Abstract
The spindle assembly checkpoint monitors the status of kinetochore–microtubule (K-MT) attachments and delays anaphase onset until full metaphase alignment is achieved. Recently, the role of spindle assembly checkpoint proteins was expanded with the discovery that BubR1 and Bub1 are implicated in the regulation of K-MT attachments. One unsolved question is whether Bub3, known to form cell cycle constitutive complexes with both BubR1 and Bub1, is also required for proper chromosome-to-spindle attachments. Using RNA interference and high-resolution microscopy, we analyzed K-MT interactions in Bub3-depleted cells and compared them to those in Bub1- or BubR1-depleted cells. We found that Bub3 is essential for the establishment of correct K-MT attachments. In contrast to BubR1 depletion, which severely compromises chromosome attachment and alignment, we found Bub3 and Bub1 depletions to produce defective K-MT attachments that, however, still account for significant chromosome congression. After Aurora B inhibition, alignment defects become severer in Bub3- and Bub1-depleted cells, while partially rescued in BubR1-depleted cells, suggesting that Bub3 and Bub1 depletions perturb K-MT attachments distinctly from BubR1. Interestingly, misaligned chromosomes in Bub3- and Bub1-depleted cells were found to be predominantly bound in a side-on configuration. We propose that Bub3 promotes the formation of stable end-on bipolar attachments.
INTRODUCTION
The genomic stability of all organisms depends on the correct segregation of chromosomes during cell division. To be accurately segregated at the onset of anaphase, chromosomes need to attach, through their sister kinetochores, to microtubules emanating from the opposite poles of the mitotic spindle, and to align at the metaphase plate (Tanaka, 2002). Current models of spindle formation go much beyond the traditional search-and-capture model and include additional assembly pathways coordinated by chromosome-driven microtubule nucleation and cooperative microtubule interactions (Kalab et al., 2006; Kapoor et al., 2006). These models clearly show that chromosome biorientation is a stochastic, error-prone process, often generating intermediate kinetochore–microtubule (K-MT) interactions that are inappropriate and must be detected and corrected. Therefore, a surveillance mechanism exists to ensure that anaphase onset is delayed until all chromatids are correctly bound to microtubules, referred to as the spindle assembly checkpoint (SAC; Gorbsky, 2001; Musacchio and Hardwick, 2002; Cleveland et al., 2003). The SAC is mediated by a set of highly conserved proteins that function as a signal transduction pathway and include Mad1, Mad2, Mad3/BubR1, Bub1, Bub3, and Mps1 (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996). These proteins localize at the kinetochores of chromosomes that have not achieved bipolar attachment, generating checkpoint signaling (Shah and Cleveland, 2000). Protein recruitment at the kinetochores occurs accordingly to a hierarchic sequence and an interdependent network that appears to govern the checkpoint pathway (Jablonski et al., 1998; Johnson et al., 2004; Vigneron et al., 2004). The downstream target of the checkpoint is Cdc20, an activator of the anaphase-promoting complex/cyclosome (APC/C). APC/C is an E3-ubiquitin ligase that marks key mitotic proteins for degradation and is the key regulator of the metaphase–anaphase transition (Nasmyth, 2002; Peters, 2002). The molecular mechanisms by which checkpoint signals are generated at kinetochores and then sent to inhibit the APC/C are not completely elucidated. The current consensus is that unattached and maloriented kinetochores activate Mad2 and BubR1, which then bind to and inhibit Cdc20 (Tang et al., 2001; Fang, 2002). Indeed, a mitotic checkpoint complex (MCC) consisting of BubR1, Bub3, Mad2, and Cdc20 has been purified from HeLa cells and shown to be a potent APC/C inhibitor (Sudakin et al., 2001). Whether kinetochores act as a catalytic platform for the formation of MCC or, alternatively, activate checkpoint proteins to form MCC at the cytosol and to sensitize APC/C for inhibition, are models presently accepted (Howell et al., 2000; de Antoni et al., 2005).
Early models for the generation of an inhibitory signal have emphasized the importance of a transient association of a Mad1/Mad2 complex with unattached kinetochores (Howell et al., 2004). Additionally to the detection of microtubule occupancy, sensing mechanisms have also been proposed to respond to physical tension generated across bioriented sister chromatids (Zhou et al., 2002; Logarinho et al., 2004). Ipl1/Aurora B activity appears to provide the link between recognition of missing tension and the signaling of unattached kinetochores. By specifically destabilizing K-MTs of mono-oriented chromosomes, Ipl1/Aurora B generates free kinetochores that maintain an active checkpoint signal (Biggins and Murray, 2001; Tanaka et al., 2002). Therefore, the checkpoint-monitoring processes are linked to an error-correcting machinery that detects reduced tension across sister kinetochores syntelically attached to microtubules emanating from a single spindle pole (Ditchfield et al., 2003; Hauf et al., 2003).
In addition to the Aurora B kinase, the checkpoint kinases BubR1 and Bub1 have also been recently proposed to act dually, linking K-MT interaction to the SAC signaling. BubR1 mediates stable K-MT interactions (Lampson and Kapoor, 2005) at the same time that performs a catalytic role at the kinetochore (Sudakin et al., 2001; Mao et al., 2003). Similarly, besides its role in the inhibitory phosphorylation of Cdc20 (Chung and Chen, 2003; Tang et al., 2004a), Bub1 is also independently required for chromosome segregation (Johnson et al., 2004; Tang et al., 2004b; Meraldi and Sorger, 2005; Vanoosthuyse and Hardwick, 2005).
Because Bub3 checkpoint protein is required for BubR1 and Bub1 kinetochore localization and forms cell cycle constitutive complexes with both kinases (Taylor et al., 1998; Brady and Hardwick, 2000), it is predictable that it might be also involved in the regulation of K-MT interactions. Therefore, we went on investigating this Bub3 function using a combination of RNA interference (RNAi)-mediated gene silencing, functional assays, and high-resolution microscopy to assess K-MT interactions and performing comparative analysis with the SAC proteins Bub1 and BubR1.
MATERIALS AND METHODS
Cell Culture and Small Interfering siRNA Transfection
HeLa cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 1% glutamax, and 1% antibiotics mixture (Invitrogen) and grown at 37°C in a 5% CO2 humidified chamber. Bub3, Bub1, and BubR1 RNAi depletions were achieved using a pool of small interfering siRNAs (siRNAs) generated from nonconserved sequences (nucleotides 422-894, 1181-1734, and 87-616 from the start codon, respectively) with the BLOCK-iT Dicer RNAi kit accordingly to the manufacturer's instructions (Invitrogen). Bub3 silencing was also ascertained using reported oligoduplexes (Meraldi et al., 2004). Oligoduplexes to repress Aurora B were as described (Hauf et al., 2003). The target sequences of the form G(N17)C, 5′-GCCAAGGATTCAGCACTAC-3′ and 5′-GTTCTTCTCATTCGGCATC-3′ were used for the preparation of T7 polymerase transcribed siRNAs against CENP-E and Mad2 transcripts, respectively (Milligan and Uhlenbeck, 1989). Cells (1 × 105) were plated in DMEM medium with 5% FBS and without antibiotics, on six-well plates (Nunc, Naperville, IL) containing poly-l-lysine–coated glass coverslips. Two hours later, cells were transfected with siRNA-oligofectamine complexes prepared in Opti-MEM I medium (Invitrogen) according to the manufacturer's instructions and analyzed 48 or 72 h after transfection. Mock transfections were used as control.
Drug Treatments
MG132 (10 μM, Sigma, St. Louis, MO) was used to inhibit the proteasome and ZM447439 (10 μM, kindly provided by Dr. Campbell Wilson, AstraZeneca Pharmaceuticals, Cheshire, United Kingdom) was used to inhibit Aurora kinase activity. Unless otherwise stated, incubation with MG132 or with ZM447439 was for 1 h. To evaluate mitotic arrest, cells were incubated with the reversible microtubule depolymerizing drug nocodazole (1 μM, Sigma) for 16 h. Mitotic index was determined by cell-rounding under phase-contrast microscopy. To analyze the cell's ability to restore the metaphase plate after microtubule depolymerization, control and siRNA-treated cells were first incubated for 1 h with nocodazole, in the presence of MG132, then released into fresh medium containing MG132, and processed for immunofluorescence 30 min later.
Western Blotting
Cells, 106, were collected by centrifugation and resuspended on SDS-sample buffer containing protease inhibitors (Sigma cocktail). Cell extracts were loaded onto a 5–20% acrylamide gradient gel and then transferred to a Hybond enhanced chemiluminescence (ECL) nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ) by semidry blotting (Hoefer, San Francisco, CA). Membrane was blocked in TBST (50 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Tween-20) plus 5% nonfat dried milk and incubated with the primary antibodies diluted in TBST plus 1% nonfat dried milk. After washing, peroxidase-conjugated secondary antibodies were used (1:1500, Vector, Burlingame, CA). All incubations were for 1 h at room temperature. Proteins were detected by ECL and imaged on Kodak biomax light film (Sigma). Signal intensities were quantified using Image J 1.34s software (http://rsb.info.nih.gov/ij/) and normalized against α-tubulin intensity levels.
Immunofluorescence
Cells were fixed for 20 min in freshly prepared 2% paraformaldehyde (Sigma) in phosphate-buffered saline (PBS), permeabilized with 0.5% Triton X-100 in PBS for 5 min, washed in PBS and blocked with 10% FBS. For the cold-stable microtubule experiments, cells were incubated in ice-cold medium during 10 min, before fixation. Primary antibodies used were as follows: mouse anti-Bub3 (1:1000 dilution; BD Transduction Laboratories, Lexington, KY); rabbit anti-Bub1 (1:1000, Abcam, Cambridge, MA); mouse anti-BubR1 (1:600; Chemicon, Temecula, CA); sheep anti-Bub1 and sheep anti-BubR1 (1:1000; gifts from S. Taylor, School of Biological Sciences, University of Manchester, United Kingdom); human anti-CREST (1:2500; gift from E. Bronze-da-Rocha, IBMC, Porto, Portugal); mouse anti-Hec1 (1:500, Abcam); rabbit anti-Spc25 (1:400; gift from T. Stukenberg, Center for Cell Signaling, University of Virginia); mouse anti-histone-phospho-H3 (1:2000; Upstate Biotechnology, Lake Placid, NY); rabbit anti-Aurora B (1:1000; Abcam); rabbit anti-Mad2 FL205 (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-Mad2 (1:1000; gift from A. Musacchio, Department of Experimental Oncology, European Institute of Oncology, Milan, Italy); mouse anti-CLIP-170 (1:100; gift from F. Perez, Department of Cell Biology Sciences III, University of Geneva, Switzerland); mouse anti-p150Glued (1:1500, BD Biosciences, San Jose, CA); mouse anti-CENP-E (1:1000; gift from T. Yen, Fox Chase Cancer Center, Philadelphia, PA); mouse anti-α-tubulin clone B-5-1-2 (1:2500; Sigma); and rabbit anti-β-tubulin (1:700; Abcam). Alexa Fluor 488–, 568–, and 647–conjugated secondary antibodies were used at 1:1500 (Molecular Probes, Eugene, OR) and DNA was stained with DAPI (Sigma).
Microscopy Analysis
Most images were acquired using a 60× apochromatic objective on a Nikon TE 2000-U microscope equipped with a DXM1200F digital camera and controlled by Nikon ACT-1 software (Melville, NY). For each image, representative focus planes were shown. Images in Figure 4, A and D, were acquired as Z-stacks with 0.2–0.3-μm spacing using a 100 × 1.35 NA objective on a Olympus BX61 microscope coupled with a DP70 digital camera (Melville, NY). Olympus CellP̂ software was used for image deconvolution and projection. Images in Figures 3A, 7A, and 8F were acquired as Z-stacks with 0.2–0.3-μm spacing using a 60 × 1.42 NA on an Olympus FluoView FV1000 confocal microscope. Maximal intensity projections of the entire Z-stack are shown, and optical sections show individual kinetochores more clearly (insets).
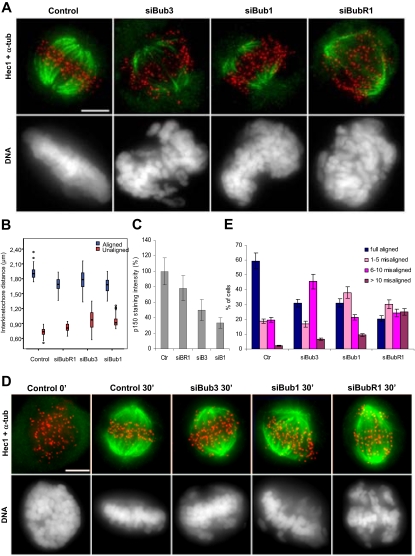
Figure 4.
Bub3 depletion leads to unstable, weak, and dysfunctional K-MT attachments. (A) Analysis of cold-stable microtubules in control and siRNA-depleted cells stained for tubulin (green), Hec1 (red), and DNA (blue); bar, 5 μm. (B) Interkinetochore distances between aligned or unaligned chromatid pairs in control and siRNA-treated cells treated with MG132. Values for unaligned chromosomes in control cells were obtained in prometaphase. Each value (median ± SD) was derived by measuring at least 50 kinetochore pairs. (C) p150Glued kinetochore fluorescence intensities were measured in control prometaphase cells and in unaligned chromosomes of siRNA-depleted cells and normalized to Spc25 kinetochore intensities. The average value (±SEM) over multiple cells (n ≥ 5, >250 kinetochores) is shown. (D) Control and siRNA-treated cells were released from a nocodazole block into fresh medium containing MG132, fixed 0 or 30 min later, and stained for tubulin (green), Hec1 (red), and DNA (blue). Immediately after nocodazole washout, microtubules were absent in all cell cultures. Thirty minutes later, both control and siRNA-treated cells formed new bipolar spindles but chromosome congression was affected in RNAi cells compared with control. (E) The number of misaligned chromosomes per cell was scored (average of three independent experiments). Scale bar, 5 μm.
Figure 3.
Misalignment defects after Bub3-depletion in comparison to those after Bub1 and BubR1 depletions. Immunofluorescence images of control and RNAi-depleted cells treated with MG132 and stained for BubR1 or Bub1 (green), Hec1 (red), tubulin (cyan), and DNA (blue), as indicated. Bar, 5 μm.
Figure 7.
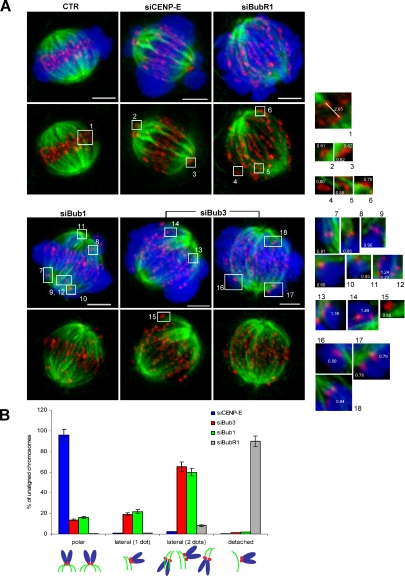
Misaligned chromosomes in Bub3-depleted cells often exhibit side-on attachments to microtubule walls. (A) Control, CENP-E, BubR1, Bub1, and Bub3 siRNA-depleted cells were incubated with MG132 and stained for CREST (red), tubulin (green), and DNA (blue). Maximal intensity projections of the entire cell Z-stack are shown. Insets are projections of 4–8 Z-stacks for individual K-MT attachments of misaligned chromosomes. Values in white indicate the interkinetochore distances between unaligned sister chromatids. Note the lateral attachments (insets 7 and 9–17) as well as end-on syntelic attachments (insets 8 and 18) of Bub3- and Bub1-depleted misaligned chromosomes. As a comparison, end-on attached kinetochores of correctly aligned sister chromatids are shown (inset 1), as well as monotelically attached CENP-E–depleted chromosomes (insets 2 and 3) and detached BubR1-depleted chromosomes (insets 4–6). Bars, 5 μm. (B) Quantitative analysis of the mode of attachment to microtubules exhibited by misaligned chromosomes of CENP-E, Bub3, Bub1, and BubR1 siRNA-depleted cells (>50 kinetochores, two independent experiments). Bars, mean ± SEM.
Figure 8.
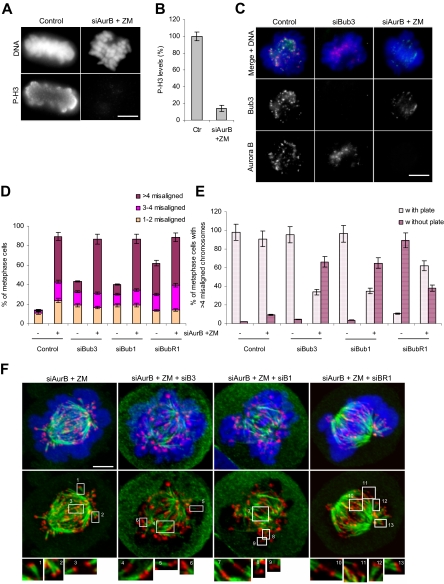
Aurora B inhibition rescues misalignment defects in BubR1- but not in Bub3- and Bub1-depleted cells. (A and B) Aurora B kinase inhibition as ascertained by the phosphorylation level of histone H3. Aurora B was repressed with both siRNA (siAurB) and kinase activity inhibitor (ZM). (A) Cells were stained with anti-phospho-H3 antibody (green) and DAPI (blue) and (B) the overall intensity was measured in 10 prometaphases (error bars, SEM). (C) Immunofluorescence images of control and siRNA-transfected cells stained for Bub3 (green), Aurora B (red), and DNA (blue). (D) Fraction of metaphase cells with misaligned chromosomes in control and single- and double-depleted cells as indicated. Misaligned chromatids were scored as described for Figure 6C. (E) Metaphase cells exhibiting >4 misaligned chromosomes were quantified for the presence or absence of a recognizable metaphase plate in control and single- and double-depleted cells as indicated. (F) Cells doubly depleted for Aurora B and Bub3, Bub1, or BubR1 were incubated with Aurora inhibitor ZM and proteasome inhibitor MG132. Cells were given a cold shock, stained for CREST (red), tubulin (green), and DNA (blue), and analyzed for the presence of stable K-MT attachments. Maximal intensity projections of the entire cell Z-stack are shown. Amphitelic (insets 3, 4, 7, and 10), syntelic (insets 1, 2, 6, 9, 12, and 13), and monotelic (insets 5 and 8) attachments are shown. Scale bars, 5 μm.
For quantification of kinetochore-staining intensities, a circular area of ∼0.6 μm diameter was drawn around each kinetochore. The pixel total brightness within the selected area was then measured using Image J 1.34s software (http://rsb.info.nih.gov/ij/). Background brightness was determined and subtracted for each measurement. Staining intensities of Bub3, Bub1, and BubR1 in the immunofluorescence and immunoblotting experiments were normalized against CREST and tubulin reference levels, respectively. Interkinetochore distances were determined using the outer kinetochore marker Hec1. Each kinetochore position was recorded as the stained centroid object. Each value was derived by measuring at least 50 kinetochore pairs in five cells.
Quantification of chromosome misalignment in siRNA-depleted cells was as adapted from (Lampson and Kapoor, 2005; Meraldi and Sorger, 2005), i.e., chromatids were considered unaligned if their kinetochores were positioned outside the area containing 40% of the central spindle. A total number of n ≥ 400 cells from three independent assays were analyzed for each RNAi experiment.
To count kinetochores with attached cold-stable microtubules, CREST staining was used to identify kinetochores in image Z-stacks. An average of 98 kinetochores was counted per cell, with three cells analyzed in each case. Each kinetochore was characterized as attached or unattached depending on whether a microtubule fiber ended at the kinetochore.
RESULTS
Bub3, Bub1, and BubR1 siRNA-directed Silencings Are Effective
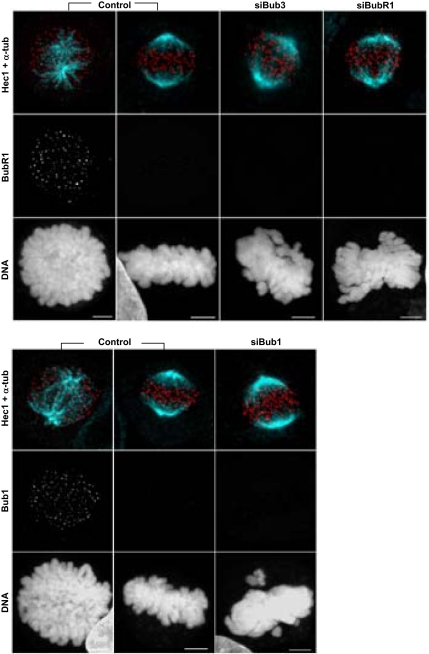
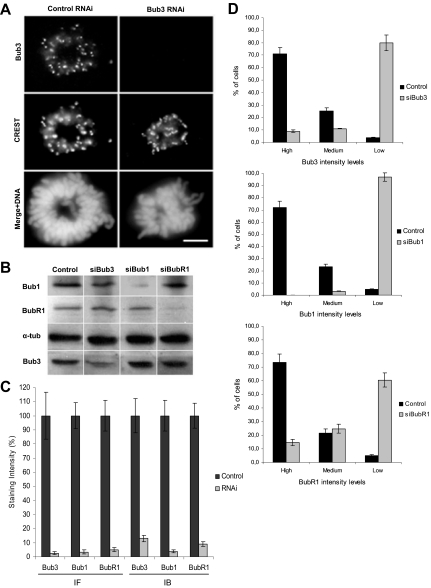
Seventy-tow hours after HeLa cell transfection with the siRNAs directed against Bub3, Bub1, or BubR1 mRNAs, efficient repressions were ascertained by immunofluorescence for kinetochore staining in prometaphase cells (Figure 1,A and Supplementary Figure 1), as well as by immunoblotting against total cell extracts (Figure 1B). The immunofluorescent levels of Bub3, Bub1, or BubR1 at the kinetochores were diminished to <5% the levels of mock-transfected cells, and the immunoblot protein levels to <15% (Figure 1C). Furthermore, we quantified the percentage of cells with high, medium, and low/absent kinetochore fluorescence intensities to determine the transfection efficiency (Figure 1D). We found efficiency values of 80, 96, and 60% for the Bub3-, Bub1-, and BubR1-RNAi transfections, respectively, when scoring as transfected only those cells that exhibited low/absent signal. Throughout subsequent analyses, only RNAi cells exhibiting low/absent levels of the depleted protein were considered, as judged by immunofluorescence with the specific antibody. BubR1 immunostaining was sometimes used alternatively to monitor the extent of Bub3 RNAi, due to incompatibility of the Bub3 antibody with other antibodies used and because BubR1 depends on Bub3 for kinetochore localization (Supplementary Figure 1; Taylor et al., 1998; Meraldi et al., 2004).
Figure 1.
Bub3-, Bub1-, and BubR1-RNAi depletion efficiency in HeLa cells. (A) Immunofluorescence images of prometaphase cells stained with Bub3 and CREST antisera showing that Bub3 is absent from kinetochores after RNAi; scale bar, 5 μm. (B) Immunoblots of cell lysates showing effective repression of Bub3, Bub1, and BubR1, with α-tubulin acting as loading control. (C) Quantification of Bub3, Bub1, and BubR1 signal intensities on prometaphase kinetochores by immunofluorescence (IF) or in whole-cell lysates by immunoblotting (IB). Staining intensities were determined relatively to CREST (for IF) or to tubulin (for IB) signal references and corrected for background noise. (D) Determination of transfection efficiency by scoring the immunofluorescent levels of kinetochore-bound Bub3, Bub1, and BubR1 as high, medium, or low. In the bar graphs, values represent the mean ± SEM of three independent experiments.
To monitor the SAC status, Bub3-depleted cells were treated with the microtubule poison nocodazole for 16 h, and the mitotic arrest was measured by cell rounding. Mock-transfected cells presented a high mitotic index (72 ± 3%) as expected for checkpoint-proficient cells, whereas Bub3- and BubR1-depleted cells had a low mitotic index (9 ± 1 and 5 ± 1%, respectively) as expected for checkpoint-defective cells (Figure 2A). Therefore, Bub3 and BubR1 repressions were specific as they abolished the checkpoint-dependent cell cycle arrest provoked by nocodazole. In contrast, Bub1-depleted cells exhibited a high mitotic index (70 ± 5%) after nocodazole treatment, suggesting that residual levels above 2–5% must be present and still accounting for checkpoint activity (Meraldi and Sorger, 2005). Nevertheless, and as shown below, we depleted Bub1 to levels (5%) at which congression is impaired.
Figure 2.
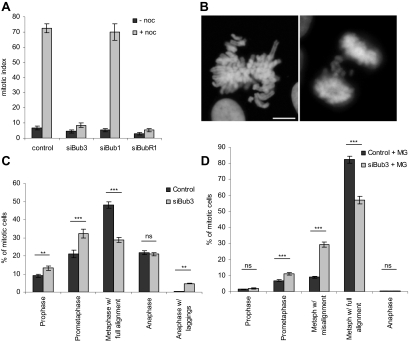
Bub3 depletion abrogates the spindle assembly checkpoint and causes chromosome misalignment. (A) Mitotic index of cells transfected with mock, Bub3, Bub1, or BubR1 siRNAs, before or after treatment with nocodazole. (B) Bub3-depleted cells exhibit abnormal mitotic figures: metaphases with misaligned chromosomes (left) and anaphases with lagging chromosomes (right); bar, 5 μm. (C) Quantification of mitotic cell fractions in mock- and siRNA-transfected cultures showing that Bub3 depletion leads to an accumulation of prometaphases and anaphases with lagging chromosomes. (D) Quantification of mitotic cell fractions treated with MG132 showing that Bub3-depleted cells still exhibit chromosome misalignment defects. In the bar graphs, values represent the mean ± SEM of three independent experiments. Total numbers of mitotic cells counted were n = 1435 in control, n = 1288 in siBub3, n = 1276 in control+MG, and n = 1045 in siBub3+MG. P values of statistical relevance were determined using a two-way ANOVA test with Bonferroni posttest; ns, not significant (p > 0.05); significant (**p < 0.01); very significant (***p < 0.001).
Bub3-depleted Cells Exhibit Chromosome Misalignment
Analysis of Bub3 siRNA-depleted mitotic cells immediately indicated a defect in chromosome alignment. To better define the nature of this defect, we set out to quantify the number of cells in the different mitotic stages. In comparison to control, we found an increase in prometaphases (32 vs. 20%, p < 0.001) and a decrease in metaphases (29 vs. 48%, p < 0.001) in the Bub3-interfered cultures. Additionally, anaphases with lagging chromosomes were also increased (4.7 vs. 0.3%, p < 0.01; Figure 2, B and C). This phenotype appeared specific, as depletion of Mad2 did not increase the number of prometaphases (Lampson and Kapoor, 2005; Orr et al., 2007; our data not shown).
Because depletion of checkpoint proteins can cause premature mitotic exit before chromosomes are properly attached to the spindle and aligned, we treated cells with the proteasome inhibitor MG132 in order to prevent anaphase onset and, thereby, to assess the effects of Bub3 depletion on chromosome alignment independently of its effects on mitotic checkpoint activity (Kops et al., 2004). After exposure to MG132, quantification of mitotic figures revealed a reduced number of prophases and anaphases (Figure 2D), indicating efficacy of the drug (Johnson et al., 2004). As expected, the number of fully aligned metaphases increased in control cell culture from 48 to 82%, whereas it was still comparatively reduced (57%, p < 0.001) in the Bub3-depleted culture (Figure 2D). Consistently, the number of metaphases with misaligned chromosomes was higher in the Bub3-depleted culture (29 vs. 10% in control, p < 0.001). These results show that the misalignment defect in Bub3-depleted cells is long-standing and not merely due to an accelerated exit from mitosis caused by the checkpoint defect.
We next quantified the extent of chromosome misalignment after exposure to MG132 in Bub3-depleted cells, in comparison to Bub1- and BubR1-depleted cells (Figure 3). Chromosomes were considered misaligned if their kinetochores were positioned outside the 40% central area of the spindle. This analysis showed that 43% of the metaphases had misaligned kinetochores in Bub3-depleted cultures, of which 18, 14, and 11% had 1–2, 3–4, and >4 misaligned chromosomes, respectively (see Figure 6C). Bub1 depletion induced a misalignment phenotype similar to that from Bub3 depletion, with 40% of metaphases with misalignment, whereas 62% of the metaphases in BubR1-depleted cell cultures had misaligned kinetochores, most of them (32%) with >4 misaligned chromosomes (siBubR1 complete in Figure 6C). The data from the quantitative analysis were tested statistically (two-way ANOVA; Supplementary Table 1), and altogether the results indicated that BubR1 depletion leads to extensive misalignment defects, more severe than those induced by Bub3 and Bub1 depletions and that Bub3 depletion phenotype is similar to Bub1.
Figure 6.
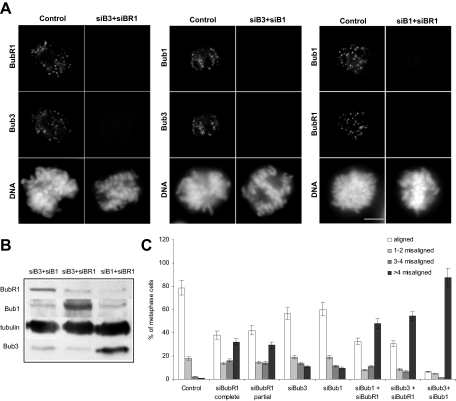
Misalignment defects in Bub3/BubR1-, Bub1/BubR1-, and Bub1/Bub3-RNAi cells. (A) Immunofluorescence images of control and siRNA-treated prometaphase cells stained with Bub3, Bub1, or BubR1 antisera as indicated and (B) immunoblots of lysates from siRNA-transfected cells, with α-tubulin acting as loading control, showing that double repressions were efficient. Scale bar, 5 μm. (C) Chromosome misalignment in cells transfected with siRNA as indicated and treated with MG132. Chromosomes were counted as misaligned if they were located outside the 40% central spindle area of the mitotic spindle. Error bars, SEM of three independent experiments (>250 kinetochores, >5 cells).
K-Fiber Stability, K-MT Occupancy and Chromosome Congression Are Defective in Bub3-depleted Cells
As misalignment usually results from chromosome-to-spindle attachment defects, we searched for the presence of such defects in the Bub3-depleted cells. We performed different assays (see descriptions below) that are commonly used to analyze specific aspects of K-MT interaction.
First, we assayed for the stability of the K-MT attachments. As kinetochore microtubules (K-fibers) are differentially stable to cooling at 4°C in comparison to nonkinetochore microtubules (Rieder, 1981), we gave cells a cold shock and checked for the presence of stable microtubules. Although in mock-depleted cells thick K-fibers were clearly attached to each kinetochore (Figure 4A), in Bub3-depleted cells only a few cold-stable K-fibers were present. Accordingly to previous observations (Lampson and Kapoor, 2005), BubR1-depleted cells also exhibited few thin cold-stable K-fibers, as did Bub1-repressed cells (our present data). Quantification of the kinetochores with cold-stable microtubule fibers attached confirmed that Bub3, Bub1, and BubR1 depletions lead to defective cold-sensitive K-MT attachments, with only 14 ± 3, 17 ± 4, and 4 ± 2% attached kinetochores, respectively, compared with 83 ± 5% in control (Table 1). Statistic analysis of these data further indicated that the defective attachments in Bub3 depletion are similar to those in Bub1 depletion (p > 0.05) but significantly different from those in BubR1 depletion (p < 0.001; Supplementary Table 2).
Table 1.
Quantification of cold stable K-fibersa
| Condition | Control | Bub3 siRNA | Bub1 siRNA | BubR1 siRNA |
|---|---|---|---|---|
| + MG132 1h | 83 ± 5% | 14 ± 3% | 17 ± 4% | 4 ± 2% |
| + ZM Aurora inhibitor + MG132 1h | 81 ± 6% | 62 ± 4% | 59 ± 3% | 79 ± 5% |
a An average of 98 kinetochores was counted per cell, with three cells analyzed in each case. Each kinetochore was characterized as attached or unattached depending on whether a microtubule fiber ended at the kinetochore.
Second, we measured the interkinetochore distances, as a failure in the establishment of stable K-MT connections usually affects centromere stretching and tension on sister kinetochores (Waters et al., 1996). In agreement, we found a significant decrease in interkinetochore distances across aligned chromatid pairs of BubR1-, Bub3-, and Bub1-depleted cells (1.68 ± 0.13, 1.78 ± 0.20, 1.66 ± 0.15 μm, respectively) compared with control (1.92 ± 0.12 μm; p < 0.001, ANOVA test followed by Scheffé multiple comparison test; Figure 4B). It is worth mentioning that although in control cells the aligned chromosomes grouped mostly into well-defined metaphase plates, in RNAi cells this was often not the case, and therefore it is possible that some incorrectly attached chromosomes were inadvertently scored as aligned because they were lying inside the area containing 40% of the central spindle. Thus, even though it is apparent that interkinetochore distances are globally decreased in the RNAi cells comparing to control, reading more into these results and try to explain the difference between Bub3 and Bub1 and BubR1 might be unreasonable. In the case of unaligned chromatid pairs, BubR1-depleted cells exhibited a mean value of 0.81 ± 0.08 μm, which was slightly higher than that of detached pairs in prometaphase control cells (0.73 ± 0.05 μm; p < 0.001, Kruskal-Wallis test followed by two-sample Kolmogorov-Smirnov multiple comparison test; Supplementary Table 3). Misaligned chromatids in Bub3- and Bub1-depleted cells had interkinetochore distances of 0.98 ± 0.18 and 0.95 ± 0.13 μm, respectively, which were significantly higher than those found for BubR1-depleted chromatid pairs (p < 0.001; Figure 4B) and intermediate between the minimum (0.73 ± 0.05 μm) and maximum (1.92 ± 0.12 μm) control levels. Thus, pulling forces on sister kinetochores were globally compromised, severely in BubR1-RNAi and moderately in Bub3- and Bub1-RNAi.
Thirdly, as the levels of centromere stretching might reflect the state of microtubule occupancy, we examined kinetochore proteins that become hardly detectable after chromosome attachment to the spindle. Even though Mad2 kinetochore levels were reported to be highly sensitive to microtubule occupancy (Waters et al., 1998), its use was precluded in our study as Bub3 is required for its kinetochore localization (Supplementary Figure 1). The use of other checkpoint proteins, such as Bub1 and BubR1, was also excluded because their kinetochore localization was reported to be compromised by Bub3 depletion and their sensitivities to attachment and/or absence of tension are still controversial (Pinsky and Biggins, 2005). Therefore, we alternatively monitored the levels of p150Glued (a subunit of the dynactin complex) and of CLIP-170 (a microtubule “plus-end–tracking” protein) because these proteins are hardly detectable on attached kinetochores and are recruited to human kinetochores independently of checkpoint proteins (King et al., 2000; Tanenbaum et al., 2005; Supplementary Figure 2A). We found that the p150Glued fluorescence intensities of unaligned Bub3- and Bub1-depleted kinetochores were reduced to 50 and 33% of control unattached chromosome levels, respectively, indicating attachment to an incomplete complement of microtubules (Figure 4C; Supplementary Figure 2B). As expected, p150Glued levels at the unaligned kinetochores of BubR1-depleted cells were as high as 78% of control levels. Similar results were found for CLIP-170 kinetochore intensities (Supplementary Figure 2, C and D).
Finally, considering the defects depicted above, we examined the ability of K-fibers to restore a metaphase plate after a nocodazole washout (adapted from Tanenbaum et al., 2005). HeLa cells were treated with nocodazole to depolymerize all microtubules, then released into fresh medium containing MG132, and stained for tubulin and CREST 0 and 30 min later (Figure 4D). Immediately after the release, the spindles were absent in cells treated with the depolymerizing drug, with chromosomes scattered throughout the cell. Thirty minutes after the washout, both control and siRNA-transfected cells had nucleated a new set of microtubules, which organized into a bipolar spindle, indicating that transfection per se or depletion of checkpoint proteins did not interfere with spindle formation. However, with respect to chromosome alignment, we found different results between the siRNA depletions in comparison to control (Figure 4D). Although 59% of the control cells had completely aligned their chromosomes within the 30 min after release, only 31% of the Bub3-depleted cells exhibited full alignment (Figure 4E). The remaining Bub3-depleted cells had several misaligned chromosomes, mostly between 6 and 10 (45% of the cells), consistent with dysfunctional K-MT attachments. In Bub1-depleted cells, we also found only 31% of fully aligned metaphases, with most of the other cells exhibiting 1–15 misaligned chromosomes. BubR1-depleted cells were the least able to recover from the washout, with only 20% of metaphases reaching full alignment, whereas the majority of the bipolarized cells did not form a defined metaphase plate. Such behavior appears in agreement with the proposed inability of these cells to stabilize K-MT attachments (Lampson and Kapoor, 2005). Hence, we conclude from this assay that chromosome misalignment in Bub3-depleted cells is due to dysfunctional K-MT interaction.
Misalignment Defects in Bub3-depleted Cells Are Not Due to CENP-E, Bub1, and BubR1 Depletion from Kinetochores
We examined whether the misalignment defects found in Bub3-depleted cells could be due to CENP-E mislocalization. CENP-E is a kinesin-like protein known to be required for chromosome congression (Wood et al., 1997; Yao et al., 1997). Its recruitment to kinetochores has been reported to depend on checkpoint proteins (Sharp-Baker and Chen, 2001; Johnson et al., 2004; Vigneron et al., 2004), even though others found recruitment to be independent (Lampson and Kapoor, 2005; Meraldi and Sorger, 2005). Therefore, we quantified the kinetochore levels of CENP-E in Bub3-, Bub1-, and BubR1-depleted cells (Figure 5). We found that depletion of these proteins affected CENP-E kinetochore localization, but with different extent (Figure 5A). In comparison to CENP-E fluorescence intensities of control prometaphase kinetochores, the CENP-E levels of Bub3-, Bub1-, and BubR1-depleted kinetochores were reduced to 59, 67, and 41%, respectively (Figure 5B). Thus, as significant levels of CENP-E are still present in Bub3-depleted cells, the misalignment defects are unlikely to be due to, or at least are not exclusively due to, a failure in CENP-E recruitment to kinetochores. The same has been proposed for BubR1 and Bub1 (Lampson and Kapoor, 2005; Meraldi and Sorger, 2005). Furthermore, the effects of CENP-E RNAi on chromosome misalignment were clearly different from those of Bub3 RNAi. CENP-E–depleted cultures exhibited a higher number of cells with misaligned chromosomes (93%) than Bub3-depleted cultures (53%; Figure 5C), and misaligned chromosomes were typically localized close to the spindle poles (Putkey et al., 2002). In addition, the number of metaphases with misalignment largely decreased in CENP-E–depleted cultures after an MG132-induced anaphase delay, whereas it remained almost unchanged in Bub3-depleted cultures. This indicates that, in contrast to the defect caused by CENP-E depletion, which can be partially corrected by delaying anaphase onset, the misalignment defect in Bub3-depleted cells is persistent and of a distinct nature (see Discussion).
Figure 5.
Misalignment defects in Bub3-depleted cells are not due to CENP-E loss from kinetochores. (A) Control and siRNA-depleted cells were stained for the CENP-E motor protein; bar, 5 μm. (B) CENP-E kinetochore fluorescence intensities (>250 kinetochores) were measured and normalized to Spc25 staining levels (average of two independent experiments). (C) Percentage of metaphases (mean ± SEM of two independent experiments) in control, Bub3, and CENP-E siRNA-depleted cells before and after incubation with MG132.
We also asked whether the defects observed in Bub3-depleted cells could be simply due to Bub1 and/or BubR1 loss from kinetochores, as these checkpoint proteins are well-known interactors of Bub3. Even though the localization interdependencies between Bub3, Bub1, BubR1, as well as other checkpoint proteins, have been extensively addressed in previous studies, we found several contradictory statements between independent reports (Johnson et al., 2004; Meraldi et al., 2004; Meraldi and Sorger, 2005), most probably due to the fact that assembly of kinetochore proteins depends on multiple branches. Therefore, we undertook localization analyses in Bub3-, Bub1-, and BubR1-depleted cells treated with the microtubule-depolymerizing drug colchicine in the presence of MG132, thus creating conditions where all the checkpoint proteins should be maximally enriched at kinetochores. In summary, our results showed that 1) Bub3 depletion abolishes BubR1 kinetochore localization but does not significantly affect Bub1 localization; 2) Bub1 depletion abolishes BubR1 kinetochore localization but does not significantly affect Bub3 localization; 3) BubR1 depletion does not affect Bub1 kinetochore localization and only slightly reduces Bub3 localization (Table 2; Supplementary Figure 1). These observations allowed us to conclude that the Bub3-RNAi misalignment phenotype cannot be explained by Bub1 mislocalization, but raised the possibility that it might be due to BubR1 loss from kinetochores. That being so, then Bub3-RNAi should present a misalignment phenotype at least as severe as that of BubR1-RNAi, and this happens not to be the case (Figures 3 and 4). Alternatively, an incomplete BubR1depletion from kinetochores in Bub3-RNAi cells could account for their less severe phenotype. To test this possibility, we analyzed the chromosome misalignment phenotype in cells partially depleted from BubR1 (these cells could be easily found 24 h after siRNA transfection). Such analysis showed that partial depletion of BubR1 generated a misalignment defect almost as severe as that caused by complete depletion and clearly more severe than that in Bub3-RNAi (Figure 6C). We also found strong p150Glued staining at the kinetochores of most misaligned chromosomes in BubR1-partially depleted cells, indicating that microtubule attachments were compromised (Supplementary Figure 3). Thus, the Bub3-RNAi misalignment defect is inconsistent with the mere loss of BubR1, Bub1, or CENP-E from kinetochores, even though we do not exclude the possibility that a combined loss of these proteins could contribute for the defect.
Table 2.
Analysis of spindle checkpoint protein interdependency for kinetochore localization
| Condition | Kinetochore binding in prometaphase cellsa |
|||
|---|---|---|---|---|
| Bub3 | Bub1 | BubR1 | Mad2 | |
| Bub3 RNAi | −/+b | ++ | −/+ | + |
| Bub1 RNAi | ++ | −/+ | −/+ | −/+ |
| BubR1 RNAi | ++ | +++ | −/+ | −/+ |
a As determined by immunofluorescence in siRNA-treated HeLa cells incubated during 1 h with both colchicine and MG132.
b Intensity levels compared with control cells: −/+, absent to highly reduced signal; +, reduced signal; ++, slightly reduced signal; and +++, normal signal.
To get insight into the relationships between the three checkpoint proteins regarding their functions in the regulation of K-MT interactions, we analyzed the misalignment defects in double RNAi-interfered cells. RNAi efficiency was ascertained both by immunofluorescence and immunoblotting (Figure 6, A and B). The double RNAi-interfered cells were arrested at metaphase–anaphase transition with MG132, and misalignment defects were quantified as described above (Figure 6C). We found Bub3/BubR1 and Bub1/BubR1 repressions to exhibit similar misalignment phenotype severity with 67 and 65% of metaphase cells showing misaligned chromosomes, respectively (p > 0.05, Supplementary Table 1). At a first sight, there appeared to be an additive effect between Bub3 and BubR1 as well as between Bub1 and BubR1 in the regulation of K-MT attachments, as Bub3/BubR1-RNAi exhibited 54% of metaphases with >4 misaligned chromosomes, which seems to be a sum of those exhibited by Bub3 and BubR1 single repressions, and Bub1/BubR1-RNAi exhibited 48% of metaphases with >4 misaligned chromosomes, which is the sum of those exhibited by Bub1 and BubR1 single depletions. However, alignment defects did not seem to be much higher than for individual BubR1-RNAi, especially as the amount of cells with full chromosome alignment did not change but rather cells with misalignment had more unaligned chromatids. Statistic analysis of the quantitative data pointed out that Bub3/BubR1- and Bub1/BubR1-RNAi are not significantly different from BubR1-RNAi when cell categories, namely fully aligned, 1–2, 3–4, and >4 misaligned, are compared one by one (Supplementary Table 1). Remarkably, chromosome alignment defects in the Bub3/Bub1-RNAi were worst than in Bub3/BubR1 and Bub1/BubR1 knockdowns, with 94% of metaphases showing misalignment, most of which (87%) with >4 unaligned chromatid pairs (Figure 6C). The number of cells with full alignment, as well as of cells with >4 misaligned chromosomes, were significantly different in Bub3/Bub1-RNAi compared with each one of the single RNAis or to the other double RNAis. Altogether, the results from double depletions strongly suggest that Bub3 and Bub1 play similar, redundant roles in K-MT attachment and that depletion of either one individually is not enough to create a dramatic effect. This idea explains the additive phenotype, which is expected for proteins with parallel function and is consistent with the initial finding of Bub3 as an extra-copy suppressor of bub1-1 mutant (Hoyt et al., 1991). Redundancy between Bub1 and Bub3 also explains why the misalignment phenotypes are similar in Bub1/BubR1 and Bub3/BubR1 double depletions and in BubR1 single depletion: in Bub1/BubR1-depleted cells, Bub3 could be compensating Bub1 absence, and in Bub3/BubR1-depleted cells, Bub1 could be compensating Bub3 absence. Nevertheless, and considering the effect of RNAi variability, we cannot exclude the possibility that Bub3 and Bub1 may act cooperatively (see Discussion).
Bub3 and Bub1 Depletions Lead to Defective K-MT Attachments That Are Distinct from Those Induced by BubR1 and CENP-E Depletions
As shown above, we observed that Bub3 and Bub1 depletions produce similar misalignment defects, which appear different from those induced by BubR1 and CENP-E depletions. Thus, we went on examine the mode of MT binding by misaligned chromosomes in all these RNAi-depleted cells using high-resolution confocal imaging. For each RNAi-depleted cell, stained with CREST and anti-α-tubulin antibodies, we acquired Z-stacks with 0.2-μm spacing (Figure 7A), and we used 4–8 Z-stacks for maximal intensity projections of individual K-MT attachments of misaligned chromosomes (insets). We found that in Bub3- and Bub1-depleted cells, misaligned chromosomes were frequently bound to microtubules emanating from the same pole. The binding configuration appeared to be lateral (side-on binding to the walls of microtubules) in most of the cases. In contrast, misaligned chromosomes of CENP-E–depleted cells were tightly associated with the spindle poles, consistently to the centrophilic configuration previously reported for CENP-E depletion (Putkey et al., 2002). Misaligned chromosomes of BubR1-depleted cells were also examined, and we found them to be mostly detached. Interkinetochore distances were measured in all the K-MT attachments analyzed, and it further informed that Bub3- and Bub1-depleted misaligned chromosomes often exhibit intermediate levels of tension, whereas those in CENP-E– and BubR1-depleted cells have low levels of tension (insets in Figure 7A and Figure 4B). We quantified the mode of MT binding by misaligned chromosomes (>50) in each of the RNAi depletions. We classified attachments as polar, side-on, and detached. As side-on attachments exhibited either one or two dots of CREST staining (reflecting different levels of centromere stretching), they were further subdivided accordingly. This quantitative analysis showed that misaligned chromosomes of Bub3- and Bub1-depleted cells were typically attached to microtubules in a side-on configuration, rarely found in CENP-E– and BubR1-depleted cells (Figure 7B). Interestingly, in BubR1-depleted cells, misaligned chromosomes despite detached had kinetochore pairs parallel to the spindle axis, with interkinetochore distances slightly higher than those produced by nocodazole treatment (Figure 4B). This is in agreement with the fact that, rather then impairing MT binding, BubR1 depletion destabilizes K-MT attachments (Lampson and Kapoor, 2005). Instability does not appear to be the nature of the attachment defect in Bub3- and Bub1-depleted cells, but further analyses will be required to determine exactly the origin of the side-on attachments found in these cells.
Aurora B Kinase Inhibition Restores K-MT Attachments in BubR1- But Not in Bub3- and Bub1-depleted Cells
An essential regulator of both K-MT attachments and the SAC response to tension defects is the conserved Ipl1- Aurora B protein kinase (Vagnarelli and Earnshaw, 2004). Ipl1-Aurora B is thought to facilitate proper attachments by destabilizing K-MT interactions that do not generate tension (Tanaka et al., 2002; Hauf et al., 2003). Previous studies have shown that inhibition of Aurora kinase activity suppresses the misalignment/attachment defects in BubR1-depleted cells (Lampson and Kapoor, 2005), whereas in Bub1-depleted cells it has an additive effect in terms of misalignment phenotype (Meraldi and Sorger, 2005). Therefore, we tested the effects of Aurora kinase activity inhibition in Bub3-depleted cells, to further determine the nature of their attachment defects in comparison to those of Bub1- and BubR1-depleted cells. To inactivate Aurora B, cells were transfected with previously validated siRNA oligos (Hauf et al., 2003) and treated with a small molecule inhibitor ZM447439 (Ditchfield et al., 2003). Aurora B was effectively inactivated under these conditions, as judged by the phosphorylation level of histone H3 substrate and by the defects in chromosome congression (Figure 8, A, B, and D). Immunostaining analysis showed that Bub3 fully localized to kinetochores in Aurora B–defective cells as well as did Aurora B in Bub3-depleted cells, demonstrating that these proteins are not interdependent for kinetochore localization (Figure 8C). Again using the same criteria as described for Figure 6C, we quantified chromosome misalignment in RNAi-depleted cells in which Aurora B kinase was repressed. In Bub3-depleted cells, Aurora repression caused an increase in the number of metaphases with >4 misaligned chromosomes, just like in Bub1-depleted cells (Figure 8D). We also observed an increase in the number of metaphases with >4 misaligned chromosomes in BubR1-depleted cells after Aurora B inhibition. However, we noticed that most of these metaphases exhibited an organized and defined plate with many aligned chromosomes compared with BubR1-depleted cells with active Aurora B kinase (Figure 8, E and F). Indeed, we quantified the number of metaphase cells with >4 misaligned chromosomes that exhibited a recognizable metaphase plate and found this number to increase in BubR1-depleted cells after Aurora B inhibition (from 10 to 62%), whereas it decreased in Aurora B inactive Bub3- and Bub1-depleted cells (Figure 8E). Examination of the stability of the K-MT attachments revealed that in BubR1-depleted cells treated with ZM447439, 79% of kinetochores had cold-stable microtubules attached compared with only 4% in cells with active Aurora kinase, as expected (Table 1). Chromosomes aligned at the center of the spindle had K-fibers extending from sister kinetochores toward opposite poles (Figure 8F, insets 10 and 11). For some kinetochores near the poles, syntelic orientation was observed (Figure 8F, insets 12 and 13). Thus, the attachment defect induced by BubR1-depletion is rescued by Aurora B inhibition. In Bub3- and Bub1-depleted cells treated with ZM447439, we interestingly found that 62 ± 4 and 59 ± 3% of kinetochores, respectively, had cold-stable microtubules attached compared with only 14 ± 3 and 17 ± 4% in cells with active Aurora. However, only few chromosomes were aligned at the center of the spindle and had K-fibers extending toward opposite poles. For most chromosomes, monotelic and syntelic attachments were observed (insets in Figure 8F). Thus, Aurora kinase inhibition has an additive effect on the misalignment phenotype of Bub3 and Bub1 depletions, most probably through stabilization of the defective attachments. This suggests that the defective attachments in Bub3- and Bub1-depleted cells are detected by active Aurora B, which acts to destabilize them, thereby creating the possibility for new correct attachments to be established. Importantly, the results further stressed the different roles between Bub3/Bub1 and BubR1 checkpoint proteins in the regulation of K-MT attachments.
DISCUSSION
Bub3 Promotes Correct K-MT Attachments
Using RNAi-mediated gene silencing, we have shown depletion of Bub3 to accelerate mitotic progression, both under normal condition and exposure to nocodazole, indicating that Bub3 is essential for SAC function consistently to previous reports (Hoyt et al., 1991; Basu et al., 1998; Kalitsis et al., 2000; Campbell and Hardwick, 2003; Lopes et al., 2005). Additionally, we have found Bub3 depletion to compromise the alignment of a significant fraction of chromosomes in the equatorial plate and have shown this phenotype to result from defective K-MT attachments as evidenced by 1) reduced interkinetochore distance, consistent with a reduction in pulling forces and tension across sister chromatid pairs; 2) loss of K-fibers after cold-treatment, consistent with unstable interactions; 3) moderate p150 levels at the kinetochores of misaligned chromosomes, consistent with partial MT occupancy; and 4) reduced K-MT ability to restore a metaphase plate after release from a nocodazole block. However, as many chromatid pairs do align on the metaphase plate in Bub3-depleted cells, it appears that Bub3 is required for efficient establishment rather than the maintenance of bipolar MT attachment.
Our data established that chromosome misalignment caused by Bub3 depletion is not a secondary consequence of checkpoint inactivation or altered mitotic timing, as misaligned chromosomes persisted even when cells were blocked in mitosis with the proteasome inhibitor MG132. Also, we demonstrated that loss of CENP-E from kinetochores, a kinesin-like motor known to play a key role in chromosome alignment, cannot explain the alignment defects observed in Bub3-depleted cells because 1) significant levels of CENP-E were still detected in Bub3-depleted kinetochores; 2) misaligned chromosomes in CENP-E–depleted cells were tightly associated with a single spindle pole, whereas those in Bub3-depleted cells were localized in between the pole and the equatorial plate; and 3) CENP-E–depleted cells extensively overrode the misalignment phenotype when anaphase was delayed after MG132 treatment. Furthermore, we also showed that Bub1 and BubR1 depletions from kinetochores are unlikely to contribute for the Bub3 phenotype as 1) significant levels of Bub1 were still found in Bub3-depleted kinetochores; 2) even though BubR1 kinetochore localization was abolished after Bub3 depletion, both partial and extensive BubR1 depletions induced misalignment/attachment defects that were very distinct from those in Bub3-depleted cells (see further discussion below); and 3) corepression of Bub3 and Bub1 (or BubR1) produced additive chromosome misalignment phenotype. Thus, like Bub1 and BubR1, Bub3 has a dual function in SAC signaling and in promoting the establishment of correct K-MT attachments.
Besides having helped to assign a specific function for Bub3, the double depletion experiments brought additional important findings. First, Bub3 and Bub1 most probably play redundant roles in the regulation of K-MT attachments. This is not only consistent with previous data from yeast where Bub3 has been identified as a copy suppressor of bub1-1 mutation (Hoyt et al., 1991), but also explains the dramatic defects found in chromosome alignment after Bub1/Bub3-RNAi. In addition, it explains why defects in Bub1/BubR1 and Bub3/BubR1 RNAis are similar and why they are less severe than in Bub1/Bub3-RNAi. Second, BubR1 function in the stabilization of K-MT attachments might be kinetochore-independent. If that was not so, BubR1 kinetochore delocalization in Bub3-depleted cells should be expected to account for a more dramatic phenotype as end-on attachments would become unstable. Thus, despite delocalized from the Bub3-depleted kinetochores, the soluble cytosolic BubR1 protein might be phosphorylating any other protein which, once modified, acts to stabilize K-MT attachments. Similarly, the role of BubR1 on checkpoint activation was previously suggested to be kinetochore-independent (Morrow et al., 2005).
Because of the transfection and repression levels variability from experiment to experiment, we do not exclude the possibility that our cellular phenotype might be affected by a remaining pool of the target protein. This is particularly true for kinases as Bub1, where even 95% of repression levels lead to mitotic checkpoint arrest in our study and others (Tang et al., 2004), whereas <5% remaining Bub1 results in an abrogated checkpoint (Meraldi and Sorger, 2005). We believe that we minimized the problem of cell heterogeneity due to variability in knockdown levels because 1) we analyzed only RNAi cells in which the target protein was undetectable by immunofluorescence, and 2) our RNAi conditions reproduced the misalignment phenotypes previously reported for BubR1 and Bub1.
MT-binding Defects in Bub3 and Bub1 Depletions Are Distinct from Those in BubR1 Depletion
In this study, we undertook a comparative analysis between the chromosome-to-spindle interaction defects induced by Bub3, Bub1, and BubR1 depletions. We found several lines of evidence suggesting that Bub3 and Bub1 act differently from BubR1 in the regulation of bipolar attachments: 1) the K-MT attachment defects in Bub3- and Bub1-depleted cells were very similar in all the assays used to assess chromosome–spindle interactions, being less severe than in BubR1-depleted cells; 2) the misaligned chromosomes in Bub3- and Bub1-depleted cells appear to bind to the sides rather than the ends of microtubules, whereas they are detached in BubR1-depleted cells; and 3) inhibition of Aurora B kinase activity reverts the attachment defect in BubR1-depleted cells, whereas in Bub3- and Bub1-depleted cells, it has an additive effect on the misalignment phenotype. Thus, our results suggest that Bub3 and Bub1 are required for the establishment of efficient end-on attachments, whereas BubR1 is necessary for their maintenance. In the budding yeast, distinct chromosome segregation roles for the SAC proteins were described, with Bub1 and Bub3 having predominant and cooperative role in the regulation of chromosome segregation (Warren et al., 2002). We have found corepression of Bub1 and Bub3 to induce the most severe chromosome misalignment phenotype, pointing to a synergistic effect. Considering that siRNA repressions were effective, the most probable explanation is that Bub3 and Bub1 have redundant roles in K-MT interaction, being therefore depletion of either one individually unable to create a dramatic effect. However, considering that even small amounts of proteins could still account for significant function on K-MT interaction, we cannot exclude the possibility that Bub3 and Bub1 may act cooperatively, as in this case corepression would also produce an additive effect on misalignment compared with single repressions.
But what is the nature of the MT-binding defect caused by Bub3 depletion? The high density of microtubules makes it difficult to image misaligned chromosomes in HeLa cells, but our data suggest that chromatids make side-on attachments to microtubules emanating from the same pole. This type of interaction is distinct from that caused by Aurora B inhibition, which is syntelic end-on. Interestingly, we observed the phenotype of simultaneous Bub3 and Aurora B depletion to be additive with respect to single depletions. Similar results were reported for Bub1 (Meraldi et al., 2005), suggesting possible cooperation of Bub3 and Bub1 in chromosome segregation as proposed for their yeast counterparts. The timing of ScBub1p recruitment to budding yeast kinetochores has also been cited as evidence that it assists in the formation of mature ends-on MT attachments (Gillett et al., 2004). Together with Mal3/EB1 MT-binding protein, Bub1 might be involved in a specific mode of spindle–kinetochore interaction, such as switching from lateral to end-on attachment (Asakawa et al., 2005; Tanaka et al., 2005). We believe our data provide further evidence for the hypothesis that Bub1, and now also Bub3, are involved in this switching.
We do not know whether Bub3 is directly responsible for the K-MT attachments or if it indirectly regulates the activity of a MT-binding protein. Recently, Bub3 was found to be a specific binding partner of cytoplasmic dynein light chain DYNLT3 (Lo et al., 2007). Depletion or inhibition of kinetochore dynein prevents the rapid poleward motion of attaching kinetochores that results from a successful search-and-capture event (Alexander and Rieder, 1991; Yang et al., 2007). In addition, after kinetochores attach to the spindle, kinetochore dynein is required for stabilizing K-fibers, which it probably does by generating tension on the kinetochore, and in its absence, chromosome congression is delayed/disrupted (Cleveland et al., 2003). It will be important to determine if these dynein-dependent steps are compromised in Bub3-depleted cells.
Understanding how chromosome–microtubule interactions are transduced into signals that can be integrated by the molecular pathway of the SAC has remained a challenge because the discovery of this mitotic surveillance mechanism. Emerging data strongly suggest that the connection relies on the dual function of checkpoint proteins. Checkpoint proteins might regulate different aspects of K-MT interactions, which include distinct kinetochore structural contributions that influence segregation, detection of different types of kinetochore status in the context of checkpoint signaling (attachment vs. tension), or communication to diverse target molecules involved in MT dynamics and attachment-correction mechanisms (Chan et al., 2005). Given the complexity of the K-MT interactions, together with the importance of ensuring accurate segregation of the genetic material in a dividing cell, it is not surprising that different proteins act redundantly or cooperatively to regulate similar and/or different aspects of such interactions. This may provide a selective advantage to multicellular organisms to increase the fidelity of chromosome segregation, reducing the possibility of aneuploidy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Cláudio Sunkel for having supported us at the beginning of this work. We thank Dr. Steff Sφrren (IBMC, Porto, Portugal) for the HeLa cells and transfection suggestions. We thank Drs. E. Bronze-Rocha, A. Musacchio, F. Perez, S. Taylor, T. Stukenberg, and T. Yen for the kindly provided primary antibodies. We thank Dr. Campbell (AstraZeneca, United Kingdom) for the ZM Aurora inhibitor, Drs. A. Almeida and C. Braga (Minho University) for statistical analyses of the data, and Dr. V. Nascimento for technical help. This work was supported by Grant POCTI/BCI/42341/2001 from Fundação para a Ciência e Tecnologia (FCT) and by Cooperativa de Ensino Superior Politécnico e Universitário (CESPU). T.R. and C.T. had a fellowship from FCT.
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- K-MT
kinetochore–microtubule
- MCC
mitotic checkpoint complex
- RNAi
RNA interference
- SAC
spindle assembly checkpoint
- siRNA
small interference RNA.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0633) on January 16, 2008.
REFERENCES
- Alexander S. P., Rieder C. L. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J. Cell Biol. 1991;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K., Toya M., Sato M., Kanai M., Kume K., Goshima T., Garcia M. A., Hirata D., Toda T. Mal3, the fission yeast EB1 homologue, cooperates with Bub1 spindle checkpoint to prevent monopolar attachment. EMBO Rep. 2005;6:1194–1200. doi: 10.1038/sj.embor.7400540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J., Logarinho E., Herrmann S., Bousbaa H., Li Z., Chan G. K., Yen T. J., Sunkel C. E., Goldberg M. L. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma. 1998;107:376–385. doi: 10.1007/s004120050321. [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A. W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady D. M., Hardwick K. G. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- Campbell L., Hardwick K. G. Analysis of Bub3 spindle checkpoint function in Xenopus egg extracts. J. Cell Sci. 2003;116:617–628. doi: 10.1242/jcs.00255. [DOI] [PubMed] [Google Scholar]
- Chan G. K., Liu S-T., Yen T. J. Kinetochore structure and function. Trends Cell Biol. 2005;11:489–598. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Chung E., Chen R. H. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat. Cell Biol. 2003;5:748–753. doi: 10.1038/ncb1022. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signalling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- de Antoni A., Pearson C. G., Cimini D., Canman J. C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E. D., Musacchio A. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S. S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett E. S., Espelin C. W., Sorger P. K. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G. J. The mitotic spindle checkpoint. Curr. Biol. 2001;11:R1001–R1004. doi: 10.1016/s0960-9822(01)00609-1. [DOI] [PubMed] [Google Scholar]
- Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., Van Meel J., Rieder C. L., Peters J. M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Hoffman D. B., Fang G., Murray A. W., Salmon E. D. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Totis L., Roberts B. T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Jablonski S. A., Chan G. K., Cooke C. A., Earnshaw W. C., Yen. T. J. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- Johnson V. L., Scott M. I., Holt S. V., Hussein D., Taylor S. S. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- Kalab P., Pralle A., Isacoff E. Y., Heald R., Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalitsis P., Earle E., Fowler K. J., Choo K. H. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T. M., Lampson M. A., Hergert P., Cameron L., Cimini D., Salmon E. D., McEwen B. F., Khodjakov A. Chromosomes can congress to the metaphase plate before orientation. Science. 2006;311:343–344. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. M., Hays T. S., Nicklas B, R. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J. Cell Biol. 2000;151:739–748. doi: 10.1083/jcb.151.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G. J., Foltz D. R., Cleveland D. W. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M. A., Kapoor T. M. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 2005;7:93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A. W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Lo K. W., Kogoy J. M., Pfister K. K. The DYNLT3 light chain directly links cytoplasmic dynein to a spindle checkpoint protein, Bub3. J. Biol. Chem. 2007;282:11205–11212. doi: 10.1074/jbc.M611279200. [DOI] [PubMed] [Google Scholar]
- Logarinho E., Bousbaa H., Dias J. M., Lopes C., Amorim I., Antunes-Martins A., Sunkel C. E. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 2004;117:1757–1771. doi: 10.1242/jcs.01033. [DOI] [PubMed] [Google Scholar]
- Lopes C. S., Sampaio P., Williams B., Goldberg M., Sunkel C. E. The Drosophila Bub3 protein is required for the mitotic checkpoint and for normal accumulation of cyclins during G2 and early stages of mitosis. J. Cell Sci. 2005;118:187–198. doi: 10.1242/jcs.01602. [DOI] [PubMed] [Google Scholar]
- Mao Y., Abrieu A., Cleveland D. W. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Sorger P. K. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;20:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V. M., Sorger P. K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Morrow C. J., Tighe A., Johnson V. L., Scott M. I., Ditchfield C., Taylor S. S. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J. Cell Sci. 2005;118:3639–3652. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Hardwick K. G. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Orr B., Bousbaa H., Sunkel C. E. Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells. Mol. Biol. Cell. 2007;18:850–863. doi: 10.1091/mbc.E06-07-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Putkey F. R., Cramer T., Morphew M. K., Silk A. D., Johnson R. S., McIntosh J. R., Cleveland D. W. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- Rieder C. L. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- Shah J. V., Cleveland D. W. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H., Chen R. H. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G. K., Yen T. J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. U., Stark M. J., Tanaka K. Kinetochore capture and bi-orientation on the mitotic spindle. Nat. Rev. Mol. Cell Biol. 2005;6:929–942. doi: 10.1038/nrm1764. [DOI] [PubMed] [Google Scholar]
- Tanaka T. U. Bi-orienting chromosomes on the mitotic spindle. Curr. Opin. Cell Biol. 2002;14:365–371. doi: 10.1016/s0955-0674(02)00328-9. [DOI] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tanenbaum M. E., Galjart N., van Vugt M., Medema R. H. CLIP-170 facilitates the formation of kinetochore–microtubule attachments. EMBO J. 2005;25:45–57. doi: 10.1038/sj.emboj.7600916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj R., Li B., Yu H. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S., Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 2004a;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Tang Z., Sun Y., Harley S. E., Zou H., Yu H. Human Bub1 protects centromeric sister-chromatids cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. USA. 2004b;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., Ha E., McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Earnshaw W. C. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 2004;113:211–222. doi: 10.1007/s00412-004-0307-3. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K. G. Bub1 and the multilayered inhibition of Cdc20-APC/C in mitosis. Trends Cell Biol. 2005;15:231–233. doi: 10.1016/j.tcb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Vigneron S., Prieto S., Bernis C., Labbe J. C., Castro A., Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C. D., Brady D. M., Johnston R. C., Hanna J. S., Hardwick K. G., Spencer F. A. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell. 2002;13:3029–3041. doi: 10.1091/mbc.E02-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Chen R. H., Murray A. W., Salmon E. D. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Skibbens R. V., Salmon E. D. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J. Cell Sci. 1996;109:2823–2831. doi: 10.1242/jcs.109.12.2823. [DOI] [PubMed] [Google Scholar]
- Weiss E., Winey M. The Saccharomyces cerevisiae spindle pole body duplication genes MPS1 is part of a mitotic checkpoint. J. Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K. W., Sakowicz R., Goldstein L. S., Cleveland D. W. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Yang Z., Tulu U. S., Wadsworth P., Rieder C. L. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Anderson K. L., Cleveland D. W. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Yao J., Joshi H. C. Attachment and tension in the spindle assembly checkpoint. J. Cell Sci. 2002;115:3547–3555. doi: 10.1242/jcs.00029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.