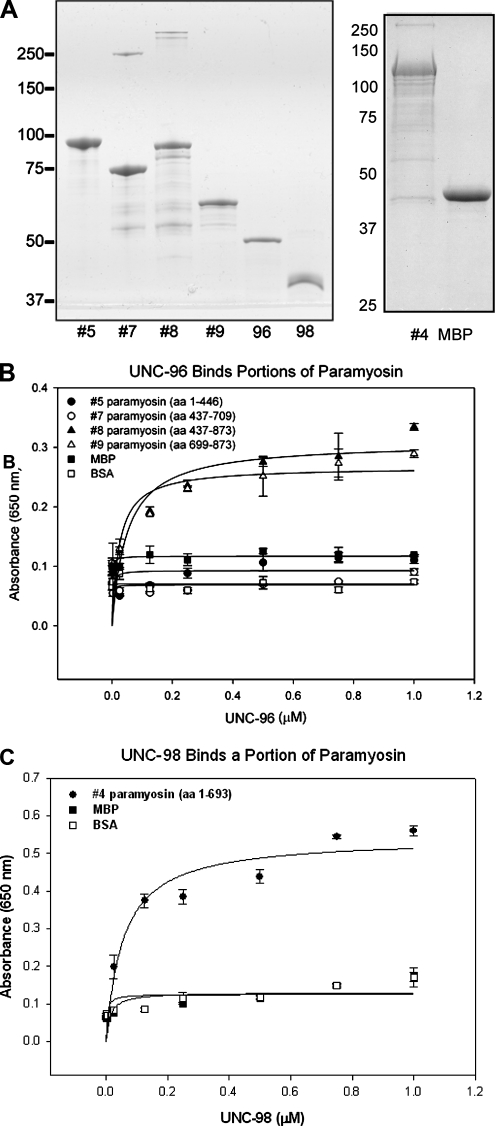
Paramyosin was purified from wild-type actomyosin (see above) using the procedure of Waterston et al. (1974). The concentrations of His-tagged UNC-98 was determined using the Bio-Rad Bradford-based protein assay (Richmond, CA). One microgram of each of these proteins was run on a 15% SDS-polyacrylamide gel, which was Coomassie-stained. Paramyosin was then coated on Corning polystyrene microtiter plates (cat. 3591) at a concentration of 0.5 μM, at 100 μl per well in the following buffer: 10 mM NaPO4, pH 7.6, 0.6 M NaCl, and incubated at 4°C overnight. The enzyme-linked immunosorbent assay (ELISA) was performed as follows: 1) Wells were incubated with block (0.2% bovine serum albumin [BSA]), 100 mM KCl, 10 mM Tris, pH 8.0, 0.05% Tween-20) for 1.5 h at room temperature. 2) Wells were then washed three times with wash buffer (the same as block, without the BSA) and vacuum aspirated. 3) The bacterially expressed His-tagged UNC-98 protein and the His-tagged N-terminal 112 amino acids of UNC-98 were then incubated at 0–1.25 μM, at 50 μl per well, for 1 h at room temperature. 4) The washing procedure was repeated as described previously. 5) Wells were coated with 75 μl of anti-6His antibody (Santa Cruz 803 anti-rabbit) at a 1:200 dilution in block for 45 min at 37°C. 6) The washing procedure was repeated. 7) Wells were incubated with 50 μl of donkey anti-rabbit HRP antibody (Amersham) at a 1:1000 dilution for 45 min at 37°C. 8) The washing procedure was repeated. 9) Wells were coated with 100 μl of mixed TMB solution (BD Biosciences, San Jose, CA), and the plate was placed in the dark for 20 min. 10) The absorbance was read at 650 nm using a Synergy HT Multi-Detection Microplate Reader with KC4 data analysis software (BIO-TEK Instruments, Winooski, VT). Microtiter plates were coated in tandem with 100 μl of 0.5 μM BSA as a control for nonspecific binding of assay components. Means and SDs of absorbance values were plotted, and the best fit ligand binding (single-site saturation) curves were plotted (SigmaPlot 9.0; Stystat, San Jose, CA).
Paramyosin Yeast Two-Hybrid Assays
Bait plasmids for expression of residues 1–200 of UNC-96 (Mercer et al., 2006), residues 201–418 of UNC-96 (Mercer et al., 2006), residues 1–112 of UNC-98 (Miller et al., 2006), and residues 1–140 of UNC-98 were generated by inserting PCR-generated cDNA into the pGDBU vector. Prey plasmids for expression of specific regions of UNC-15 (paramyosin) were generated by inserting amplified cDNA into the pGAD-C1 vector. Primers for amplification of these inserts are available upon request. UNC-15 cDNAs harboring nine amino acids changes (R759A, K762A, E763A, R773A, K776A, E777A, D794A, D797A, and R798A) were prepared by using PCR with oligonucleotides containing point mutations. Yeast two-hybrid assays were performed as described in Mackinnon et al. (2002).
Paramyosin MBP Fusions and ELISAs
Truncation derivatives of paramyosin were expressed as MBP fusion proteins by first removing the corresponding inserts from the pGAD-C1 recombinant plasmids using the BamHI and PstI restriction sites. The inserts were ligated into the pBluescript vector using the same sites. The inserts were removed from pBluescript and inserted into pMAL-KK-1 (kindly provided by Dr. K. Kaibuchi, Nagoya University, Japan) using the BamHI and EcoRI restriction sites. These proteins were expressed in BL21-CodonPlus (DE3)-RIL bacteria (Stratagene) and purified as described in Mercer et al. (2006). Two to 5 μg of these proteins along with UNC-96 His and UNC-98 His were separated on 7.5 or 10% SDS-PAGE gels and Coomassie-stained. ELISAs were performed using methods described above.
Transgenic Animals Expressing Full-Length or C-Terminally Deleted Paramyosin Fused to GFP
The full length unc-15 coding sequence was amplified from cDNA using primers unc-15-hs-5′ (gactctgcagatgtcattgtatcgttcgccatcc) and unc-15-hs-3′ (ctgaggatccataatcgtcttccgtgacgaaaatc). The unc-15 coding sequence lacking the nucleotides that encode the C-terminal 174 residues of paramyosin was amplified from cDNA using primers unc-15-hs-5′ and N unc-15 hs-3′ (ctgaggatccttcctcatgaagttgttcaacggc). The PCR products were digested with BamHI and PstI and were inserted into the pPD95.77 plasmid using the same restriction sites. The unc-15 genes in fusion with the coding region for GFP were cut from the pPD95.77 plasmid using restriction sites PstI and EcoRI, and T4 polymerase was used to blunt the staggered ends. pPD49.83 (contains heat-shock promoter) was digested with EcoRV, and the unc-15 gfp constructs were inserted to generate unc-15 gfp under the control of a heat-shock promoter. The plasmids were injected into wild-type animals in combination with rol-6 DNA to generate transgenic lines. Multiple transgenic lines were generated for each construct. To achieve unc-98 knockdown by RNA interference (RNAi), a full-length unc-98 injection construct utilized in Mercer et al. (2003) was inserted into the L4440 vector using restriction sites SstI and HindIII. This RNAi plasmid was transformed into HT115 (DE3) cells. One hundred Roller animals expressing either the full-length or C-terminally deleted paramyosin GFP were fed the RNAi feeding bacteria (Kamath and Ahringer, 2003) beginning at the L4 stage. The following day these animals were transferred to 10 RNAi feeding plates and were allowed to lay embryos. After 1 d, these adult animals were transferred off of the plates, and the embryos laid on the plate were allowed to grow for 4 d at 15°C. Transgenic animals with a wild-type background and those fed unc-98 RNAi bacteria were heat-shocked at 30°C for 5 h. One hundred to 400 adult rollers in each group were picked into M9 buffer, washed, and then fixed by the picric acid procedure (Nonet et al., 1993). These worms were costained with anti-paramyosin (5-23) monoclonal at 1:200 and affinity-purified rabbit anti-green fluorescent protein (GFP; Invitrogen) at 1:500 dilutions. (Anti-GFP rather than GFP fluorescence was used because the Nonet method, which is convenient for small numbers of worms, does not allow detection of GFP fluorescence.) Anti-GFP was visualized by anti-rabbit antibodies conjugated with Alexa 488 (Molecular Probes, Eugene, OR), and the paramyosin monoclonal was visualized by anti-mouse antibodies conjugated with Cy3 (Jackson Immunochemicals, West Grove, PA). Images were captured with a Carl Zeiss LSM 510 confocal microscopy system.
RESULTS
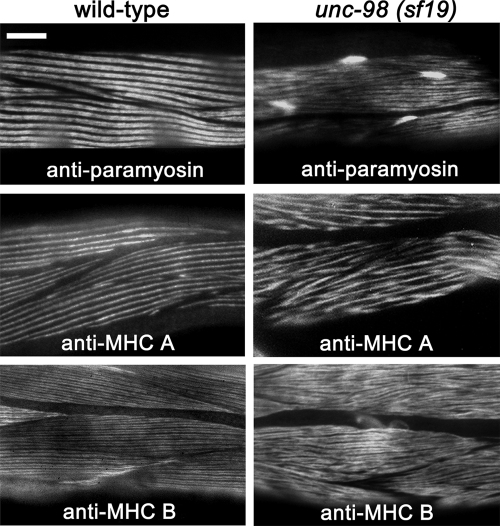
By polarized light microscopy, unc-98 mutants show discrete birefringent needles at the ends of their body wall muscle cells (Zengel and Epstein, 1980; Mercer et al., 2003, 2006). These needles are somewhat similar in appearance to the paramyosin accumulations found in paramyosin (unc-15) mutants (Waterston et al., 1977; our unpublished data). Therefore, unc-98(sf19) mutant animals were stained with antibodies against paramyosin. As shown in Figure 1, in wild-type muscle, paramyosin is found in A-bands, whereas in unc-98(sf19) muscle, some paramyosin is found in A-bands, but much of it is found in accumulations at the ends of the muscle cells. unc-98(sf19) mutants were also stained with antibodies against MHC A and B (Figure 1). Compared with wild-type muscle, MHC A is not organized into as sharply a defined pattern, but it is not found in accumulations. Likewise, in unc-98(sf19), MHC B has a less organized staining pattern and is also excluded from the accumulations at the ends of the cells. Similar results were obtained when a second unc-98 allele, su130, was examined (data not shown). Therefore, of the major components of thick filaments, unc-98 mutations have their greatest effect on the organization of paramyosin. unc-96 mutants also have birefringent needles at the ends of their body wall muscle cells by polarized light microscopy (Zengel and Epstein, 1980) and have paramyosin localized to accumulations at the ends of muscle cells (Mercer et al., 2006). Because the paramyosin accumulations in unc-96 and -98 mutants have a location and size similar to the birefringent needles seen by polarized light, we suggest that the needles are accumulations containing paramyosin.
Figure 1.
Body wall muscle cells from unc-98 mutants contain accumulations of paramyosin. Immunofluorescent localization of paramyosin, MHC A and B in wild-type and unc-98(sf19) mutant muscle are shown. Compared with wild-type, unc-98 mutant muscle shows only mild disruption in the localization of MHC A and B, but a dramatic disruption in the localization of paramyosin. In unc-98(sf19), some paramyosin is located in A-bands, but much of it is found in accumulations at the ends of the muscle cells. Bar, 10 μm.
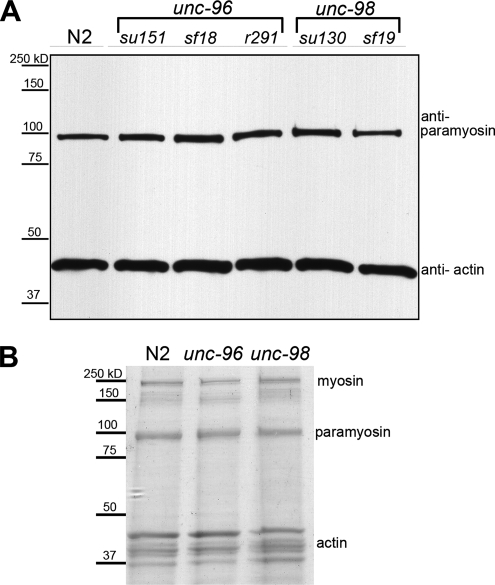
Because mutations in unc-96 and -98 result in paramyosin accumulations, the steady-state level of paramyosin was compared between wild-type and the unc-96 and -98 mutants. When equal quantities of total Laemmli soluble proteins were compared by Western blot for paramyosin, no significant differences were seen (Figure 2A). Western blots using extracts from unc-96 mutants exposed to unc-96 RNAi and unc-98 mutants exposed to unc-98 RNAi, to enhance the nonnull mutations, also showed no appreciable change in the paramyosin levels (data not shown). As an independent assessment of paramyosin levels, actomyosin was prepared from wild-type and these mutants. As shown in Figure 2B, the level of paramyosin by Coomassie staining within these actomyosin preparations is also similar between wild-type and the unc-96 and -98 mutants.
Figure 2.
The level of paramyosin is similar among wild-type (N2) and unc-96 and -98 animals. (A) Western blot analysis showing that the level of paramyosin does not vary substantially within protein extracts from wild-type and unc-96 and -98 animals. Actin is used as a loading control. (B) Actomyosin was purified from wild-type, unc-96(sf18), and unc-98(sf19) animals. The levels of paramyosin are similar within these preparations, as visualized by Coomassie staining of an SDS-PAGE gel.
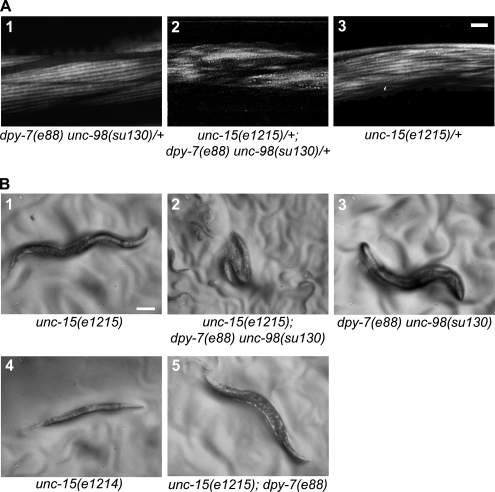
To obtain in vivo evidence that UNC-98 and paramyosin are functionally linked in building the contractile apparatus, we looked for possible genetic interactions. First, unc-15(e1215)/+; unc-98(su130)/+ double heterozygotes were generated. By polarized light microscopy, unc-98(su130)/+ heterozygotes have a normal appearing myofilament lattice, with alternating bright A-bands and dark I-bands (Figure 3A1). unc-15(e1215) is a semidominant mutation (Gengyo-Ando and Kagawa, 1991), and the polarized light banding pattern within the unc-15(e1215)/+ heterozygote is mildly disrupted (Figure 3A3). The body wall muscle of unc-15(e1215)/+; unc-98(su130)/+ double heterozygotes is highly disorganized, with indistinguishable A- and I-bands (Figure 3A2). Similarly, unc-15 and -96 show a genetic interaction as double heterozygotes: unc-15/+; unc-96/+ have a disorganized myofilament lattice by polarized light microscopy (Mercer et al., 2006).
Figure 3.
Genetic interaction between unc-98 and -15, the structural gene for paramyosin. (A) By polarized light microscopy, unc-15(e1215)/+; dpy-7(e88) unc-98(su130)/+ double heterozygotes (2) have a highly disorganized myofilament lattice in which the A- and I-banding is indistinguishable, whereas, dpy-7(e88) unc-98(su130)/+ heterozygotes (1) and unc-15(e1215)/+ heterozygotes (3) maintain an ordered A (bright) and I (dark) banding pattern. Bar, 10 μm. (B) unc-15(e1215) (1) and dpy-7(e88) unc-98(su130) (3) homozygous mutant worms are motile on a plate, whereas unc-15 (e1215); dpy-7(e88) unc-98(su130) homozygotes (2) are unable to unfold and are paralyzed as adults. The marker dpy-7(e88) has an obvious short and fat phenotype and is closely linked to unc-98, allowing us to track the unc-98 mutation in crosses. dpy-7 has no effect on myofibril organization and does not cause paralysis when combined with either unc-98 (3) or unc-15 (5). The paramyosin null mutant, unc-15(e1214) (4) is paralyzed, but does not have the folded-up posture of the unc-15(e1215); dpy-7(e88) unc-98(su130) double mutant. Bar, 0.1 mm.
Additional evidence for a genetic interaction between unc-98 and -15 was obtained by comparing homozygous unc-98 and homozygous unc-15 mutants with unc-15; unc-98 double homozygotes. Neither the unc-15(e1215) homozygote (Figure 3B1) nor the unc-98(su130) homozygote (Figure 3B3) has dramatic motility defects on an agar plate independently. However, mutants homozygous for both unc-15(e1215) and unc-98(su130) are paralyzed and unable to unfold themselves (Figure 3B2). A similar paralysis is seen in the unc-15; unc-96 double homozygote (Mercer et al., 2006).
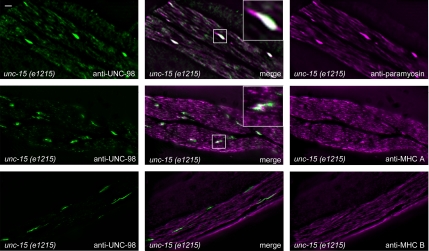
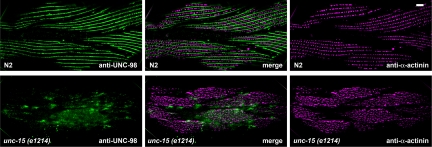
To obtain further in vivo evidence for a physical interaction between paramyosin and UNC-98, we examined the localization of UNC-98 in unc-15 missense mutants that contain multifilament assemblages consisting of central paramyosin paracrystals and polar thick filament-like structures (Epstein et al., 1987, 1993). unc-15(e1215) is a mild missense allele of unc-15 (Gengyo-Ando and Kagawa, 1991). unc-15(e1215) mutants were costained with UNC-98 antibodies in combination with antibodies against paramyosin, MHC A or B. Accumulations of paramyosin contain UNC-98. In addition, these UNC-98 containing accumulations also contain MHC A but not MHC B (Figure 4, rows 2 and 3). Similar results were obtained with the stronger unc-15 missense allele, e73 (data not shown). Similarly, in unc-15(e1215), UNC-96 can be found in accumulations that also contain paramyosin and to a lesser extent, MHC A, but not MHC B (data not shown).
Figure 4.
Immunofluorescence localization of UNC-98, paramyosin, and MHC A and B within a paramyosin missense mutant. The paramyosin mutant, unc-15(e1215), displays characteristic accumulations of paramyosin within its body wall muscle cells, as seen in the top right image. These accumulations also contain UNC-98 (top, left). Additionally, accumulations of UNC-98 contain some MHC A (middle row of images), but not MHC B (bottom row of images).
Based on the mislocalization of UNC-98 within the unc-15 missense mutants, the effect of total absence of paramyosin on UNC-98 localization was examined. Wild-type and unc-15(e1214) null mutants were costained with antibodies against UNC-98 and α-actinin (as a muscle cell marker). Within unc-15(e1214) paramyosin nulls, UNC-98 is highly mislocalized and the overall level of UNC-98 appears reduced (Figure 5). The localization of UNC-98 is, therefore, dependent on the presence of paramyosin. UNC-96 localization also depends on the presence of paramyosin (Mercer et al., 2006).
Figure 5.
UNC-98 mislocalizes in a paramyosin null mutant. As shown in the top left image, UNC-98 localizes to M-lines (Mercer et al., 2003) within wild-type C. elegans muscle cells. However, as seen in the bottom left image, when paramyosin is absent from muscle cells as in the null mutant unc-15(e1214), UNC-98 no longer localizes to the M-lines and is randomly dispersed within the cells. In the right column, α-actinin staining in the dense bodies can be seen in both wild-type and unc-15(e1214) mutant muscle, although there is some secondary disorganization of the dense bodies within unc-15(e1214) mutant cells. The α-actinin staining was utilized to locate cells microscopically because muscle cells within the paramyosin null mutant were difficult to identify given the highly disrupted localization of UNC-98.
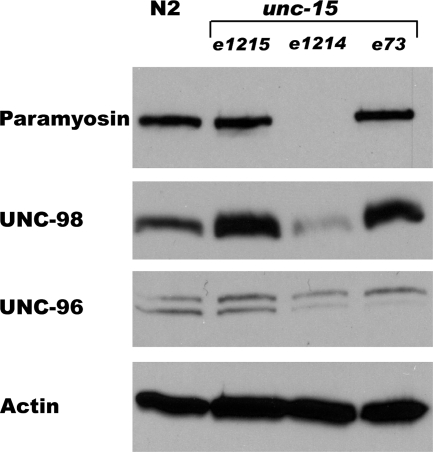
We next examined the effect of mutations in the paramyosin gene on the levels of UNC-98 and -96 by quantitative Western blotting. Equal amounts of total Laemmli-soluble proteins were Western-blotted and examined with antibodies to paramyosin, actin (as a loading control), and UNC-98 and -96. As shown in Figure 6, the absence of paramyosin (in the unc-15 null allele, e1214) results in a dramatic decrease in the level of UNC-98. The effect of abnormal paramyosin carrying missense mutations (unc-15 alleles e1215 and e73), although not causing a change in total paramyosin levels, results in the elevation of the level of UNC-98. The effects of mutations in unc-15 on UNC-96 levels were minimal, although there is a hint that at least the absence of paramyosin results in a decline in the level of UNC-96 (Figure 6).
Figure 6.
The levels of UNC-98 and -96 in wild-type and paramyosin mutant worms. Extracts containing equal amounts of total Laemmli-soluble proteins from wild-type (N2) and the three unc-15 mutants were separated on a gel, blotted, and reacted with antibodies to the indicated proteins. The level of UNC-98 is depressed in the absence of paramyosin (unc-15(e1214)) and elevated in the presence of paramyosin containing missense mutations (e1215 and e73). The levels of actin serve as loading controls.
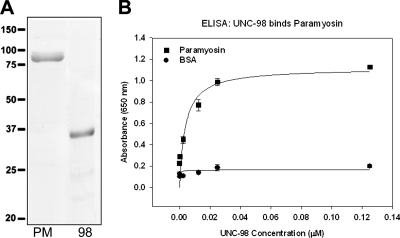
Given the evidence that UNC-98 and paramyosin interact in vivo, we tested for a direct interaction between these two proteins in vitro. Paramyosin was purified from wild-type worms using methods described in Epstein et al. (1974) and Waterston et al. (1974). A full-length His-tagged UNC-98 protein was expressed and purified from E. coli. An SDS-PAGE gel with each of the proteins is shown in Figure 7A. As shown in Figure 7B, by ELISA, UNC-98 binds paramyosin in a saturable manner, but does not bind to BSA. This paramyosin/UNC-98 interaction is strong, with an apparent Kd = 3.7 nM. UNC-96 has also been shown to saturably bind paramyosin in an ELISA (Mercer et al., 2006).
Figure 7.
An ELISA assay demonstrates that UNC-98 has a saturable interaction with paramyosin in vitro. (A) The proteins used in the ELISA assay include paramyosin purified from wild-type worms and bacterially expressed His-tagged full-length UNC-98 (310 amino acids). These purified proteins were visualized on an SDS-PAGE gel by Coomassie staining. (B) When the bacterially expressed UNC-98 (concentrations from 0 to 1 μM) was exposed to purified paramyosin that had been adsorbed to the plate, it bound increasingly to paramyosin until it reached a saturation level, with an apparent Kd = 3.7 nM. UNC-98 did not bind BSA within this range of concentrations.
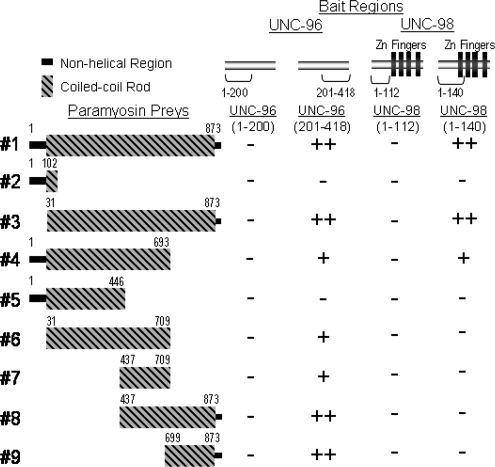
Yeast two-hybrid analysis was used to determine which portions of each protein were required for paramyosin to interact with UNC-98 and -96. Portions of UNC-96 and -98 were tested as two-hybrid baits to portions of paramyosin as two-hybrid preys (Figure 8). Two UNC-96 baits were used (Mercer et al., 2006): one is composed of the N-terminal 200 amino acids, and the other is composed of the C-terminal 218 amino acids. The N-terminal half of UNC-96 did not bind any portion of paramyosin. The C-terminal half of UNC-96 interacted with several paramyosin preys. There are two regions of paramyosin involved in its interaction with UNC-96: amino acids 437–709 of paramyosin (fragment 7) are sufficient for yeast growth, and amino acids 699–873 of paramyosin (fragment 9) are sufficient for stronger yeast growth. Two UNC-98 baits were used: one is composed of the N-terminal 112 amino acids (Miller et al., 2006), and the other is composed of the N-terminal 140 amino acids. The C-terminal portion of UNC-98 has a high background in yeast two-hybrid assays, and therefore, it does not make a good bait. The N-terminal 112 amino acids of UNC-98 do not interact with any portion of paramyosin, despite the fact that this portion does interact with MHC A (Miller et al., 2006). However, the N-terminal 140 amino acids of UNC-98 interact with several of the paramyosin preys. The strongest yeast growth was seen when either the full-length paramyosin (fragment 1), or a derivative lacking the N-terminal nonhelical 30 residues (fragment 3) was used. Weaker yeast growth was observed when the N-terminal 693 residues of paramyosin (fragment 4) were used.
Figure 8.
The C-terminal half of UNC-96 and the N-terminal 140 residues of UNC-98 interact with specific portions of paramyosin. Four yeast two-hybrid baits were used to test for interaction with portions of paramyosin: the N-terminal 200 residues of UNC-96, the C-terminal 218 residues of UNC-96, the N-terminal 112 residues of UNC-98, and the N-terminal 140 residues of UNC-98. The N-terminal half of UNC-96 does not interact with any portion of paramyosin. The C-terminal half of UNC-96 interacts with several portions of paramyosin. Specifically, two regions of paramyosin are sufficient. Preys containing residues 437–709 (numbers 1, 3, 4, 6, 7, and 8), allow some yeast growth, whereas preys containing residues 699–873 (numbers 1, 3, 8, and 9) allow more yeast growth, indicating a stronger interaction. The N-terminal 112 residues of UNC-98 do not interact with any portion of paramyosin. However, the N-terminal 140 residues of UNC-98, which include the first zinc finger, interact most strongly with preys containing the entire coiled-coil portion of paramyosin (numbers 1 and 3). The N-terminal 30 residue nonhelical region is not necessary for its interaction with UNC-98. A prey encoding residues 1–693 (number 4) allows weak yeast growth.
To obtain additional evidence for the interactions identified by yeast two-hybrid assay, an ELISA was conducted with bacterially expressed proteins. His-tagged UNC-96 and UNC-98 and maltose binding protein (MBP) fusions to portions of paramyosin were generated (Figure 9A). For the UNC-96 ELISAs, paramyosin fragment 5 (residues 1-446; the region that does not interact with UNC-96 in two-hybrid), fragment 7 (residues 437-709), fragment 8 (residues 437-873), and fragment 9 (residues 699-873), MBP and BSA were adsorbed to the ELISA plate. As shown in Figure 9B UNC-96 bound in a saturable manner to paramyosin fragment 8 (437-873) with an apparent Kd = 49 nM and to paramyosin fragment 9 (699-873) with an apparent Kd = 24 nM but did not bind any other region of paramyosin, MBP, or BSA. Similarly, the smallest region of paramyosin (4, 1-693) shown to interact with UNC-98 by two-hybrid was tested in an ELISA. As shown in Figure 9C, this portion of paramyosin binds to UNC-98 in a saturable manner with an apparent Kd = 56 nM. Thus, by ELISAs, the minimal regions of paramyosin that interact with UNC-98 and -96 are nonoverlapping: the N-terminal 80% of paramyosin (residues 31-693) can interact with UNC-98, whereas the C-terminal 20% of paramyosin (residues 699-873) can interact with UNC-96.
Figure 9.
ELISA confirmation that UNC-96 and -98 interact with specific portions of paramyosin. (A) Bacterially expressed MBP, MBP-tagged portions of paramyosin (residues 1-446, 437-709, 437-873, 699-873, and 1-693), and His-tagged UNC-96 and -98 were purified and separated by SDS-PAGE. The MBP-tagged portions of paramyosin, MBP, and BSA were bound to an ELISA plate. (B) UNC-96 bound residues 437-873 (apparent Kd = 49 nM) and 699-873 (apparent Kd = 24 nM) of paramyosin in a saturable manner in a range from 0 to 1.0 μM, but did not bind MBP or BSA. (C) Similarly, UNC-98 bound residues 1-693 (apparent Kd = 56 nM) of paramyosin in a saturable manner in a range from 0 to 1.0 μM, but did not bind MBP or BSA.
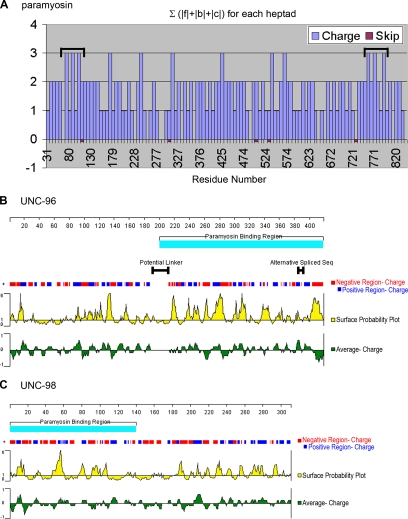
By sequence analysis, we attempted to explain how these portions of paramyosin interact with UNC-96 and -98. A cardinal feature of α-helical coiled-coil dimers, like paramyosin, is the heptad repeat (the amino acids arbitrarily designated a–g). Residues at positions a and d form a hydrophobic core, with the a residue of one polypeptide interacting with the d residue of the other polypeptide. Residues e and g, which border the hydrophobic core, are frequently charged and often provide salt bridges to define the chain alignments and orientation. Importantly, residues f, b, and c reside on the surface of the coiled coil cylinder and are usually charged or polar (Branden and Tooze, 1999). Although the basic topology of the coiled-coil surface is uniform, we wondered whether the f, b, and c “coat residues” might show some variation that might explain binding to UNC-98 and -96. Figure 10A is a plot of the sum of the absolute values of charges for f, b, and c at each heptad. (These were calculated using “1” for each E, D, R, and K). Inspection of this graph reveals two regions with particularly high charge density, each having a cluster of peaks of charge 3. One of these regions spans residues 73-107, the other spans residues 757-798. Interestingly, the first region ends with a skip residue and the second region begins near a skip residue. Significantly, the second region of increased charge density (757-798) overlaps the region we have identified as the strongest binding region for UNC-96 (699-873).
Figure 10.
Characteristics of the paramyosin, UNC-96 and -98 proteins that may influence their interactions. (A) Plot of the sum of the absolute values of charges for f, b, and c of each heptad (These were calculated using 1 for each E, D, R, and K). Two regions, residues 73-107 and 757-798 (indicated by brackets), have a higher charge density. Blue bars indicate the absolute values of the charges, and red boxes indicate positions of the skip residues. (B) Characteristics of the UNC-96 amino acid sequence. The turquoise bar represents the 218-residue region that binds paramyosin. The black bar on the left represents a region of highly flexible residues with very little charge. The black bar on the right represents the alternative splice forms of UNC-96. The red (negative) and blue (positive) banding represents the distribution of charge along the polypeptide. The yellow plot represents the surface probability of the residues. The green plot represents the average charge of the residues. (C) Characteristics of the UNC-98 amino acid sequence. The turquoise bar represents the 140-residue region that binds paramyosin. The red (negative) and blue (positive) banding represents the distribution of charge along the polypeptide. The yellow plot represents the surface probability of the residues. The green plot represents the average charge of the residues.
Analysis by the Protean programs of DNASTAR on UNC-96 and -98 is also revealing (Figure 10, B and C). A prediction of surface probability indicates that the region of UNC-96 (residues 201-418) that binds paramyosin has a different pattern of surface probability than the region that does not bind paramyosin: the frequency and magnitude of peaks indicating surface probability is increased in the binding region. In addition, throughout nearly all of the UNC-96 sequence short patches of positive charge alternate with short patches of negative charge, but in the very middle (residues 190-213) there is a paucity of charge. This 23-residue region has a high proportion of amino acids that are turn-forming or flexible (3Gs, 2Ps, 4Ss, 3Ts, 4As, and 2Ns); programs predict that this region is random coil. Thus, UNC-96 may consist of two domains separated by a flexible loop or linker, with the C-terminal domain interacting with paramyosin. In the case of UNC-98, a surface probability plot also shows a distinct pattern in the paramyosin binding region (residues 1-140): although the frequency of peaks is reduced, the magnitude of the peaks is increased. Overall, this analysis suggests that the surface of the coiled-coil rod of paramyosin might interact with UNC-96 and -98 through electrostatic or polar interactions.
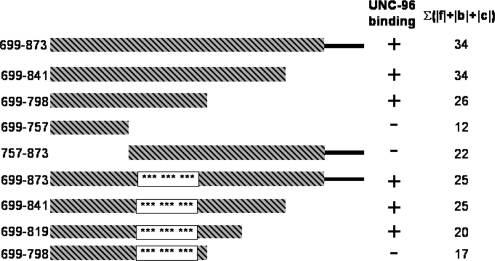
Additional two-hybrid experiments were conducted to more finely map the region of paramyosin that interacts with UNC-96 and to test the idea that the cluster of three heptads with high surface charge are important for this interaction. As shown in Figure 11, the minimal region of paramyosin required to interact with UNC-96 is 99 amino acids long, 699-798. As noted above, this region contains the cluster of three heptads each having a sum of 3 for the absolute values of coat residues, f, b, and c. To test the contribution of the surface charge from these heptads, paramyosin fragments were constructed in which the coat residues of each of these three heptads were converted to alanines. When these mutant fragments were tested for interaction with UNC-96, all showed interaction except for the smallest fragment (residues 699-798). This demonstrates the critical importance of the charge peaks to the binding of this fragment. In fact, tabulation of the total charge of coat residues for each of the paramyosin fragments tested, either of wild-type or mutant sequence, suggests that for interaction to occur, a minimum total surface charge of 23–25 is required (see right-most column of Figure 11).
Figure 11.
Two-hybrid experiments more finely map the UNC-96 binding site in paramyosin and show the influence of total charge on the surface of the coiled-coil rod for this interaction. Beginning with the smallest portion of paramyosin that was shown to interact with UNC-96 in Figure 8 (construct 9) deletion derivatives were tested. The smallest region that can interact with UNC-96 is a 99 residue segment, residues 699-798. Sequence analysis of this region (Figure 10) indicated a cluster of three heptads each with high charge density from coat residues f, b, and c. Several paramyosin preys were created in which all nine residues had been converted to alanines. The mutated paramyosin fragments showed interaction, except when residues 699-798 were used. The right-most column gives the total charge of coat residues for each paramyosin fragment. For interaction to occur the minimum total surface charge is 23-25. The hatched bar represents coiled-coil structure, and the thin black bar represents the nonhelical tail piece. The white bar denotes the region in which the nine coat residues were mutated to alanines (asterisks).
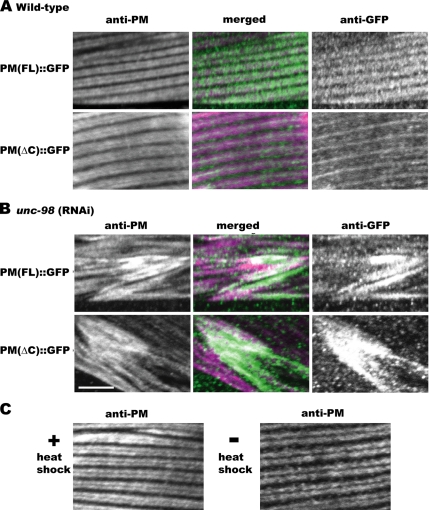
To determine whether the UNC-96–interacting region of paramyosin (amino acids 699-873) is important in vivo, truncated paramyosin lacking this region was expressed in wild-type animals. Transgenic animals were generated that express from a heat-shock promoter, GFP fused to either full-length paramyosin or paramyosin missing its C-terminal 174 residues. Expression of each protein was induced by heat shock in adults carrying the transgenes. Heat shock alone was shown not to affect the organization of endogenous paramyosin (Figure 12C). Immunofluorescence localization of paramyosin antibodies and GFP antibodies in a wild-type background shows that transgenically expressed full-length paramyosin, marked by GFP antibodies, colocalizes with endogenous paramyosin in the A-bands, marked by paramyosin antibodies (Figure 12 A, top). However, C-terminally deleted paramyosin, marked by GFP antibodies, is not distributed throughout the A-band, but rather is restricted to the middle of the A-band (Figure 12A, bottom). From this experiment, we conclude that the C-terminal 174 residues of paramyosin, which bind to UNC-96, are crucial for paramyosin's normal localization throughout the A-band.
Figure 12.
The C-terminal 174 residues of paramyosin are required for the incorporation of paramyosin into A-bands of wild-type muscle, but not for its localization to paramyosin accumulations in unc-98 RNAi muscle. (A) Full-length paramyosin fused to GFP expressed via a heat-shock promoter colocalizes with endogenous paramyosin throughout the A-band of wild-type muscle (top). C-terminally deleted paramyosin fused to GFP expressed via a heat-shock promoter is not distributed throughout the A-band, but rather is limited to the middle of the A-band (bottom). (B) Both full-length (top; GFP antibodies) and C-terminally deleted (bottom; GFP antibodies) localize within paramyosin accumulations at the ends of muscle cells in unc-98 RNAi animals similar to that of endogenous paramyosin (top and bottom; paramyosin antibodies). (C) Wild-type animals stained with anti-paramyosin, with or without heat shock. The localization of paramyosin is not affected by heat shock. Bar, 10 μm.
GFP-tagged full-length paramyosin and C-terminally deleted paramyosin were also expressed from transgenes in animals in which unc-98 expression had been knocked down by RNAi, which phenocopies the muscle defects of unc-98 mutants as seen by polarized light (Mercer et al., 2003). Both the transgenically expressed full-length paramyosin::GFP fusion and the C-terminally deleted paramyosin::GFP fusion localized with paramyosin accumulations at the ends of muscle cells (Figure 12B). We conclude that although the UNC-96 binding region of paramyosin is required for the normal distribution of paramyosin in A-bands, it is not required for its incorporation into accumulations.
DISCUSSION
Among the ∼40 Unc genes, when mutant show a muscle structural defect, unc-98 and -96 have a similar and unique phenotype: By polarized light microscopy, there are bright needle-like structures at the ends of the body wall muscle cells. So far, we know three components of these needles. In unc-98 mutants, the needles contain UNC-96 protein, and in unc-96 mutants, the needles contain UNC-98 protein (Mercer et al., 2006). In either mutant, the needles contain paramyosin (Figure 1; Mercer et al., 2006). It should be noted that within unc-98 or -96 mutant muscle, these proteins (paramyosin and UNC-98 and -96) are also found in their normal locations, the A-bands and M-lines. As noted above, the birefringent needles in unc-98 and -96 mutants look somewhat similar to needle-like structures that are multifilament assemblages found in most unc-15 mutant alleles (Waterston et al., 1977; Epstein et al., 1987, 1993; Gengyo-Ando and Kagawa, 1991), although the unc-15 accumulations appear longer and are not just restricted to the ends of the muscle cells. The reason why the needles in unc-98 and -96 are located at the ends of muscle cells (usually only one or two needles per cell) is a mystery. Nevertheless, this implies that the paramyosin located in the needles of unc-98 and -96 mutants is in an abnormal assembly state. We have also shown that the paramyosin accumulations of unc-15 mutants contain UNC-98 (Figure 4) and UNC-96 (data not shown). Thus, in any condition in which paramyosin accumulates in an ordered assemblage outside of myofibrils, these accumulations contain UNC-98 and/or -96. This suggests that paramyosin interacts directly with UNC-98 and -96, and indeed we have demonstrated such interactions by ELISA with purified proteins (Figures 7 and 9; Mercer et al., 2006) and by yeast two-hybrid assay (Figures 8 and 11).
We have also demonstrated a genetic interaction between a mild mutation in unc-15 and either unc-98 (Figure 3) or unc-96 (Mercer et al., 2006), suggesting that mutations in unc-98 or -96 further reduce the function of paramyosin to its null state. Indeed, the folded paralysis or “jack-knife” phenotype of the double homozygotes is also seen in the unc-15(e1214) null animals that hatch from laid eggs rather than inside the parent worm (P. Hoppe, personal communication). Although UNC-98 and -96 affect, at least partially, the localization of paramyosin, they do not seem to affect the total level of paramyosin: As shown in Figure 2, the total amount of paramyosin does not change in either unc-96 or -98 loss-of-function mutants. Nevertheless, the state of paramyosin does affect the localization and total amount of UNC-98 and possibly UNC-96: As shown in Figure 4, UNC-98 (and UNC-96, data not shown) colocalize with paramyosin aggregates, and as shown in Figure 5, in the absence of paramyosin, UNC-98 (and UNC-96; Mercer et al., 2006) is diffusely localized. Indeed, by quantitative immunoblot, the levels of UNC-98 follow the paramyosin state: in the absence of paramyosin (in the unc-15 null mutant), the level of UNC-98 is greatly diminished, and in paramyosin missense mutants (which form aggregates), the level of UNC-98 is increased (Figure 6). The dependence of UNC-98, and possibly UNC-96, levels on the state of paramyosin might be due to a possible chaperone function for UNC-98 and -96 to prevent aggregation of paramyosin. Colocalization of aggregated paramyosin and UNC-96 or -98 further supports this “chaperone” model: For example, in unc-15 missense mutants, paramyosin aggregates colocalize with UNC-98 (Figure 4). In addition, needles in unc-96 mutants contain both paramyosin and UNC-98 (Mercer et al., 2006). Future biochemical studies will be required to explore whether UNC-98 and/or UNC-96 have chaperone-like activities toward paramyosin. As shown in Figure 12, paramyosin lacking the C-terminal UNC-96 binding region might be poorly incorporated into thick filaments (at least A-bands). This in vivo experiment suggests that UNC-96 chaperone activity is required for efficient incorporation of paramyosin into thick filaments.
By ELISA we have demonstrated a direct interaction of high affinity (5–10 nM Kd) between UNC-98 and paramyosin (Figure 7). We had previously reported similar results showing a direct interaction between UNC-96 and paramyosin (Mercer et al., 2006). Recently, we have realized that the method we used to prepare paramyosin results in paramyosin that is missing one or both termini (Epstein and Liu, 1995). Nevertheless, this paramyosin preparation does bind to UNC-98 and -96 by ELISA, and also, paramyosin lacking the N-terminal 30 residues still interacts with both UNC-98 and -96 by two-hybrid analysis (Figure 8, #3). By two-hybrid experiments (Figures 8 and 11), and later confirmed by ELISAs (Figure 9), we mapped the UNC-98–binding region to paramyosin residues 31-693 and the UNC-96–binding region to a 99-residue region of paramyosin, residues 699-798. Because UNC-98 and -96 bind to separate portions of paramyosin, this might at least partially explain why each protein is required and thus not redundant (further discussed in Mercer et al., 2006): loss of function of either unc-98 or -96 results in a muscle phenotype. Our sequence analysis of paramyosin, UNC-96 and -98 suggests that the interaction of the surface of the coiled-coil rod of paramyosin with UNC-96 or -98 occurs primarily through electrostatic or polar interactions. Experimental evidence for this model was given for the paramyosin/UNC-96 interaction: conversion of the nine charged residues of the heptads with highest surface charge eliminated binding of the smallest binding region of paramyosin (residues 699-798; Figure 11).
We have provided multiple lines of evidence for interaction between UNC-98 and -96 with paramyosin, which is located in two concentric layers beneath the surface of thick filaments (Epstein et al., 1995; Deitiker and Epstein, 1993). We reported previously that UNC-98 (Miller et al., 2006) and UNC-96 (Qadota et al., 2007) interact with MHC A, which is located on the surface of thick filaments. Moreover, we have shown that UNC-98 interacts with UNC-97 (Mercer et al., 2003), which is one of the components associated with integrin adhesion complexes. Also, UNC-96 interacts with LIM-9 and -8, which bind to UNC-97 (Qadota et al., 2007). These interactions of UNC-96 and -98 with proteins other than paramyosin suggest that UNC-96 and -98 are located on the surface of thick filaments. Thus, the most likely explanation for these discrepant results is that UNC-96 and -98 are located both on the surface and below the surface (perhaps even in the cores) of thick filaments. Future experiments will be required to determine whether this hypothesis is correct. Also, given the immunolocalization of UNC-98 and -96 at the M-lines, these proteins are likely located only in the middle portion of the long thick filaments. However, one of their binding partners, paramyosin is not restricted to the middle and likely spans the entire length of thick filaments. We can speculate that there are different paramyosin binding proteins depending on the longitudinal position in the thick filament: UNC-98 and -96 and α-filagenin (Liu et al., 2000) in the middle, and β- and γ-filagenins in the flanking regions (Liu et al., 1998, 2000).
ACKNOWLEDGMENTS
We gratefully acknowledge Pam Hoppe (Western Michigan University) for very helpful comments on the manuscript. We also thank Andy Fire (Stanford University) for the heat-shock promoter-GFP vector, Robert Barstead (Oklahoma Medical Research Foundation) for the random primed nematode cDNA library, and Kozo Kaibuchi (Nagoya University) for vector pMAL-KK-1. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is supported by the National Center for Research Resources of the National Institutes of Health. These studies were supported by grant AR052133 from the National Institutes of Health to G.M.B. and a predoctoral fellowship 0415274B from the American Heart Association Southeast Affiliate to R.K.M.
Abbreviations used:
- MHC
myosin heavy chain
- MBP
maltose-binding protein
- GFP
green fluorescent protein
- ELISA
enzyme-linked immunosorbent assay.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/10.1091/mbc.E07-07-0723) on February 6, 2008.
REFERENCES
- Benian G. M., Tinley T. L., Tang X., Borodovsky M. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branden C., Tooze J. Introduction to Protein Structure. New York: Garland Publishing; 1999. [Google Scholar]
- Deitiker P. R., Epstein H. F. Thick filament substructures in C. elegans: evidence for two populations of paramyosin. J. Cell Biol. 1993;123:303–311. doi: 10.1083/jcb.123.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H. F., Waterston R. H., Brenner S. A mutant affecting the heavy chain of myosin in Caenorhabditis elegans. J. Mol. Biol. 1974;90:291–300. doi: 10.1016/0022-2836(74)90374-x. [DOI] [PubMed] [Google Scholar]
- Epstein H. F., Ortiz I., Berliner G. C. Assemblages of multiple thick filaments in nematode mutants. J. Muscle Res. Cell Motil. 1987;8:527–536. doi: 10.1007/BF01567911. [DOI] [PubMed] [Google Scholar]
- Epstein H. F., Casey D. L., Ortiz I. Myosin and paramyosin of Caenorhabditis elegans embryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J. Cell Biol. 1993;122:845–858. doi: 10.1083/jcb.122.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H. F., Liu F. Proteins and protein assemblies. In: Epstein H. F., Shakes D. C., editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego, CA: Academic Press; 1995. pp. 437–450. [Google Scholar]
- Epstein H. F., Lu G. Y., Deitiker P. R., Oritz I., Schmid M. F. Preliminary three-dimensional model for nematode thick filament core. J. Struct. Biol. 1995;115:163–174. doi: 10.1006/jsbi.1995.1041. [DOI] [PubMed] [Google Scholar]
- Francis G. R., Waterston R. H. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K., Kagawa H. Single charge change on the helical surface of the paramyosin rod dramatically disrupts thick filament assembly in Caenorhabditis elegans. J. Mol. Biol. 1991;219:429–441. doi: 10.1016/0022-2836(91)90184-8. [DOI] [PubMed] [Google Scholar]
- Hannak E., Oegema K., Kirkham M., Gonczy P., Habermann B., Hyman A. A. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J. Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H., Gengyo K., McLachlan A. D., Brenner S., Karn J. Paramyosin gene (unc-15) of Caenorhabditis elegans. Molecular cloning, nucleotide sequence and models for thick filament structure. J. Mol. Biol. 1989;207:311–333. doi: 10.1016/0022-2836(89)90257-x. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Liu F., Bauer C. C., Ortiz I., Cook R. G., Schmid M. F., Epstein H. F. β-filagenin, a newly identified protein coassembling with myosin and paramyosin in Caenorhabditis elegans. J. Cell Biol. 1998;140:347–353. doi: 10.1083/jcb.140.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Ortiz I., Hutagalung A., Bauer C. C., Cook R. G., Epstein H. F. Differential assembly of α- and γ-filagenins into thick filaments in Caenorhabditis elegans. J. Cell Sci. 2000;113:4001–4012. doi: 10.1242/jcs.113.22.4001. [DOI] [PubMed] [Google Scholar]
- Mackinnon A. C., Qadota H., Norman K. R., Moerman D. G., Williams B. D. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr. Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- Mercer K. B., Flaherty D. B., Miller R. K., Qadota H., Tinley T. L., Moerman D. G., Benian G. M. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol. Biol. Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer K. B., Miller R. K., Tinley T. L., Sheth S., Qadota H., Benian G. M. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol. Biol. Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. M., 3rd, Ortiz I., Berliner G. C., Epstein H. F. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Miller R. K., Qadota H., Landsverk M. L., Mercer K. B., Epstein H. F., Benian G. M. UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J. Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamide L. S., Bamburg J. R. A filter paper dye-binding assay for quantitative determination of protein without interference from reducing agents or detergents. Anal. Biochem. 1990;190:66–70. doi: 10.1016/0003-2697(90)90134-u. [DOI] [PubMed] [Google Scholar]
- Moerman D. G., Fire A. Muscle: structure, function and development. In: Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R., editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 417–470. [PubMed] [Google Scholar]
- Moerman D. G., Williams B. D. The C. elegans Research Community, WormBook, editors. Sarcomere assembly in C. elegans muscle. WormBook. 2006 doi: 10.1895/wormbook.1.81.1. doi/ 10.1895/wormbook.1.81.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Muller S. A., Haner M., Ortiz I., Aebi U., Epstein H. F. STEM analysis of C. elegans muscle thick filaments: evidence for microdifferentiated substructures. J. Mol. Biol. 2001;305:1035–1044. doi: 10.1006/jmbi.2000.4363. [DOI] [PubMed] [Google Scholar]
- Nonet M. L., Grundahl K., Meyer B. J., Rand J. B. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Norman K. R., Cordes S., Qadota H., Rahmani P., Moerman D. G. UNC-97/PINCH is involved in the assembly of integrin cell adhesion complexes in Caenorhabditis elegans body wall muscle. Dev. Biol. 2007;309:45–55. doi: 10.1016/j.ydbio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Qadota H., Mercer K. B., Miller R. K., Kaibuchi K., Benian G. M. Two LIM domain proteins and UNC-96 link UNC-97/PINCH to myosin thick filaments in C. elegans muscle. Mol. Biol. Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H. Muscle. In: Wood W. B., editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 281–335. [Google Scholar]
- Waterston R. H., Epstein H. F., Brenner S. Paramyosin of Caenorhabditis elegans. J. Mol. Biol. 1974;90:285–290. doi: 10.1016/0022-2836(74)90373-8. [DOI] [PubMed] [Google Scholar]
- Waterston R. H., Fishpool R. M., Brenner S. Mutants affecting paramyosin in Caenorhabditis elegans. J. Mol. Biol. 1977;117:679–697. doi: 10.1016/0022-2836(77)90064-x. [DOI] [PubMed] [Google Scholar]
- Waterston R. H., Thomson J. N., Brenner S. Mutants with altered muscle structure of Caenorhabditis elegans. Dev. Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Weiner O. D., Servant G., Welch M. D., Mitchison T. J., Sedat J. W., Bourne H. R. Spatial control of actin polymerization during neutrophil chemotaxis. Nat. Cell Biol. 1999;1:75–81. doi: 10.1038/10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel J. M., Epstein H. F. Identification of genetic elements associated with muscle structure in the nematode C. elegans. Cell Motil. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]