Abstract
Mitosis is a fundamental feature of all cellular organisms. It must be tightly regulated to allow normal tissue growth and to prevent cancer formation. Here, we identify a new protein that is required for mitosis. We show that the Ras association (RA) domain–containing protein, RASSF7, is part of an evolutionarily conserved group of four proteins. These are RASSF7, RASSF8, and two new RASSF proteins P-CIP1/RASSF9 and RASSF10. We call this group the N-terminal RASSF family. We analyzed the function of Xenopus RASSF7. RASSF7 was found to be expressed in several embryonic tissues including the skin, eyes, and neural tube. Knocking down its function led to cells failing to form a mitotic spindle and arresting in mitosis. This caused nuclear breakdown, apoptosis, and a striking loss of tissue architecture in the neural tube. Consistent with a role in spindle formation, RASSF7 protein was found to localize to the centrosome. This localization occurred in a microtubule-dependent manner, demonstrating that there is a mutually dependant relationship between RASSF7 localization and spindle formation. Thus RASSF7, the first member of the N-terminal RASSF family to be functionally analyzed, is a centrosome-associated protein required to form a spindle and complete mitosis in the neural tube.
INTRODUCTION
Mitosis is a fundamental biological process found in all cellular organisms. During mitosis in animal cells the centrosomes organize microtubule networks to establish the mitotic spindle. This requires the cooperation of several processes including microtubule nucleation, stability, and anchorage. It involves a network of proteins that are closely associated with the centrosome, including γ-tubulin, which forms a γ-tubulin ring complex (γ-TuRC) and initiates microtubule nucleation (Moritz et al., 1995; Zheng et al., 1995). Without this control of microtubules, correct mitosis cannot occur. A multitude of signaling molecules are also found clustered at the centrosome. Prominent examples include the mitotic kinases such as polo-like kinase (PLK) 1, 2, and 4 (Barr et al., 2004), AuroraA (Carmena and Earnshaw, 2003), and Nek2 (Hayward and Fry, 2006). Active Cdk1, the kinase that controls mitotic initiation, also first appears at the centrosome (Jackman et al., 2003). This suggests that the centrosome, in addition to regulating microtubules, functions as a signaling platform which regulates different aspects of mitosis (Basto and Pines, 2007). A link between aberrant control of mitosis and cancer progression has been suspected for almost 100 years (Balmain, 2001), and many regulators of mitosis are now known to be tumor suppressors or oncogenes (Malumbres and Barbacid, 2007). Thus identifying the centrosomal proteins responsible for controlling mitosis is crucial for explaining this key biological process but also for understanding tumor progression.
One group of proteins that are linked to mitosis and cancer progression is the Ras-association domain (RASSF) family. Classically, the vertebrate RASSF family (reviewed in the introduction of Avruch et al., 2005), comprises six members (RASSF1-6), which are expressed as multiple splice variants. Epigenetic-induced silencing of these genes occurs at high frequencies in tumors (Agathanggelou et al., 2005). Five of these have been shown to exhibit functions compatible with tumor suppressor properties (Khokhlatchev et al., 2002; Vos et al., 2003; Eckfeld et al., 2004; Agathanggelou et al., 2005; Allen et al., 2007). Structurally, the RASSF proteins are characterized by the presence of two domains: a Ras-association (RA) domain (Ponting and Benjamin, 1996) and a protein–protein interaction Sav/RASSF/Hippo (SARAH) domain (Scheel and Hofmann, 2003). In addition, two further RA domain–containing proteins have been identified, called RASSF7 and RASSF8, so the RASSF family currently contains eight members (van der Weyden and Adams, 2007). RASSF8 (also known as “carcinoma associated HoJ-1” and “C12orf2”) has been implicated in a chromosomal translocation event, which leads to synpolydactyly (Debeer et al., 2002). It has also been proposed to be a tumor suppressor (Falvella et al., 2006). Transcript levels are reduced in lung cancer cells, and ectopic expression leads to inhibition of anchorage-independent growth. However, polymorphisms in the human RASSF8 gene are not associated with adenocarcinoma risk (Falvella et al., 2007). RASSF7, originally called HRC1, was identified because it lies close to HRAS1 in the genome (Weitzel et al., 1992). It has not been studied except for our previous work showing that Xenopus RASSF7 (then called carcinoma associated) is expressed in the epithelial cells that surround the early embryo (Chalmers et al., 2006). Currently the biological functions of RASSF7 and RASSF8 are unknown.
We have analyzed the sequence of human RASSF7 and RASSF8 and found two additional RASSF proteins, P-CIP1/RASSF9 and RASSF10. Surprisingly, these four proteins should not be considered part of the classical RASSF family. Instead they are members of a new family of four RA domain–containing proteins, which we call the N-terminal (NT) RASSF family. The family appears to be evolutionarily conserved, because we identified NT RASSF homologues in the lower vertebrate Xenopus and the invertebrate Drosophila. To begin to understand the role of this family we analyzed the function of one of its members, Xenopus RASSF7. We show that RASSF7 is expressed in several embryonic tissues including the brain, where it is required for completing mitosis. Knocking down its function blocks spindle formation and triggers mitotic arrest, nuclear breakdown, apoptosis, and developmental defects including a loss of tissue architecture in the neural tube. Finally, consistent with a role in regulating spindle microtubules, RASSF7 protein was found to localize to centrosomes in a microtubule-dependent manner.
MATERIALS AND METHODS
DNA Constructs and Protein Alignments
The following human proteins were used for the domain analysis: hsRASSF1A (NP_009113), hsRASSF2 (NP_055552), hsRASSF3 (NP_835463), hsRASSF4 (NP_114412), hsRASSF5/splice variant NORE1A (NP_872604), hsRASSF6 (NP_958834), hsRASSF7 (NP_003466), hsRASSF8 (NP_009142), P-CIP1/RASSF9 (AAD03250), and RASSF10 (NP_001073990). The Xenopus NT RASSF proteins were identified using BLAST searches, xlRASSF7 (ABR21988), xlRASSF8 (ABR21989), xlP-CIP1/RASSF9 (ABZ80615), and xlRASSF10 (ABZ80616). The Drosophila proteins used were dmRASSF (NP_651126) and dmRASSF7/8 (NP_651411/CG5053). The Drosophila dmRASSF7/8 protein was identified by BLAST searches using vertebrate RASSF7. There are two other Drosophila proteins that may be homologues of P-CIP1/RASSF9 and RASSF10 (RE30146P and CG32150); however, these are not predicted to have a RA domain. The Simple Modular Architecture Research Tool (SMART) was used to identify all protein domain architectures (Letunic et al., 2004), except the SARAH domains that have previously been identified (Agathanggelou et al., 2005). This information was used to construct the protein domain diagrams. The genomic location of human Ras and RASSF genes was investigated using the Ensembl genome browser database (Hubbard et al., 2007). Human chromosome 11 and 12 schematics were adapted from Ensembl.
Xenopus RASSF7 pBlueScript SK(−) (Xl095b08) was obtained from the XDB website and fully sequenced. This clone was used instead of the clone identified previously Xl038l06, then called carcinoma associated (Chalmers et al., 2006), as the latter appeared truncated. The coding sequence for Xenopus laevis RASSF7 was PCR amplified from Xl095b08 and cloned into a gateway entry clone using the pENTR Directional TOPO Cloning Kit (Invitrogen, Carlsbad, CA), and used to generate an N-terminal GFP-RASSF7 pCS2, a C-terminal RASSF7-HA, and a RASSF7 pCS110K vector using gateway cloning (Invitrogen). We generated the destination vector for making N-terminal green fluorescent protein (GFP) fusions using a pCS2 GFP vector and the Gateway vector conversion kit (Invitrogen).
RT-PCR
RNA extraction, RT reaction, and PCR were carried out as described previously (Chalmers et al., 2002). PCR primers were as follows: RASSF7 forward: 5′-TTCAGGCCAGGAATCAGG-3′; RASSF7 reverse: 5′- GACACCATTGGGTTCTGC -3′; ODC forward: 5′-CAGCTAGCTGTGGTGTGG-3′; and ODC reverse: 5′-CAACATGGAAACTCACACC-3′.
Standard Growth Conditions of Embryos
X. laevis eggs were fertilized using standard procedures (Sive et al., 2000). Embryos were cultured in 0.1× Marc's modified Ringer's solution (MMR), pH 7.4 (100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES). Later stage embryos (stage 23 onward) were cultured in 0.1× MMR supplemented with 25 μg/ml gentamicin. Embryos were staged by cell number or as described previously (Nieuwkoop and Faber, 1967). For microtubule disruption assays, stage 10 embryos were treated with 20 μg of nocodazole (Sigma, St. Louis, MO) for 2 h. Whole mount pictures were obtained with a Nikon DXM1200C digital camera (Melville, NY) on a Leica MZFL III microscope (Deerfield, IL).
In Situ Hybridization
RASSF7 pBlueScript SK(−) (Xl095b08) was used to make a Dig-labeled probe, and in situ hybridization was carried out using a previously described method (Harland, 1991).
Immunoblotting
Extracts were obtained using FREON (Riedel-de Haën, Honeywell, Berlin, Germany) as previously described (Regad et al., 2007). Proteins were resolved by SDS-PAGE gel and detected by Western blot with anti-hemagglutinin (HA), clone 12CA5 (Roche; 1 in 400), or anti-α-tubulin, clone DM1A (Sigma; 1 in 5000). Horseradish peroxidase–conjugated goat anti-mouse immunoglobulin G (Sigma) was used as a secondary antibody at a dilution of 1 in 5000.
Morpholino Antisense Knockdown and RNA Overexpression
Morpholino oligonucleotide (MO; Gene Tools, Philomath, OR) sequences were as follows: Standard Control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; RASSF7 MO1, 5′-CATCCACCCACACCTTCAGCTCCAT-3′; and RASSF7 MO2, 5′-ATTGAACACGAGGAATGAGGTCGGC-3′. The control MO (Con MO) was directed against a human reticulocyte β-globin mutation. RASSF7 MO1 was directed against the translational start site of RASSF7, whereas RASSF7 MO2 was directed against the sequence immediately 5′ of the translational start site of RASSF7. Twenty nanograms of each of the MOs were injected into both cells of embryos at the two-cell stage.
The RASSF7 pCS110K plasmid was used to generate RASSF7 RNA for overexpression experiments, using the message machine kit (Ambion, Austin, TX). Two nanograms of RASSF7 RNA was injected into both cells of two-cell stage embryos. GFP-RASSF7 CS2 and RASSF7-HA were used to make RNA for localization experiments, by expression from the SP6 promoter using the message machine kit (Ambion). GFP-RASSF7 RNA, 2.5 ng, was injected into one cell of embryos at the two-cell stage, and embryos were cultured until stage 10. Once cultured until the required stage, embryos were fixed in MEMFA (0.1 M MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde) for 2 h at room temperature and where required, processed for histology or immunohistochemistry.
Histology and Immunohistochemisty
Embryos for histological staining were serial dehydrated using a washing series of 70, 90, 95, and 100% ethanol followed by histoclear (Raymond A. Lamb, Eastbourne, East Sussex, United Kingdom). The dehydrated embryos were then embedded in paraffin wax (Raymond A. Lamb) heated to 60°C. These were sectioned on a microtome in 10-μm-thick sections. Samples were stained with hematoxylin and eosin according to the manufacturer's instructions (Sigma), and examined on a Leica DMRB microscope.
Embryos used to visualize GFP or for antibody staining were embedded in fish gelatin as described previously (Chalmers et al., 2003), except tadpole-stage embryos, which were incubated in 20% sucrose for 2 h, washed several times in PBS, and then embedded in 15% fish gelatin for freezing. The embryos were cryosectioned and antibody-stained as previously described (Chalmers et al., 2003). The following antibodies were used: anti-γ-tubulin, clone GTU-88 (Sigma; 1 in 100); anti-laminin (Abcam; 1 in 100); anti-α-tubulin, clone DM1A (Sigma; 1 in 100), and clone YL1/2 (Abcam; 1 in 100); anti-active caspase 3 (Abcam; 1 in 100); and anti-histone H3 phospho S10 (Abcam; 1 in 500). The following secondary antibodies were used: anti-mouse Alexa 568 (Molecular Probes, Eugene, OR); anti-rabbit Alexa 488 (Molecular Probes); anti-rat Alexa 488 (Molecular Probes); and anti-mouse Alexa 488 (Molecular Probes). All secondary antibodies were used at a 1 in 300 dilution. The nuclear stains, DAPI (Sigma) and Sytox Green (Molecular Probes) were used at dilutions of 1 in 10,000 and 1 in 5000, respectively. Phalloidin (Molecular Probes) was used at a concentration of 1 U/ml. TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling) staining was performed using the in situ cell death detection kit, TMR red (Roche Applied Science, Indianapolis, IN), according to the manufacturer's instructions. Stained sections were mounted in Vectasheild (Vector Laboratories, Burlingame, CA) and imaged on a Zeiss LSM 510 META confocal microscope (Thornwood, NY).
Statistics
Means and SDs were calculated and plotted using GraphPad Prism 4 (San Diego, CA) or Microsoft Excel (Redmond, WA). Each experiment was repeated in triplicate. p < 0.05 was considered significant. Statistical analysis was carried out using unpaired t tests.
RESULTS
RASSF7 Is a Member of the NT RASSF Family, a Structurally Distinct and Evolutionarily Conserved Group of Proteins
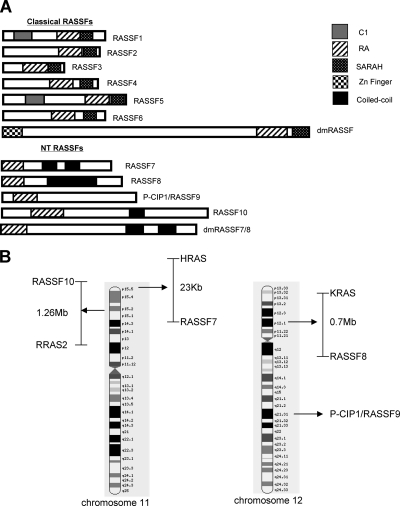
RASSF7 and RASSF8 have been assigned to the RASSF family because of their RA domain. However, the SMART database currently contains 61 human RA domain–containing proteins, the vast majority of which are not part of the RASSF family. We analyzed the structure of RASSF7 and RASSF8 to see if they are genuine members of the RASSF family. They contain a RA domain; however, it is located at the extreme N-termini of these proteins. This is in contrast to the C-terminal position seen in classical RASSF members (Figure 1A). Furthermore RASSF7 and RASSF8 lack the characteristic SARAH domain. Given the differences between their polypeptide structures and those of the other RASSFs, it appears that they should not be considered members of the classical RASSF family. We suggest they represent a distinct family, which we term the NT RASSF family.
Figure 1.
RASSF7 belongs to a novel family of proteins that co-localize with Ras genes in the human genome. (A) Schematic representations of the classical and N-terminal (NT) RASSF families: Classical human family RASSF1-6 and classical Drosophila RASSF (dmRASSF), NT family RASSF7–10 and Drosophila RASSF7/8-like (dmRASSF7/8). See materials and methods for accession numbers. C1, protein kinase C conserved region 1 (grey); SARAH, Sav/RASSF/Hippo (spotted); zinc finger (checked); coiled-coil (black); and RA, Ras-association domain (striped). The NT RASSF represent a distinct family of proteins from the classical RASSFs. (B) Graphical representation of human chromosomes 11 and 12 (modified from Ensembl), showing that the NT RASSF genes map near various Ras isoforms. The exception is P-CIP1/RASSF9, where we can not find a Ras gene nearby.
BLAST analysis of RASSF7 and RASSF8 identified two further proteins that have similar domain architectures to these two proteins: peptidylglycine alpha-amidating monooxygenase (PAM) COOH-terminal interactor 1 (P-CIP1), which we term P-CIP1/RASSF9, and a protein with structural similarity to RASSF7–9, which we call RASSF10 (Figure 1A). No other proteins with a high degree of similarity were identified, so there appears to be 10 vertebrate RASSF proteins, four of which are part of the NT RASSF family. We have identified homologues of the four NT RASSF genes in the lower vertebrate Xenopus (see Materials and Methods) and a homologue in the invertebrate Drosophila melanogaster, dmRASSF7/8 (Figure 1A). dmRASSF7/8 is closely related to vertebrate RASSF7/RASSF8 and distinct from the previously identified Drosophila RASSF gene (Polesello et al., 2006). We were unable to find a homologue in Saccharomyces cerevisiae, suggesting that brewing yeast either does not have NT RASSF proteins or that it has highly divergent versions.
Human RASSF7 and RASSF8 are located close to members of the Ras superfamily in the genome (Weitzel et al., 1992; Falvella et al., 2006). We investigated the genomic location of P-CIP1/RASSF9 and RASSF10 and found that RASSF10 also maps close to a Ras gene (Figure 1B). Thus three of four NT RASSF members exhibit close association to Ras genes. No consistent association between human classical RASSF and Ras genes could be found (data not shown), highlighting another difference between the two families. We conclude that the presence of distinct NT and classical RASSF families is an evolutionarily conserved feature, found in species ranging from Drosophila to Xenopus and humans.
RASSF7 Is Expressed in Several Embryonic Tissues
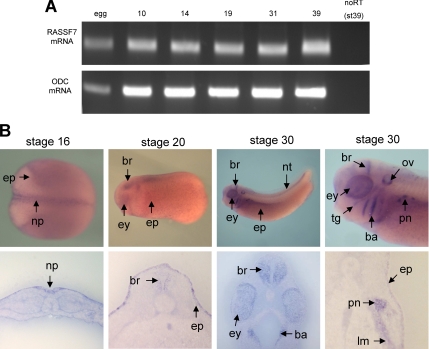
To begin to understand the role of the NT RASSF family we investigated the function of one member, Xenopus RASSF7. First we examined expression patterns of the gene in developing Xenopus embryos. Xenopus RASSF7 transcripts were detected by RT-PCR in eggs, embryos, and tadpole stages, showing that RASSF7 has both maternal and zygotic expression (Figure 2A). In situ hybridization indicated that RASSF7 had prominent neural and epidermal expression at neural plate stages (Figure 2B). This expression was restricted to the superficial layer of the neural plate and epidermis, as described previously (Chalmers et al., 2006). The neural and epidermal expression was maintained in tadpole stages. Several other tissues also expressed RASSF7 at this stage including the eye, ear (otic vesicle), branchial arches, and embryonic kidney (pronephros).
Figure 2.
RASSF7 expression in developing Xenopus embryos. (A) RT-PCR analysis of early Xenopus development for RASSF7 and as a control, the ornithine decarboxylase (ODC) gene. Thirty cycles were used in the PCR. RASSF7 exhibits both maternal and zygotic expression. (B) Tissue-specific expression patterns of RASSF7 analyzed by in situ hybridization at stages 16, 20, and 30. A wholemount and transverse section is shown for each stage. ep, epidermis; np, neural plate; ey, eye; br, brain; nt, neural tube; pn, pronepheros; lm, lateral plate mesoderm; ba, branchial arches; ov, otic vesicle; and tg, trigeminal ganglion. There is prominent expression of RASSF7 in many embryonic tissues.
RASSF7 Is Required for Normal Development
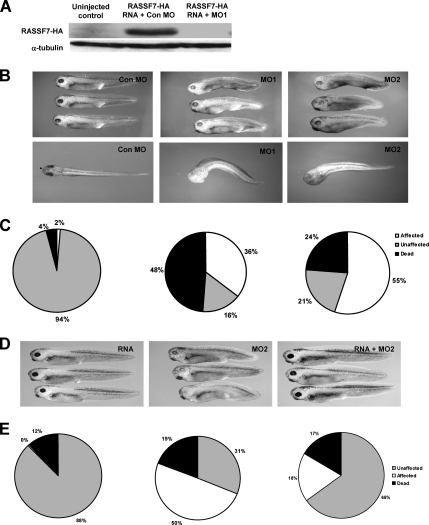
To determine a biological role for RASSF7 in vivo, we performed knockdown studies in Xenopus embryos. MOs were used to knockdown RASSF7 during Xenopus development. RASSF7 MO1 but not the control MO (Con MO) completely inhibited the translation of injected RASSF7 RNA (Figure 3A). MO1-injected embryos displayed phenotypic defects compared with those embryos injected with the control MO. RASSF7 knockdown embryos developed a shortened antero-posterior axis, reduced eye pigmentation, developmental delay, slight arching of the back, and a left-right bend in the body-axis (Figure 3B). There was also more lethality than seen with the control MO (Figure 3C). To confirm the specificity of the phenotype, we used a second MO (MO2) targeted to a different site in the RASSF7 RNA. This gave a phenotype very similar to that of MO1 (Figure 3, B and C). To further test the specificity of this phenotype, we performed a rescue experiment using RASSF7 RNA that lacks the MO2-binding site. Interestingly, overexpression of RASSF7 produced no discernable effect on Xenopus development, but the RNA could rescue the phenotype induced by MO2 (Figure 3, D and E).
Figure 3.
Knockdown of RASSF7 expression causes developmental defects. (A) Immunoblot of HA-tagged RASSF7 (56 kDa) from uninjected and MO injected embryos at stage 16. Injected embryos were injected with 20 ng of MO and 2 ng of HA-RASSF7 RNA into both cells at the two-cell stage. α-tubulin is shown as a loading control (50 kDa). MO1 is able to eliminate tagged RASSF7 in the embryos. (B) Lateral and dorsal views of Xenopus embryos injected with morpholinos (MOs), two RASSF7 MOs (MO1 and MO2), and a control MO (Con MO). RASSF7 knockdown embryos display a short and bent body axis, with reduced eye pigmentation and developmental delay. (C) Percentages of affected embryos for each MO at stage 39, calculated from four independent experiments (Con MO, n = 375; MO1, n = 359; MO2, n = 345). (D) Phenotypic rescue was achieved for MO2 by injection of RASSF7 RNA. (E) Quantification of the rescue experiment (percentages calculated from three independent experiments; RNA, n = 157; MO2, n = 171; RNA plus MO2, n = 166).
Given that a RASSF7-specific MO is capable of eliminating tagged RASSF7 protein, that two MOs targeting different sequences of the RASSF7 transcript both gave similar phenotypes (a phenotype not observed with the control MO and a variety of other Xenopus-specific MOs; data not shown), and that RASSF7 RNA was capable of rescuing the phenotype, we are confident that the phenotypes observed were specific for knocking down RASSF7 expression.
RASSF7 Knockdown Causes Severe Neural Tube Defects
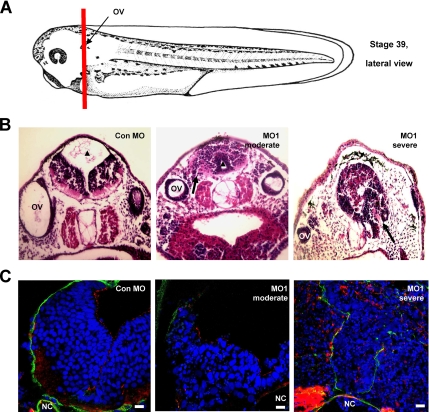
Anterior sections of the knockdown embryos (Figure 4A) revealed striking morphological abnormalities in the neural tube of the RASSF7 MO-injected embryos (Figure 4B). In RASSF7 knockdown embryos, the neuroepithelial cell layer extended away from the pial regions of the developing brain into zones normally occupied by connective tissue and often also into the ventricular cavity. The spreading of the neuroepithelial layer occurred from the forebrain through to the hindbrain (data not shown). There was a range of developmental defects from moderately affected, where the neuroepithelial cell layer extended but the ventricular cavity was still present or reduced (seen with both RASSF7 MOs), to severely affected, where the ventricular cavity was completely lost (only seen with MO1). The strong phenotype seen in the neural tube is consistent with the high level of RASSF7 expression seen in this tissue (Figure 2B).
Figure 4.
RASSF7 knockdown causes pronounced neural tube defects. (A) Schematic representation of a stage 39 Xenopus tadpole indicating the anterior/posterior level that sections were taken. Diagram adapted from a Xenbase image, based on previous work (Nieuwkoop and Faber, 1967). The sections cut through the otic vesicle (OV). (B) Stage 39 embryos injected with either Con MO or MO1 were sectioned and stained with hematoxylin and eosin. The ventricular cavity (▴) is reduced in MO1 injected embryos compared with control sections and is termed “moderately affected” or is completely lost and is therefore termed “severely affected.” In MO1-injected embryos, cells spread from the pial regions of the brain into areas normally occupied by connective tissue (arrows). (C) Developing brain tissue was stained with polarity markers: anti-laminin (green), phalloidin (red), and counterstained with DAPI (blue). Images were from an AP position similar to that in A and focus on the neural tube, just above the notochord (NC). Tissue polarity is disrupted in both the moderately and severely affected tissues. All bars, 10 μm.
Molecular markers were used to establish if the knockdown caused a breakdown in the normal polarized structure of the neural tube (Figure 4C). The apical marker actin was dispersed throughout the neural tissue of severely affected embryos. RASSF7 knockdown tissues also had a reduced and disrupted staining of the basement membrane marker laminin, with many cells spreading beyond the original laminin boundary. RASSF7 knockdown caused a breakdown of the normal molecular boundaries of the neural tube and spreading of neural cells away from the tissue.
RASSF7 Knockdown Triggers Nuclear Fragmentation and Apoptosis
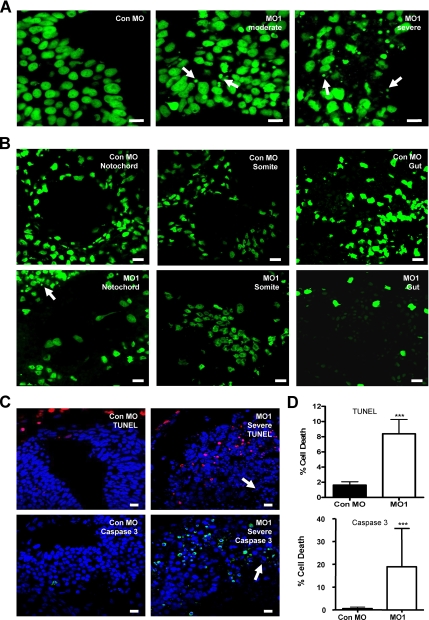
In addition to breakdown of tissue architecture there was another prominent phenotype of the RASSF7 MO-injected embryos. The RASSF7 knockdown tissues exhibited extensive nuclear fragmentation (Figure 4C; DAPI staining). A second DNA stain, Sytox Green, confirmed the presence of nuclear fragments in both moderately and severely affected tissues (Figure 5A). Fragments were detected in other tissues of RASSF7 knockdown embryos, particularly in the eye and skin (Supplementary Figure 1). These sites also express RASSF7 (Figure 2B). In contrast, in the somites, notocord and endoderm, tissues where we did not see RASSF7 expression, the nuclei appeared normal (Figure 5B). Importantly, RASSF7 RNA is capable of rescuing this nuclear fragmentation (Supplementary Figure 2A), confirming that this phenotype is caused by the reduction in RASSF7 expression in the developing neural tube. To test if these fragments represented apoptotic bodies, we carried out TUNEL (Hensey and Gautier, 1998) and active caspase 3 staining (Tseng et al., 2007). The number of apoptotic cells was increased by approximately fivefold for TUNEL staining and 40-fold for caspase 3 staining in the MO1 affected tissues compared with control (Figure 5, C and D). Despite the big increase in apoptosis, a large number of the nuclear fragments were not apoptotic (Figure 5C, arrows). This suggests that in addition to apoptosis there must be another cause of the nuclear fragmentation.
Figure 5.
RASSF7 knockdown results in nuclear fragmentation and cell death in the developing neural tube. (A) Sytox Green staining of neural cells in MO injected embryos. Nuclear fragments of varying sizes (arrows) are observed in RASSF7 knockdown tissues, which are not present in the control tissues. (B) Nuclear fragments are not observed in the notochord, somites, and developing gut (endoderm) of RASSF7 knockdown embryos, as indicated by sytox green staining. These are tissues where RASSF7 expression was not observed. Affected neural tissue spreading toward the notochord is highlighted by an arrow in the MO1 notochord image. (C) Sections of neural tissue were TUNEL stained (red) or anti-active caspase 3 (green), and DAPI counterstained (blue). Arrows indicate fragmented nuclei that are not apoptosis-positive. (D) Graphs indicate percentage of cell death. *** p< 0.001, n = 9 from three experiments for both MOs (TUNEL); and n = 3 from three independent experiments for both MOs (active caspase 3). More than 7000 cells were counted for each individual specimen. Error bars, SD. All bars, 10 μm.
RASSF7 Knockdown Does Not Cause a Reduction in the Number of Centrosomes
The phenotype of the RASSF7 knockdown is similar to that described for the process of cell death by mitotic catastrophe (Roninson et al., 2001). In this process mitotic arrest leads to nuclear fragmentation, which produces cells with multiple, small nuclei (Bunz et al., 1998). Conventional apoptosis is often observed after mitotic catastrophe. However, this only occurs in a minority of cells, whereas the vast majority remain TUNEL-negative (Nabha et al., 2002). Mitotic catastrophe has been associated with defective centrosomes so we tested whether RASSF7 knockdown led to centrosome defects. In control neural tissues, centrosomes (marked by γ-tubulin staining) lined the ventricular cavity (Figure 6A) because they are localized apically in neuroepithelial cells (Astrom and Webster, 1991). In contrast, centrosomes were dispersed throughout the neural tissue in severely affected RASSF7 knockdown embryos (Figure 6A), reflecting disruption to the neuroepithelial tissue architecture noted above (Figure 4, B and C). However, the centrosomes of the affected tissue were neither fragmented nor lost; indeed comparable numbers of centrosomes were found between control and MO1 tissues (Figure 6B).
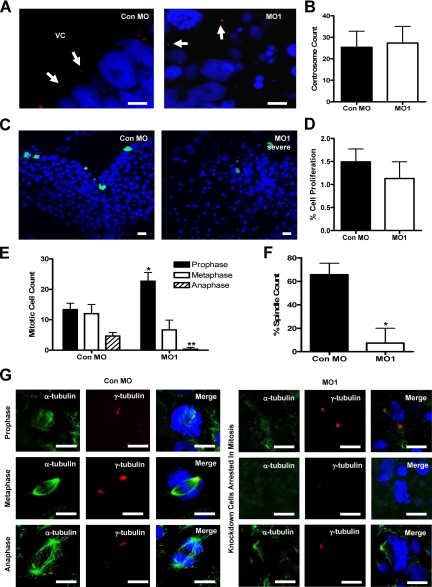
Figure 6.
RASSF7 is required for spindle formation and completion of mitosis in the neural tube. (A) Centrosome number is not affected by RASSF7 knockdown. γ-tubulin staining (red) was used to examine the centrosomes (highlighted by arrows) of the neural tube of Con MO– and MO1-injected embryos and were counterstained with DAPI (blue). VC, ventricular cavity. (B) Centrosome number in the neural tube is unaffected by RASSF7 knockdown. No significant difference was found between Con MO and MO1; n = 6 specimens for each MO from three independent experiments. (C) Cellular proliferation was examined in the neural tube of MO-injected embryos by staining with anti-phospho-S10 histone H3 (green) and DAPI (blue). (D) The number of phospho-histone H3 proliferating cells as a percentage of the total number of cells counted (DAPI stain). Proliferation is similar between the two MO tissues and is not significantly different. n = 9 from three injection experiments for both MOs, >7000 cells were counted for each individual specimen. (E) Counts of dividing cells in the following phases: prophase (also includes prometaphase), * p< 0.05; metaphase, no significant difference; and anaphase (also includes telophase), ** p< 0.01. Mitotic phases were distinguished by DAPI staining using the following criteria: prophase/prometaphase, condensed DNA; metaphase, genetic material aligned on the metaphase plate; and anaphase/telophase, chromosome separation. Counts were from three experiments (Con MO, n = 90; MO1, n = 90). All error bars, SD. Knockdown cells arrest in early mitosis. (F) The total number of spindles counted as a percentage of the total number of dividing cells. * p< 0.05, n = 6 specimens for each MO, from three experiments. The number of mitotic spindles was greatly reduced in the dividing RASSF7 knockdown cells. (G) α-tubulin staining (green) was used to visualize spindles in mitotic cells of the neural tube, whereas γ-tubulin staining was used to mark the centrosomes (red). Sections were counterstained with DAPI (blue). Mitotic spindles were missing, or occasionally abnormal, in the dividing RASSF7 knockdown cells. All bars, 10 μm. RASSF7 knockdown cells fail to progress through mitosis because of a deficiency in spindle formation.
RASSF7 Knockdown Cells Fail to Form a Spindle and Arrest in Mitosis
RASSF7 knockdown did not cause a reduction in the numbers of centrosomes, so we investigated whether it affected progression through mitosis. First, we established the percentage of cells in mitosis, using an anti-histone H3 phospho S10 antibody (Saka and Smith, 2001). The rate of proliferation was not changed in the knockdown tissues, suggesting that entry into mitosis occurred normally (Figure 6, C and D). The number of cells in each stage of mitosis was then analyzed using the DAPI nuclear stain. This showed large differences between control and RASSF7 knockdown embryos (Figure 6E). In RASSF7 knockdown tissues, cells were largely arrested in prophase/prometaphase, with less than one-third of the population in metaphase and almost no cells reaching anaphase. Once cells enter mitosis they require a spindle to separate sister chromosomes. In knockdown embryos, spindles failed to form in the vast majority of dividing cells of the neural tube (Figure 6, F and G). Importantly, RASSF7 RNA was capable of restoring mitotic spindle assembly (Supplementary Figure 2B). In the rare occasions where a spindle formed in knockdown cells it was weakly stained and monopolar (Figure 6G). A subset of the knockdown cells also appeared to lack centrosomes (Figure 6G). We do not see an overall reduction in centrosome numbers so this may be due to the centrosome moving away from the nucleus. However, almost half of the dividing cells observed did have centrosomes, indicating that in these cells the spindle failed to form or was abnormal, despite the presence of a centrosome (Figure 6G and Supplementary Figure 2B). Thus RASSF7 is required for cells to form a spindle and complete mitosis in the neural tube.
RASSF7 Localizes to the Centrosomes in a Microtubule-dependent Manner
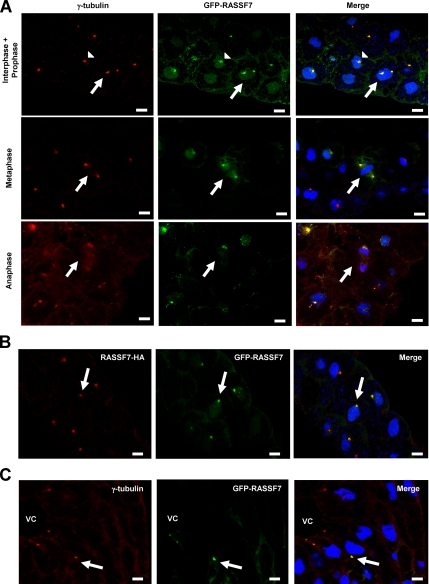
To establish why RASSF7 might be required for mitosis, we investigated its subcellular localization. We used a Xenopus RASSF7 N-terminal GFP-, and a C-terminal HA-fusion construct. Both were found to localize to discrete dots, adjacent to the nuclei of expressing cells (Figure 7, A and B). This staining colocalized with γ-tubulin, indicating that the fusion proteins localize to the centrosomes. We analyzed the localization of GFP-RASSF7 at interphase, prophase, metaphase, and anaphase (Figure 7A). In each case RASSF7 showed consistent localization to the centrosomes, although often weaker at anaphase. In addition the centrosome localization of GFP-RASSF7 was conserved in later stage embryos (Figure 7C).
Figure 7.
RASSF7 localizes to the centrosomes. (A) Embryos were injected with GFP-RASSF7 at the two-cell stage, cultured until stage 10, fixed, sectioned, and stained as described in materials and methods. GFP-RASSF7 (green) colocalized with γ-tubulin (red) during interphase and prophase (arrow head and arrow, respectively) and during metaphase and anaphase (arrows). Sections were counterstained with DAPI (blue). The localization has been repeated in more than three independent experiments. (B) RASSF7-HA (red) and GFP-RASSF7 (green) exhibit the same localization in stage 10 embryos. Interphase cells shown. (C) Later stage embryos (stage 20) also have centrosome localized GFP-RASSF7 (green), as indicated by γ-tubulin (red). Image shows the neural tube of a stage 20 embryo. VC, ventricular cavity. All bars, 10 μm. RASSF7 colocalizes with γ-tubulin throughout the cell cycle.
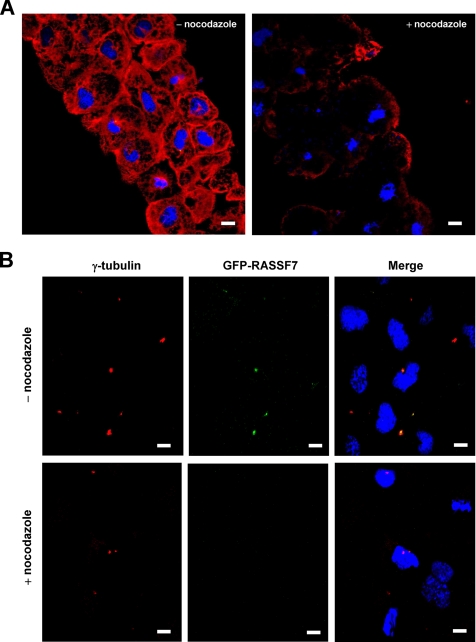
RASSF7 could be an intrinsic component of the centrosome or alternatively its localization may be mediated by microtubules. To see if the centrosome localization of RASSF7 was maintained when microtubules were lost, we treated the GFP-RASSF7 injected embryos with the microtubule depolymerizing drug, nocodazole. We found that in the cells of the animal cap of stage 10 embryos, nocodazole was capable of depolymerizing most of the microtubules (Figure 8A). This loss of microtubules resulted in the complete loss of the centrosome localization of RASSF7 (Figure 8B). This indicates that RASSF7 can only localize to the centrosome in the presence of minus-end microtubules. These results suggest that RASSF7 function is mediated from the centrosome, a structure crucial for the control of microtubules and mitosis.
Figure 8.
The centrosome localization of RASSF7 requires microtubules. (A) Microtubules are lost in cells treated with 20 μg/ml of nocodazole for 2 h. Microtubules of the animal cap from stage 10 embryos were stained using α-tubulin (red), and the cell nuclei were visualized using DAPI (blue). (B) Nocodazole treatment resulted in a loss of GFP-RASSF7 (green) localization at the centrosome. γ-tubulin was used to stain the centrosomes (red) and nuclei counterstained with DAPI (blue). All bars, 10 μm.
DISCUSSION
Mitosis is an essential process for all cellular organisms that is comprised of a tightly controlled series of events. Understanding how these steps are regulated is vital, as aberrant control of the system can lead to oncogenesis (Hanahan and Weinberg, 2000), and as such, many proteins involved in mitosis can act as tumor suppressors or oncoproteins. Here we show that Xenopus RASSF7, the first member of the novel NT RASSF family to be functionally analyzed, is a centrosome-associated protein that is required for completion of mitosis. This study significantly extends our understanding of RASSF7 function during cell division and provides new insights into mitotic spindle assembly at the centrosome. Our findings demonstrate that RASSF7 has an essential role in M phase microtubule organization and mitotic progression in dividing cells.
We show that the NT RASSF family is an evolutionarily conserved novel family of RA domain–containing proteins that comprise of four vertebrate members: RASSF7, RASSF8, and two new RASSF proteins, P-CIP1/RASSF9 and RASSF10 (Figure 1A). The conserved domain structure of human, Xenopus and Drosophila NT RASSF proteins raises the possibility that other members of the NT RASSF family may play a role in controlling mitosis. It could be that in the RASSF7 negative tissues other members of the family play a similar role. A recent genome-wide screen in Drosophila provides some support for this conserved function hypothesis (Goshima et al., 2007). It showed that in the first round of the screen, inhibiting dmRASSF7/8 (NP_651411/CG5053) produced a weak effect on the mitotic spindle. This experiment requires confirmation because weak effects identified in the first round of the screen were not followed up. Little is known about the biological function of the other vertebrate members. Ectopic expression of RASSF8 leads to inhibition of anchorage-independent growth (Falvella et al., 2006). This might suggest an involvement in mitosis but would also be consistent with other possible roles. P-CIP1/RASSF9 has been shown to interact with the secretory granule modifying enzyme PAM and associate with recycling endosomes (Chen et al., 1998), which does not suggest a role in spindle formation. No work has been carried out on RASSF10. A lot of future work is needed to establish if the NT RASSF members share similar functions as well as similar structures.
The major phenotype of RASSF7 knockdown in Xenopus embryos, identified by histology, was in the neural tube and we focused on this tissue. However, RASSF7 does not only function in the neural tube, as there were fragmented nuclei and apoptotic cells in other tissues that express RASSF7, including the eye and the skin (Supplementary Figure 1). The external developmental effects noted in the knockdown embryos, such as bent axis and reduced eye pigmentation, can probably be attributed to the incorrect development of a range of tissues expressing RASSF7. An intriguing question is why the neural tube showed the biggest defect in tissue architecture. One possibility is that cell divisions may differ in the neural tube to other tissue types. It has previously been noted that when undergoing proliferative divisions, the neuroepithelial cells of the developing brain are particularly susceptible to spindle abnormalities (Gotz and Huttner, 2005), although this is more often due to spindle positioning rather than loss (Fish et al., 2006). Alternatively, other NT RASSF proteins may be able to compensate for RASSF7 loss in certain tissues, but not the neural tube. This would also explain why a gene that is required for mitosis is not expressed in all mitotic tissues. It will be interesting to investigate other NT RASSF expression patterns in developing Xenopus tissues.
Our results show that RASSF7 is essential for progression through mitosis and mitotic spindle formation. We found that during RASSF7 depletion, dividing cells arrested early in mitosis. We did not distinguish between cells in prophase and prometaphase. However it is likely the majority of the arrested cells were in a prometaphase-state, given that after inhibition of spindle formation by nocodazole treatment, dividing cells often arrest in prometaphase (Kimura et al., 2000; Eskelinen et al., 2002). Aberrant spindle formation is thought to drive chromosomal instability in cells predispositioned to aneuploidy, such as in cancer cells. It is likely that the nuclear fragments we observed in the neural tube of RASSF7 deficient embryos (Figure 5) represented the products of failed divisions and resulting breakdown in the nuclei. It is also possible that some cells managed to divide but the chromosomes were not equally distributed in the daughter cells. Thus a reduction in RASSF7 expression appears to lead to aneuploidy in proliferating cells.
Previous observations have shown that animal cell types lacking centrosomes are still able to form a spindle, albeit less efficiently (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001; Basto et al., 2006). The loss of centrosomes would not be expected to produce the phenotype we describe. Consistent with this, we see cells where spindles were not formed despite the presence of the centrosome. This shows that the arrest in mitosis is not caused by losing centrosomes but by a more specific defect in forming a mitotic spindle such as in bipolar organization, microtubule nucleation, stability, or anchoring at the centrosome during M phase. Knockdown of proteins that function in bipolar organization or anchoring produce different phenotypes (Young et al., 2000; Cullen and Ohkura, 2001; Cassimeris and Morabito, 2004; Toya et al., 2007), arguing that RASSF7 knockdown produces a defect in microtubule nucleation or stability.
The mechanisms responsible for microtubule nucleation and stability involve a complex array of centrosome-associated proteins. Our results suggest that RASSF7 represents a novel part of this process. However, unlike the majority of centrosomal proteins, such as gamma tubulin, pericentrin, Cep 192, and TACC proteins (Gergely et al., 2000; Young et al., 2000; Andersen et al., 2003; Gomez-Ferreria et al., 2007), RASSF7 localization requires microtubules. This shows that there is a reciprocal interaction between RASSF7 and the microtubules. RASSF7 is required for microtubules to form a spindle, and microtubules are required for RASSF7 to localize. It will be interesting to investigate how RASSF7 interacts with microtubules and whether this is by direct or indirect binding. An intriguing possibility is that RASSF7 may interact with microtubules via other microtubule dependent components of the centrosome. These include components of the cytoplasmic dynein motor, where the dynactin component is required for anchoring of microtubules at the centrosomes (Quintyne et al., 1999).
It has been proposed that RASSF8 is a potential tumor suppressor, with reduced expression in lung cancer cells (Falvella et al., 2006). This suggests that RASSF7 may also be a tumor suppressor. However elevated levels of RASSF7 expression have recently been shown to be a marker of pancreatic islet cell tumors (Lowe et al., 2007). In fact RASSF7 was found to be up-regulated 87-fold in tumors compared with normal tissues. This argues against RASSF7 having tumor-suppressing role and raises the possibility it may even promote cancer formation. Our data shows that inhibiting RASSF7 causes an arrest in mitosis that is followed by cell death. Inhibiting the kinase PLK1 also produces an arrest in mitosis and cell death (Cogswell et al., 2000; Liu and Erikson, 2003). This phenotype offers the opportunity for therapeutic intervention, and PLK1 inhibitors are currently being developed for the clinic (Plyte and Musacchio, 2007). There is clearly a long way to go, but further analysis of RASSF7 will show if it also represents a potential therapeutic target.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. Curtis Altmann and Joanna Argasinska (Prof. Jim Smith's laboratory, Gurdon Institute, University of Cambridge) for providing plasmids. We also thank Dr. Andrew Ward and Dr. Iwan Evans for their manuscript comments. The RASSF7 cDNA clone was donated by the National Institute for Basic Biology X. laevis EST project run by Professor Nateo Ueno. Finally, we thank anonymous reviewers for their helpful suggestions. This work was supported by Medical Research Council Career Development and RCUK academic fellowships for A.C.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0652) on February 13, 2008.
REFERENCES
- Agathanggelou A., Cooper W. N., Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- Allen N. P., Donninger H., Vos M. D., Eckfeld K., Hesson L., Gordon L., Birrer M. J., Latif F., Clark G. J. RASSF6 is a novel member of the RASSF family of tumor suppressors. Oncogene. 2007;26:6203–6211. doi: 10.1038/sj.onc.1210440. [DOI] [PubMed] [Google Scholar]
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Astrom K. E., Webster H. D. The early development of the neopallial wall and area choroidea in fetal rats. A light and electron microscopic study. Adv. Anat. Embryol. Cell Biol. 1991;123:1–76. [PubMed] [Google Scholar]
- Avruch J., Praskova M., Ortiz-Vega S., Liu M., Zhang X. F. Nore1 and RASSF1 regulation of cell proliferation and of the MST1/2 kinases. Methods Enzymol. 2005;407:290–310. doi: 10.1016/S0076-6879(05)07025-4. [DOI] [PubMed] [Google Scholar]
- Balmain A. Cancer genetics: from Boveri and Mendel to microarrays. Nat. Rev. Cancer. 2001;1:77–82. doi: 10.1038/35094086. [DOI] [PubMed] [Google Scholar]
- Barr F. A., Sillje H. H., Nigg E. A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C. G., Khodjakov A., Raff J. W. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Basto R., Pines J. The centrosome opens the way to mitosis. Dev. Cell. 2007;12:475–477. doi: 10.1016/j.devcel.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Carmena M., Earnshaw W. C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell. Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 2004;15:1580–1590. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers A. D., Lachani K., Shin Y., Sherwood V., Cho K.W.Y., Papalopulu N. Grainyhead-like 3, a transcription factor identified in a microarray screen, promotes the specification of the superficial layer of the embryonic epidermis. Mech. Dev. 2006;123:702–718. doi: 10.1016/j.mod.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Chalmers A. D., Strauss B., Papalopulu N. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development. 2003;130:2657–2668. doi: 10.1242/dev.00490. [DOI] [PubMed] [Google Scholar]
- Chalmers A. D., Welchman D., Papalopulu N. Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev. Cell. 2002;2:171–182. doi: 10.1016/s1534-5807(02)00113-2. [DOI] [PubMed] [Google Scholar]
- Chen L., Johnson R. C., Milgram S. L. P-CIP1, a novel protein that interacts with the cytosolic domain of peptidylglycine alpha–amidating monooxygenase, is associated with endosomes. J. Biol. Chem. 1998;273:33524–33532. doi: 10.1074/jbc.273.50.33524. [DOI] [PubMed] [Google Scholar]
- Cogswell J. P., Brown C. E., Bisi J. E., Neill S. D. Dominant-negative polo-like kinase 1 induces mitotic catastrophe independent of cdc25C function. Cell Growth Differ. 2000;11:615–623. [PubMed] [Google Scholar]
- Cullen C. F., Ohkura H. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 2001;3:637–642. doi: 10.1038/35083025. [DOI] [PubMed] [Google Scholar]
- Debeer P., Schoenmakers E. F., Twal W. O., Argraves W. S., De Smet L., Fryns J. P., Van De Ven W. J. The fibulin-1 gene (FBLN1) is disrupted in a t(12;22) associated with a complex type of synpolydactyly. J. Med. Genet. 2002;39:98–104. doi: 10.1136/jmg.39.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckfeld K., Hesson L., Vos M. D., Bieche I., Latif F., Clark G. J. RASSF4/AD037 is a potential ras effector/tumor suppressor of the RASSF family. Cancer Res. 2004;64:8688–8693. doi: 10.1158/0008-5472.CAN-04-2065. [DOI] [PubMed] [Google Scholar]
- Eskelinen E. L., Prescott A. R., Cooper J., Brachmann S. M., Wang L., Tang X., Backer J. M., Lucocq J. M. Inhibition of autophagy in mitotic animal cells. Traffic. 2002;3:878–893. doi: 10.1034/j.1600-0854.2002.31204.x. [DOI] [PubMed] [Google Scholar]
- Falvella F. S., Manenti G., Spinola M., Pignatiello C., Conti B., Pastorino U., Dragani T. A. Identification of RASSF8 as a candidate lung tumor suppressor gene. Oncogene. 2006;25:3934–3938. doi: 10.1038/sj.onc.1209422. [DOI] [PubMed] [Google Scholar]
- Falvella F. S., Spinola M., Manenti G., Conti B., Pastorino U., Skaug V., Haugen A., Dragani T. A. Common polymorphisms in D12S1034 flanking genes RASSF8 and BHLHB3 are not associated with lung adenocarcinoma risk. Lung Cancer. 2007;56:1–7. doi: 10.1016/j.lungcan.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Fish J. L., Kosodo Y., Enard W., Paabo S., Huttner W. B. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F., Karlsson C., Still I., Cowell J., Kilmartin J., Raff J. W. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. USA. 2000;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria M. A., Rath U., Buster D. W., Chanda S. K., Caldwell J. S., Rines D. R., Sharp D. J. Human cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 2007;17:1960–1966. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M., Huttner W. B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harland R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hayward D. G., Fry A. M. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237:155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Hensey C., Gautier J. Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev. Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Miller F. J., Cham M., Khodjakov A., Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Hubbard T. J., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M., Lindon C., Nigg E. A., Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhlatchev A., Rabizadeh S., Xavier R., Nedwidek M., Chen T., Zhang X. F., Seed B., Avruch J. Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 2002;12:253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- Kimura K., Tsuji T., Takada Y., Miki T., Narumiya S. Accumulation of GTP-bound RhoA during cytokinesis and a critical role of ECT2 in this accumulation. J. Biol. Chem. 2000;275:17233–17236. doi: 10.1074/jbc.C000212200. [DOI] [PubMed] [Google Scholar]
- Letunic I., Copley R. R., Schmidt S., Ciccarelli F. D., Doerks T., Schultz J., Ponting C. P., Bork P. SMART 4.0, towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Erikson R. L. Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:5789–5794. doi: 10.1073/pnas.1031523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe A. W., Olsen M., Hao Y., Lee S. P., Taek Lee K., Chen X., van de Rijn M., Brown P. O. Gene expression patterns in pancreatic tumors, cells and tissues. PLoS ONE. 2007;2:e323. doi: 10.1371/journal.pone.0000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. Cell cycle kinases in cancer. Curr. Opin. Genet. Dev. 2007;17:60–65. doi: 10.1016/j.gde.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Moritz M., Braunfeld M. B., Sedat J. W., Alberts B., Agard D. A. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Nabha S. M., Mohammad R. M., Dandashi M. H., Coupaye-Gerard B., Aboukameel A., Pettit G. R., Al-Katib A. M. Combretastatin-A4 prodrug induces mitotic catastrophe in chronic lymphocytic leukemia cell line independent of caspase activation and poly(ADP-ribose) polymerase cleavage. Clin. Cancer Res. 2002;8:2735–2741. [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J. Amsterdam: North Holland; 1967. Normal Table of Xenopus laevis. [Google Scholar]
- Plyte S., Musacchio A. PLK1 inhibitors: setting the mitotic death trap. Curr. Biol. 2007;17:R280–R283. doi: 10.1016/j.cub.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Polesello C., Huelsmann S., Brown N. H., Tapon N. The Drosophila RASSF homolog antagonizes the Hippo pathway. Curr. Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Benjamin D. R. A novel family of Ras-binding domains. Trends Biochem. Sci. 1996;21:422–425. doi: 10.1016/s0968-0004(96)30038-8. [DOI] [PubMed] [Google Scholar]
- Quintyne N. J., Gill S. R., Eckley D. M., Crego C. L., Compton D. A., Schroer T. A. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T., Roth M., Bredenkamp N., Illing N., Papalopulu N. The neural progenitor-specifying activity of FoxG1 is antagonistically regulated by CKI and FGF. Nat. Cell Biol. 2007;9:531–540. doi: 10.1038/ncb1573. [DOI] [PubMed] [Google Scholar]
- Roninson I. B., Broude E. V., Chang B. D. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist. Update. 2001;4:303–313. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- Saka Y., Smith J. C. Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev. Biol. 2001;229:307–318. doi: 10.1006/dbio.2000.0101. [DOI] [PubMed] [Google Scholar]
- Scheel H., Hofmann K. A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr. Biol. 2003;13:R899–R900. doi: 10.1016/j.cub.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M., Harland R. M. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. Early Development of Xenopus laevis: A Laboratory Manual. [Google Scholar]
- Toya M., Sato M., Haselmann U., Asakawa K., Brunner D., Antony C., Toda T. Gamma-tubulin complex-mediated anchoring of spindle microtubules to spindle-pole bodies requires Msd1 in fission yeast. Nat. Cell Biol. 2007;9:646–653. doi: 10.1038/ncb1593. [DOI] [PubMed] [Google Scholar]
- Tseng A. S., Adams D. S., Qiu D., Koustubhan P., Levin M. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev. Biol. 2007;301:62–69. doi: 10.1016/j.ydbio.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L., Adams D. J. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim. Biophys. Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos M. D., Ellis C. A., Elam C., Ulku A. S., Taylor B. J., Clark G. J. RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor. J. Biol. Chem. 2003;278:28045–28051. doi: 10.1074/jbc.M300554200. [DOI] [PubMed] [Google Scholar]
- Weitzel J. N., Kasperczyk A., Mohan C., Krontiris T. G. The HRAS1 gene cluster: two upstream regions recognizing transcripts and a third encoding a gene with a leucine zipper domain. Genomics. 1992;14:309–319. doi: 10.1016/s0888-7543(05)80221-6. [DOI] [PubMed] [Google Scholar]
- Young A., Dictenberg J. B., Purohit A., Tuft R., Doxsey S. J. Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol. Biol. Cell. 2000;11:2047–2056. doi: 10.1091/mbc.11.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wong M. L., Alberts B., Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.