Abstract
The transcriptional regulator CONSTANS (CO) promotes flowering of Arabidopsis under long summer days (LDs) but not under short winter days (SDs). Post-translational regulation of CO is crucial for this response by stabilizing the protein at the end of a LD, whereas promoting its degradation throughout the night under LD and SD. We show that mutations in CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), a component of a ubiquitin ligase, cause extreme early flowering under SDs, and that this is largely dependent on CO activity. Furthermore, transcription of the CO target gene FT is increased in cop1 mutants and decreased in plants overexpressing COP1 in phloem companion cells. COP1 and CO interact in vivo and in vitro through the C-terminal region of CO. COP1 promotes CO degradation mainly in the dark, so that in cop1 mutants CO protein but not CO mRNA abundance is dramatically increased during the night. However, in the morning CO degradation occurs independently of COP1 by a phytochrome B-dependent mechanism. Thus, COP1 contributes to day length perception by reducing the abundance of CO during the night and thereby delaying flowering under SDs.
Keywords: CONSTANS, flowering, photomorphogenesis, ubiquitin ligase
Introduction
Exposure to light influences many aspects of the plant life cycle, a process referred to as photomorphogenesis. Light promotes seed germination and seedling growth, thereby ensuring that young plants are exposed to an optimal environment for photosynthesis. Photomorphogenesis also has important functions in the development of adult plants (Neff et al, 2000). The mechanisms controlling adult photomorphogenic traits such as control of flowering in response to day length are less well understood than those that occur in the seedling. However, a genetic pathway that promotes flowering of Arabidopsis in response to long days (LDs) has been defined (Searle and Coupland, 2004; Imaizumi and Kay, 2006). Within this pathway, the transcriptional regulator CONSTANS (CO) has an important function by promoting flowering specifically under LDs. Here, we demonstrate that the ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), a major regulator of seedling photomorphogenesis (Deng et al, 1992), negatively regulates CO protein abundance in the vascular tissue of adult plants as part of the mechanism by which Arabidopsis discriminates between LD and SD during flowering-time control.
CO is a major regulator of photoperiodic flowering. Mutations in CO delay flowering specifically under LD, whereas its overexpression from a viral promoter causes extreme early flowering under LD and SD. CO contains two B-box-type zinc-finger motifs near its N terminus and a CCT (CONSTANS, CONSTANS-LIKE, TOC1) domain at its C terminus (Putterill et al, 1995). The latter domain is plant specific, but shows similarity to the DNA-binding domain of the HAP2 subunit of the CCAAT box-binding complex, suggesting that CO might bind to DNA directly (Wenkel et al, 2006). The closely related genes FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF) are highly and rapidly increased in expression in response to CO expression (Samach et al, 2000; Wigge et al, 2005; Yamaguchi et al, 2005). These genes encode RAF kinase inhibitor-like proteins that exert an effect as potent inducers of flowering (Kardailsky et al, 1999; Kobayashi et al, 1999). CO activates FT in the companion cells of the phloem within the vascular tissue, and FT protein is then proposed to move through the phloem sieve elements to the shoot apical meristem (An et al, 2004; Corbesier et al, 2007; Jaeger and Wigge, 2007; Mathieu et al, 2007), where it changes gene expression patterns and induces flowering (Abe et al, 2005; Wigge et al, 2005; Searle et al, 2006).
The mechanism by which CO activity is controlled by day length involves both transcriptional and post-translational regulation. CO transcription is regulated by the circadian clock so that its expression rises around 12 h after dawn and stays high until the following dawn (Suarez-Lopez et al, 2001). Exposure to light between 10 and 14 h after dawn further promotes CO transcription through the activity of the photoreceptor FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) and its interacting partner GIGANTEA (GI) (Suarez-Lopez et al, 2001; Imaizumi et al, 2003; Sawa et al, 2007). At the post-translational level, CO protein is stabilized when plants are exposed to light, whereas in darkness CO protein is rapidly degraded through ubiquitination and the activity of the proteasome. These mechanisms combine to ensure that a peak in CO protein abundance occurs under LDs when plants are exposed to light between 10 and 16 h after dawn, whereas under SD, when plants are exposed to darkness during this interval, CO protein does not accumulate (Valverde et al, 2004). The importance of ubiquitination and degradation of CO protein by the proteasome in these processes was demonstrated by use of proteasome inhibitors. These regulatory steps ensure that transcription of FT and TSF occurs under LDs but not under SDs.
The photoreceptors required for post-translational regulation of CO have been characterized. Mutations in the genes encoding the photoreceptors phytochrome A (phyA) and cryptochrome 2 (cry2) delay flowering, and these mutations also reduce the accumulation of CO protein (Valverde et al, 2004). Similarly, far-red light or blue light promotes flowering and stabilizes CO protein, and these regions of the spectrum activate phyA and cry2, respectively. In contrast, red light delays flowering and reduces the accumulation of CO protein. This response appears to be mainly controlled by phytochrome B (phyB), because phyB mutations cause early flowering and allow increased accumulation of CO protein.
COP1 is a major negative regulator of photomorphogenic responses, so that cop1 mutants undergo photomorphogenesis in darkness in the absence of photoreceptor activation (Deng et al, 1991). COP1 encodes a RING finger protein with a coiled-coil motif and WD40 repeats (Deng et al, 1992), and exerts an effect as a ubiquitin ligase that promotes the degradation of transcription factors implicated in seedling photomorphogenesis (Osterlund et al, 2000). In mammalian and plant cells, COP1 seems to exert an effect as part of a complex that also includes DEETIOLATED 1 (DET1), DAMAGED DNA-BINDING PROTEIN 1 (DDB1), cullin 4A and RING BOX 1 (RBX1) (Chory et al, 1989; Schroeder et al, 2002; Wertz et al, 2004; Hoecker, 2005; Chen et al, 2006). However, the SUPPRESSOR OF PHYTOCHROME A-105 1 (SPA) family of proteins is plant specific, related in sequence to COP1 and modulates the ubiquitin ligase activity of COP1. SPA proteins contain a coiled-coil domain and WD40 repeats related to those of COP1 as well as a kinase-like domain not present in COP1 (Hoecker et al, 1999). Quadruple mutants in which the four SPA genes are mutated exhibit a phenotype similar to that of cop1 mutants (Laubinger et al, 2004). Furthermore, SPA1 and COP1 physically interact and SPA1 modulates COP1 activity in vitro (Hoecker and Quail, 2001; Saijo et al, 2003; Seo et al, 2003).
Several protein targets for COP1 are transcription factors that regulate seedling photomorphogenesis (Osterlund et al, 2000; Seo et al, 2003; Duek et al, 2004; Jang et al, 2005; Yang et al, 2005). Each of these proteins was shown based on mutagenesis studies to have a function in the regulation of seedling growth in response to light. COP1 targets each of these proteins for degradation in the dark, but in the light COP1 activity is suppressed allowing these transcription factors to accumulate and promote seedling photomorphogenesis.
In addition to these roles in seedling development, COP1 also influences photomorphogenesis of adult plants. Although null mutant alleles of COP1 cause seedling lethality, plants homozygous for weaker cop1 alleles are viable. These plants are early flowering, particularly under SDs, indicating that COP1 is required for the suppression of flowering (McNellis et al, 1994). Furthermore cop1 mutants, but not wild-type (WT) plants, flower in darkness if provided with sugar (Nakagawa and Komeda, 2004). In addition, spa1 mutants flower early and SPA proteins modulate CO abundance so that in spa1 spa3 spa4 triple mutants 16 h after dawn under LDs increased levels of CO protein were detected (Ishikawa et al, 2006; Laubinger et al, 2006). Here, we analysed the role of COP1 in the light regulation of flowering time by genetic and molecular studies. We show that COP1 represses CO activity in the vascular tissue, and reduces CO protein levels particularly under SDs and in the dark, thereby facilitating a flowering response to day length.
Results
Genetic and spatial interactions between COP1 and CO in the regulation of flowering time
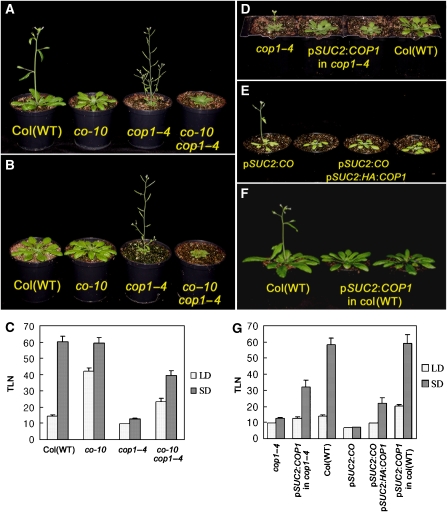
Previously cop1 mutants were shown to flower earlier than WT plants under short days (SDs) and at a similar time to WT plants under LDs (Mcnellis et al, 1994). Under our conditions, cop1–4 mutants flowered dramatically earlier than WT plants under SDs, as shown previously, but in addition flowered earlier than WT plants under LDs. The cop1–4 mutant produced around 53 leaves fewer than WT plants before flowering under SDs, whereas under LDs the difference between mutant and WT was around 5 leaves (Figure 1A–C; Supplementary Table 1). Therefore, the photoperiod response of cop1–4 mutants was severely reduced so that they flowered after forming only 3 leaves more under SDs than LDs, whereas WT plants formed around 45 leaves more under SDs.
Figure 1.
Genetic characterization of the interaction between CO and COP1. (A, B) cop1–4 mutants flowered earlier than wild-type Columbia plants irrespective of photoperiod, and the co-10 mutation suppresses the extreme effect of the cop1–4 mutation on flowering time under 16 h LD (A) and 8 h SD (B). (C) Flowering times in LD and SD of genotypes shown in (A, B). Flowering time is expressed as total leaf number (TLN) at flowering. (D) COP1 expression under the phloem-specific promoter SUC2 largely rescued the early-flowering cop1–4 mutant phenotype. The plants were grown under SD. (E) Simultaneous expression of CO and COP1 in the phloem tissue. SUC2:CO SUC2:HA:COP1 transgenic plants flowered later than SUC2:CO transgenic plants. (F) SUC2:COP1 caused late flowering of wild-type Columbia plants under LD. (G) Flowering times expressed as TLN at flowering under LD and SD of genotypes shown in (D–F).
In WT plants, CO promotes early flowering under LDs but not SDs. To test whether the early flowering of cop1–4 mutants under SDs was caused by activation of CO under these conditions, the cop1–4 co-10 double mutant was constructed and its flowering time was measured. The double mutant flowered after forming around 30 leaves more than cop1–4 mutants under SDs, demonstrating that CO has an important role in the early flowering of cop1–4 mutants under SDs (Figure 1B and C). Nevertheless, cop1–4 co-10 plants formed 20 leaves fewer than co-10 plants under these conditions, indicating that part of the early flowering of the cop1–4 mutant occurs independently of CO. Under LDs, cop1–4 co-10 plants also flowered at a time intermediate between co-10 and cop1–4 (Figure 1A and C). These genetic results suggest that COP1 exerts an effect as a negative regulator of CO under SDs, so that CO promotes flowering of cop1–4 mutants but not WT plants under SDs.
CO is expressed only in the vascular tissue and exerts an effect in the phloem companion cells to activate the transcription of the flowering-time gene FT (Takada and Goto, 2003; An et al, 2004). To test whether COP1 also regulates flowering from the phloem, COP1 or HA:COP1 was expressed from the SUC2 promoter, which is active specifically in the phloem companion cells (Imlau et al, 1999). The SUC2:COP1 transgene was introduced into WT Columbia plants and into cop1–4 mutants, whereas SUC2:HA:COP1 was introduced into SUC2:CO plants. SUC2:COP1 delayed flowering of cop1–4 mutants under LDs and SDs, and of WT plants under LDs (Figure 1D–G). Therefore, COP1 exerts an effect in the companion cells, where CO is expressed, to delay flowering. However, SUC2:COP1 cop1–4 plants still flower earlier than WT plants under SDs, suggesting that COP1 expression in companion cells is not sufficient to completely rescue the early-flowering phenotype of cop1–4 mutants, and therefore that COP1 probably also exerts an effect in additional cell types to delay flowering. The observation that SUC2:HA:COP1 delays flowering of SUC2:CO plants under LDs and SDs supports the idea that the delay of flowering associated with SUC2:COP1 is at least in part caused by reduction of CO activity (Figure 1G). Taken together, the flowering-time phenotypes of plants misexpressing COP1 in the phloem are consistent with the idea that COP1 exerts an effect in the phloem companion cells to repress the promotion of flowering by CO.
COP1 reduces FT mRNA levels
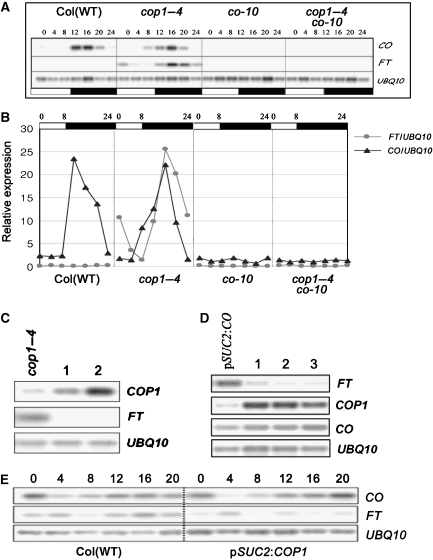
FT transcription is activated by CO and likely represents a direct target of CO protein (Samach et al, 2000; Wigge et al, 2005). Therefore, if COP1 exerts an effect to repress CO activity this should be reflected in reduced FT mRNA levels. In WT plants grown under SDs, CO mRNA is present during the night but FT mRNA is not expressed, because CO protein is rapidly degraded in the dark (Suarez-Lopez et al, 2001; Valverde et al, 2004). The effects of COP1 on CO transcription and CO activity were tested by analysing CO and FT mRNA levels at 4-h intervals for 24 h in SD-grown plants of different genotypes (Figure 2A and B). In WT Columbia, CO mRNA was detected during the night under SDs, but was absent in co-10 and cop1–4 co-10 plants as expected due to the T-DNA insertion present in the CO gene in the co-10 allele (Materials and methods). In cop1–4 mutants, the CO mRNA pattern is similar to that observed in WT plants but rises earlier, appearing weakly 8 h after dawn, whereas in WT plants CO mRNA was first detected 12 h after dawn. In contrast, FT mRNA was detected in cop1–4 mutants but not WT plants, consistent with the early flowering of these mutants under SDs. Similarly, under LDs, FT mRNA levels were much higher in cop1–4 plants than in WT plants consistent with the earlier flowering of the mutants under these conditions (Supplementary Figure 1). However, under LDs, CO mRNA was consistently detected at lower levels in cop1–4 mutants than in WT plants (Supplementary Figure 1). The expression of FT mRNA in cop1–4 mutants requires CO activity, as demonstrated by the absence of FT mRNA in cop1–4 co-10 plants (Figure 2A). These results are consistent with the idea that COP1 delays flowering of WT plants under SDs, and to a lesser extent under LDs, by repressing CO activity and thereby preventing FT expression.
Figure 2.
Effect of COP1 on CO and FT mRNA levels. (A) CO and FT mRNA analysis in wild-type (WT) Columbia, cop1–4 mutants, co-10 mutants and cop1–4 co-10 double-mutant plants under 8 h SDs. (B) Quantification of the mRNA levels shown in (A). Expressed as a ratio between UBQ10 mRNA level and FT or CO mRNA level. (C) COP1 and FT mRNAs in cop1–4 mutants and two SUC2:COP1 cop1–4 transformants. All plants were grown under SD and harvested 16 h after dawn. (D) FT, COP1 and CO mRNAs in SUC2:CO and three SUC2:CO SUC2:HA:COP1 transformants. All plants were grown under SD and harvested 8 h after dawn. (E) CO and FT mRNAs in WT Columbia plants and in a SUC2:COP1 Columbia transformant. All plants were grown under LD and harvested at 4-h intervals. All genotypes are in the accession Columbia, and in (C, D) the numbers represent independent transgenic plants. In (A, E) 2-week-old seedlings were sampled, whereas in (C, D) rosette leaves of 3-week-old plants were harvested.
The abundance of FT mRNA was also tested in transgenic plants expressing COP1 or HA:COP1 mRNAs at high levels in the phloem companion cells from the SUC2 promoter (Figure 2C–E). CO and FT mRNA levels were compared through a LD time course in SUC2:COP1 and WT plants. CO mRNA levels were very similar in both genotypes at all times, whereas FT mRNA levels were severely reduced in SUC2:COP1 plants (Figure 2E), consistent with the overexpression of COP1 in phloem companion cells leading to a reduction in CO activity at the post-transcriptional level. Similarly, 16 h after dawn under SDs, when FT mRNA reaches peak levels in cop1–4 mutants (Figure 2A), SUC2:COP1 cop1–4 plants displayed severely reduced levels of FT mRNA (Figure 2C). Finally, in SUC2:HA:COP1 SUC2:CO plants the level of FT mRNA was lower than in the SUC2:CO progenitor plants, but the level of CO mRNA was unaffected (Figure 2D). Therefore, analysis of FT mRNA in these transgenic plants supports the conclusion that COP1 delays flowering by repressing at the post-transcriptional level the capacity of CO to promote FT transcription in the phloem companion cells.
COP1 and CO physically interact in vitro and in vivo
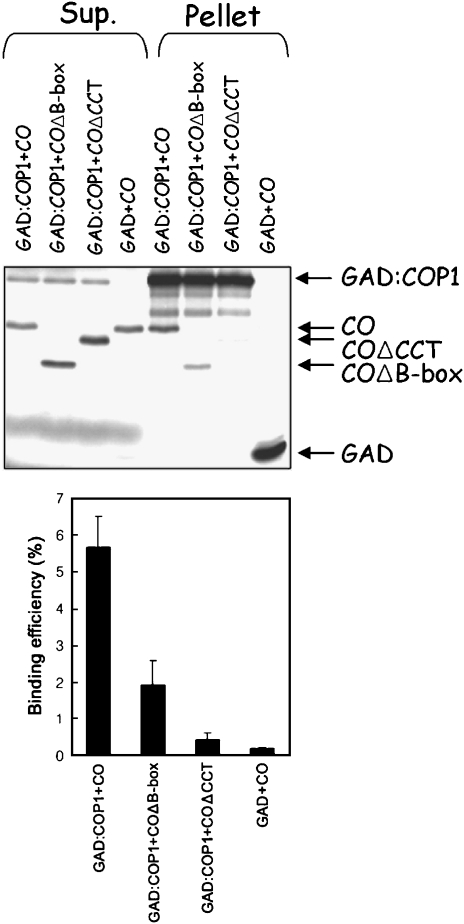
The observation that the ubiquitin ligase COP1 represses CO-mediated activation of FT suggested that CO might be a substrate for COP1. To test this hypothesis, we first investigated whether COP1 was able to physically interact with CO. In the yeast two-hybrid system, we detected no interaction between CO and COP1, although an interaction between COP1 and the CO-related protein CO-LIKE3 (COL3) was previously detected by this method (Datta et al, 2006), and we were able to confirm this interaction. Therefore, whether COP1 and CO interact in vitro was tested using a co-immunoprecipitation assay (Figure 3). COP1 attached to the GAL4 activation domain (GAD:COP1) and CO were made in an in vitro transcription/translation system and combined. GAD:COP1 was precipitated with anti-GAD antibody and CO was co-precipitated with GAD:COP1 (Figure 3). In contrast, CO was not co-immunoprecipitated with GAD alone. These experiments suggest that CO interacts with COP1 in the GAD:COP1 fusion protein. Fragments of CO were also combined with GAD:COP1 to determine which regions of CO are required for the interaction with COP1. Two segments of CO were tested: one containing the region between amino acids 107 and 373, which was called COΔB-box because it did not contain the zinc-finger B-boxes found at the N terminus of CO, and a second containing the region between amino acids 1 and 331, which was named COΔCCT, because the CCT domain near the C terminus of CO was removed. In vitro precipitation experiments demonstrated that COΔB-box was co-immunoprecipitated with GAD:COP1, whereas COΔCCT was not. Therefore, the N-terminal region containing the B-boxes is not required for interaction with COP1, suggesting that the interaction with COP1 is mediated by the C-terminal region of CO that contains the CCT domain.
Figure 3.
In vitro interaction between CO and COP1 detected by co-immunoprecipitation. 35S-methionine-labeled CO, COΔB-box or COΔCCT was incubated with 35S-methionine-labeled GAD:COP1 or GAD and co-immunoprecipitated with anti-GAD antibodies. Supernatant fractions and pellet fractions were resolved by SDS–PAGE and visualized by autoradiography using a phosphorimager. Quantification of the fractions of prey proteins that were co-immunoprecipitated by the indicated bait proteins GAD:COP1 or GAD. Error bars denote the standard error of the mean of two replicate experiments.
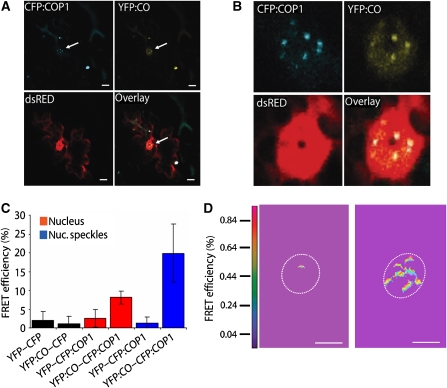
Whether the interaction between CO and COP1 also occurred in vivo in plant cells was tested using fluorescent resonance energy transfer (FRET). Microprojectile bombardment was used to co-express cyan fluorescent protein (CFP):COP1 and yellow fluorescent protein (YFP):CO in leaf epidermal cells of Arabidopsis. CFP:COP1 and YFP:CO colocalized to the nucleus and also colocalized in speckles within the nucleus (Figure 4A and B). Physical interaction of CFP:COP1 and YFP:CO was tested by measuring FRET using photoacceptor bleaching, as previously described (Wenkel et al, 2006) (Figure 4C and D). Quantification of FRET signals demonstrated that FRET occurred between YFP:CO and CFP:COP1 both in the nucleus and specifically in nuclear speckles (Figure 4C and D). In control experiments using YFP and CFP, YFP:CO and CFP or YFP and CFP:COP1 FRET was detected at significantly lower levels (Figure 4C). These experiments demonstrate that YFP:CO and CFP:COP1 colocalize and physically interact in the nuclei of plant cells.
Figure 4.
CO protein physically interacts with COP1 in plant cells. (A) Transient co-expression of 35S:YFP:CO and 35S:CFP:COP1 constructs. A 35S:dsRED construct was cotransformed to highlight the transformed cell. The arrows represent the nucleus in which CO and COP1 are colocalized. (B) Enlargement of the nucleus shown in each of the panels represented in (A). (C) Quantification of FRET in vivo between CFP:CO and YFP:COP1. YFP:CO detected as an increase in CFP fluorescence after photobleaching of YFP. Quantification of FRET efficiencies after acceptor photobleaching measured in nuclei and nuclear speckles. Data are mean±s.d. of 10–20 cells from three separate experiments. (D) Visualization of increase in CFP fluorescence after YFP photobleaching. Left-hand panel, cells expressing CFP:COP1 and YFP, which exerts an effect as a negative control. Right-hand panel, cells expressing CFP:COP1 and YFP:CO. Scale bar: 6 μm in (A) and 8 μm in (D).
COP1 and phyB have complementary roles in repressing CO protein levels under LDs and SDs
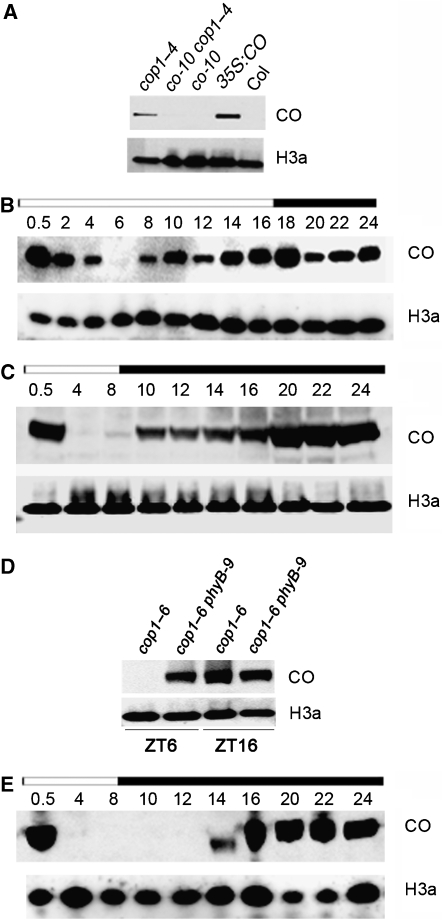
The genetic and molecular experiments described earlier supported the hypothesis that COP1 negatively regulates CO activity at the post-transcriptional level. Therefore, we tested the effect of COP1 on CO protein levels. First, CO protein abundance was examined in nuclei of WT Columbia, co-10, transgenic 35S:CO and cop1–4 plants harvested 16 h after dawn under LDs, when CO protein is expected to be at highest abundance (Valverde et al, 2004) (Figure 5A). As shown previously, CO was clearly detectable in 35S:CO transgenic plants that overexpress the protein, but was below the level of detection in nuclei of WT plants. However, in cop1–4 mutants, CO was clearly detected, suggesting that in WT plants COP1 has a major function in reducing CO protein levels at this time. Strong support that the protein detected in cop1–4 mutants was indeed CO protein came from the analysis of cop1–4 co-10 double mutants, in which the protein detected in cop1–4 mutants was no longer present (Figure 5A).
Figure 5.
Detection of CO protein in cop1–4, cop1–6 phyB-9 and spa1–7 plants. (A) CO protein was detected in 35S:CO transgenic plants and cop1–4 mutants, but not in WT Columbia, co-10 or cop1–4 co-10 mutants. Plants were grown under 16 h LDs and harvested 16 h after dawn. (B, C) CO protein in cop1–4 mutants under 16 h LD or 8 h SD. Numbers above each lane represent hours after dawn that the sample was harvested. Light bar represents day; dark bar represents night. (D) CO protein detection in cop1–6 and cop1–6 phyB-9 plants grown under SDs. Samples were harvested 6 and 16 h after dawn. The reduction in CO protein at 6 h in cop1–6 plants (see also (C)) does not occur in cop1–6 phyB-9 plants. (E) CO protein detection in spa1–7 mutants under 8 h SD. Numbers and bars as described for (B, C). In WT plants, CO protein could not be detected and therefore is not included as control ((A); Valverde et al, 2004). For all panels, histone 3a was used as a loading control.
CO mRNA shows a diurnal rhythm in abundance in WT plants and in cop1–4 mutants, therefore the diurnal pattern of CO protein abundance was tested under LDs and SDs in cop1–4 mutants (Figure 5B and C). Under SDs of 8 h light, cop1–4 mutants flower dramatically earlier than WT plants (Figure 1) and CO protein was present for most but not all of the diurnal cycle (Figure 5C). CO was strongly detected soon after dawn, was absent or present at much lower abundance 4 and 8 h after dawn, and then was present strongly for the remainder of the night from 10 to 24 h after dawn. The appearance of CO protein from 10 h after dawn is likely due to an increase in CO mRNA levels, as the abundance of CO mRNA increased steeply between 4 and 14 h after dawn in the same plants used for the protein analysis (Supplementary Figure 2). In contrast, CO mRNA abundance fell between 14 and 24 h after dawn, whereas CO protein levels were high throughout this time. This comparison suggests that impairing COP1 function causes CO protein to be relatively stable in the dark. However, the steep decline in CO protein abundance between 0.5 and 4 h after dawn suggests that a second post-translational mechanism, independent of COP1, might negatively regulate CO protein levels in the morning.
Under LDs of 16-h photoperiods, CO protein was detected from dawn until 4 h into the photoperiod, was undetectable 6 h after dawn and then was present for the remainder of the photoperiod and throughout the night (Figure 5B). This pattern was similar to that detected under SDs, but the protein was detectable for longer and was only absent at one time point, 6 h after dawn. The broader peak in CO protein under LDs is likely due to CO mRNA being expressed for longer under LDs, as previously described (Suarez-Lopez et al, 2001; Imaizumi et al, 2003).
The photoreceptor phyB was previously shown to promote the degradation of CO protein early in the day in 35S:CO plants, and this was proposed to contribute to the inhibitory effect of phyB on flowering time (Valverde et al, 2004). To test whether phyB is responsible for the reduction in CO protein levels early in the day in cop1 mutants, the phyB-9 cop1–6 double mutant was tested for flowering time and CO protein levels. Under SDs, phyB-9 cop1–6 plants flowered at a very similar time to cop1–6 mutants, demonstrating that the early flowering of cop1–6 mutants is not enhanced by loss of function of phyB (Supplementary Figure 3). Under LDs, the double mutant flowered significantly later than either single mutant, which indicates a complexity in the interaction between COP1 and phyB under these conditions that cannot be simply explained by regulation of CO protein levels (see Discussion). To test whether phyB is responsible for the reduction in CO protein early in the day in cop1 mutants, protein was extracted from phyB-9 cop1–6 and cop1–6 plants 6 and 16 h after dawn under SDs. In the cop1–6 plants, CO protein was undetectable 6 h after dawn, as observed for cop1–4 mutants, but in phyB-9 cop1–6 plants CO protein accumulated strongly 6 h after dawn (Figure 5D). In contrast, CO mRNA was present at similar levels in cop1–6 and phyB-9 cop1–6 plants 6 h after dawn (Supplementary Figure 3). These results indicate that phyB is required for post-transcriptional regulation of CO expression early in the day and independently of COP1. However, in the samples harvested 16 h after dawn CO protein was present at similar levels in cop1–6 and phyB-9 cop1–6 plants, indicating that phyB does not influence CO protein levels at that time of day (Supplementary Figure 3).
SPA1 interacts with COP1 and is implicated in the degradation of some COP1 substrates. Recently, CO protein was shown to be more abundant in spa1 spa3 spa4 triple mutants 16 h after dawn under LDs (Laubinger et al, 2006). The diurnal pattern of CO protein abundance in spa1–7 mutant plants was tested to compare with that described for cop1–4 mutants (Figure 5C and E). A similar pattern of CO protein accumulation was observed in spa1–7 and cop1–4 mutants between 14 h after dawn and the following morning, but the rise in the abundance of the protein was delayed in the spa1–7 mutant, so that it could not be detected until 14 h after dawn. In contrast, in the cop1–4 mutant CO protein was strongly detected 10 h after dawn. These results suggest a functional relationship between COP1 and SPA1 proteins in the degradation of CO, and that of the four SPA proteins SPA1 has the major role in regulating CO levels. The delayed increase in CO abundance in spa1–7 compared with cop1–4 mutants might be due to the activity of other SPA proteins.
Degradation of CO protein in red light is not impaired by the cop1–4 mutation
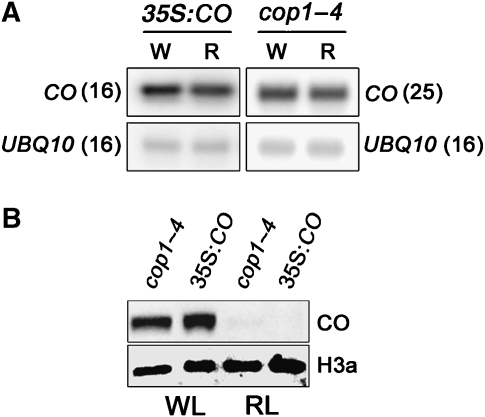
Arabidopsis plants flower later under red light and previously this was proposed to be at least partly due to degradation of CO protein under these conditions (Valverde et al, 2004). To test whether red light-mediated degradation of CO protein is also impaired in the cop1–4 mutants, CO protein abundance was compared in 35S:CO and cop1–4 plants grown under white and red light (Figure 6). Similar levels of CO protein were detected in both lines grown under 16 h of white light (Figure 6). Furthermore, when both genotypes were exposed to 16 h of red light, CO protein abundance fell sharply in both 35S:CO plants and cop1–4 mutants (Figure 6). The reduced levels of CO protein observed in cop1–4 mutants under red light compared with white light are not due to lower levels of CO mRNA, which were identical under both conditions (Figure 6). This result demonstrates that a COP1-independent mechanism is required for CO protein degradation under red light.
Figure 6.
Comparison of CO protein and mRNA in plants exposed to white or red light. 35S:CO or cop1–4 seedlings (12-day old) grown in LD were divided into two groups and exposed to 16 h red or white light, respectively. Samples were harvested for RNA and protein analysis at the end of the 16 h light period under both conditions. (A) CO and ubiquitin mRNA levels in 35S:CO or cop1–4 plants exposed to white (W) or red (R) light. Numbers in parentheses represent the numbers of cycles used to amplify the cDNA prior to separation on a gel. (B) CO and histone protein levels in the same plants used for (A). In both genotypes, CO is detected in white light (WL)-grown plants but not in red light (RL)-grown plants.
Discussion
We demonstrated that COP1 ubiquitin ligase is required to shape the diurnal pattern of CO protein accumulation as part of the flowering response of Arabidopsis to photoperiod. COP1 delays flowering of WT plants under SDs by preventing CO protein accumulation during the night and thereby ensuring that FT transcriptional activation does not occur. Under LDs WT plants flower early, but even under these conditions COP1 modulates CO protein levels, lowering the abundance of the protein towards the end of the day and during the night. These effects on CO are consistent with the extreme early flowering of cop1–4 mutants under LDs and SDs, the largely day length-insensitive phenotype of cop1–4 mutants and suppression of these phenotypes to a large extent by co mutations. However, in cop1–4 mutants CO protein abundance is still reduced in the early morning and in red light, indicating that a second mechanism independent of COP1 regulates CO protein abundance under these conditions. The mechanism that exerts an effect in the early morning is shown to depend on the phyB photoreceptor. Our observations place COP1 within the regulatory network for photoperiod perception and regulation of flowering in Arabidopsis (Figure 7), and extend the characterized functions of COP1 beyond those previously described in seedling photomorphogenesis.
Figure 7.
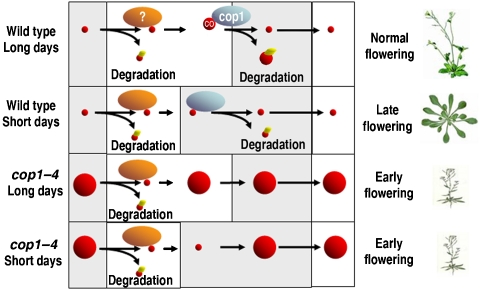
Model for regulation of CO stability during photoperiodic flowering control in wild-type plants and cop1–4 mutants. Photoperiodic flowering in Arabidopsis involves two mechanisms of CO protein degradation: a phyB-dependent mechanism occurs early in the day or in response to red light and a second mechanism involving COP1 occurs late in the day and during the night. (top) In wild-type plants under LDs, CO accumulates in the evening due to an increase in CO mRNA and photoreceptor-mediated repression of COP1. CO can promote FT expression at this time and thereby flowering. During the night, COP1 is active and causes rapid degradation of CO protein by ubiquitination and activity of the proteasome. (second from top) Under SD, CO mRNA is expressed during the night and the protein is degraded through COP1 activity. CO protein does not accumulate and FT mRNA is absent, resulting in late flowering. (second from bottom) In cop1–4 mutants under LDs, CO is not degraded in the dark and accumulates to high levels. CO also accumulates to high levels at the end of the day, consistent with COP1 targeting CO for degradation at that time. However, CO protein still disappears early in the day, suggesting a COP1-independent mechanism of degradation at that time. (bottom) Under SDs in cop1–4 mutants, CO accumulates to a high level during the night and promotes FT expression at higher level than in wild-type plants. Enhanced CO activity at these times is responsible for the early flowering of cop1–4 mutants under SDs. In the morning, CO protein is degraded by a COP1-independent mechanism. The symbols represent CO protein abundance (red circles), COP1 (blue spheres), ubiquitin (small yellow circles on CO) and an unknown red light-activated degradation mechanism that is also active in the morning (dark orange spheres). A full-colour version of this figure is available at The EMBO Journal Online.
COP1 reduces CO protein abundance to confer a photoperiodic flowering response
COP1 represses seedling photomorphogenesis by catalysing the ubiquitination and therefore degradation of proteins that promote seedling photomorphogenesis (Osterlund et al, 2000; Seo et al, 2003; Duek et al, 2004; Jang et al, 2005; Yang et al, 2005). In addition to their effects on seedling development, cop1 mutations severely impair the development of adult plants, although the mechanisms by which this occurs are less well understood. Altered adult traits include photoperiodic flowering so that cop1 mutants flower at similar times under LDs and SDs (Mcnellis et al, 1994). In addition, cop1 mutants flower in constant darkness in the presence of sucrose, whereas WT seedlings do not (Nakagawa and Komeda, 2004). Under these conditions, cop1 mutants exhibit higher expression of FT and SOC1 mRNAs than WT plants. We showed that the early flowering of cop1–4 mutants under LDs or SDs largely depends on CO function and that in these mutants CO protein persists in the dark under SDs and LDs. These results suggest that post-translational regulation of CO is impaired in cop1–4 mutants.
Under LDs, CO protein levels are high in cop1–4 mutants throughout most of the day and rise earlier after dawn than under SDs, as shown for CO mRNA (Suarez-Lopez et al, 2001; Imaizumi et al, 2003). This effect on CO mRNA levels is at least in part due to FKF1- and GI-mediated degradation of the transcriptional repressor CYCLING DOF FACTOR 1 (CDF1), which is triggered by light, allowing CO mRNA abundance to rise earlier during the day under LDs (Imaizumi et al, 2005; Sawa et al, 2007). Perhaps surprisingly, our results indicate that under LDs, when CO promotes flowering of WT plants, COP1 also has a strong negative effect on CO protein levels. At the end of the day and during the night, CO protein accumulates to much higher levels in cop1–4 mutants than WT plants, although CO mRNA abundance is actually lower than in WT plants. These results indicate that under LDs COP1 has an unexplained role in increasing CO mRNA abundance, and a major function in lowering CO activity by reducing CO protein abundance.
Under SDs, CO mRNA is expressed and rises during the night. Our demonstration of COP1-mediated degradation of CO protein in the dark under SDs provides a molecular explanation for why CO does not promote flowering under these conditions, and is consistent with previous demonstrations that application of proteasome inhibitors led to stabilization of the CO protein. The importance of this process in conferring a photoperiodic response is illustrated by the extreme early flowering of cop1–4 mutants under SDs, which is responsible for almost abolishing the response to photoperiod. COP1 therefore have an important function in turning over CO protein in the light and dark under these conditions.
COP1 activity is higher in the dark than light. One mechanism by which this light regulation occurs is through exclusion of COP1 from the nucleus in the light (Von Arnim and Deng, 1994), whereas in addition COP1 activity is repressed by direct interaction with activated cryptochromes (Wang et al, 2001; Yang et al, 2001). Our observation that CO protein levels are very high in the dark under LD or SD in cop1–4 mutants is consistent with COP1 activity being high in the dark under both day lengths and rapidly turning over CO protein. The increase in CO protein at the end of the day under LD indicates that even in the light COP1 contributes to keeping CO protein levels low. COP1 activity might only be reduced and not fully suppressed at the light intensities used in these experiments. If so, then higher light intensities might promote flowering at least partly by more effectively repressing COP1 activity and allowing CO protein levels to rise higher. This would suggest a role for COP1 and CO in the regulation of flowering by light intensity.
Degradation of CO protein in the morning or in red light does not require COP1
Previously, two distinct post-transcriptional mechanisms were postulated to shape the diurnal pattern of CO protein accumulation. One of these occurred early in the day and involved a phyB-mediated signal and another occurred in the dark during the night (Valverde et al, 2004). Degradation of CO protein in red light was proposed to involve the same phyB pathway that caused rapid turnover of the protein early in the day. We observed that in cop1–4 mutants the CO protein was still effectively degraded in red light and that there was a strong diurnal trough in CO protein levels early in the day. Degradation of CO early in the day was shown to require phyB but not COP1. The degradation of CO in red light likely occurs by the same phyB-dependent mechanism acting early in the day, as was shown by Valverde et al (2004), and this could be tested by comparing cop1–6 phyB-9 and cop1–6 plants under red light. Also, we cannot exclude that other phytochromes related to phyB may also have a function in CO regulation. In particular, phyC and phyE were demonstrated to influence flowering time (Halliday and Whitelam, 2003; Monte et al, 2003; Balasubramanian et al, 2006). Nevertheless, our data suggest that a second ubiquitin ligase may be responsible for phyB-mediated turnover of CO early in the day and in continuous red light (Figure 7). Interestingly, the bHLH transcription factor PHYTOCHROME INTERACTING FACTOR 3, which is involved in seedling photomorphogenesis and phytochrome signalling, was also recently shown to be degraded by a red light-activated ubiquitin-mediated process soon after dawn (Al-Sady et al, 2006). There might be a common mechanism promoting the degradation at dawn of several transcription factors involved in light signalling. Alternatively, a set of ubiquitin ligases might exist that specifically promote the degradation of individual transcription factors at this time. Further genetic and biochemical approaches are required to understand the mechanisms underlying CO protein degradation at dawn.
Spatial regulation of photoperiodic response by COP1
CO and FT are expressed in the vascular tissue and their expression in the phloem companion cells is sufficient to promote flowering (Takada and Goto, 2003; An et al, 2004). Furthermore, reducing FT expression specifically in the phloem companion cells delays flowering (Mathieu et al, 2007). Thus, the perception of photoperiod that is mediated through transcriptional and post-transcriptional regulation of CO likely takes place in the companion cells. Similarly, Cry2, which positively regulates CO accumulation, exerts an effect in the companion cells to regulate flowering (Endo et al, 2007). In contrast, phyB, a photoreceptor that delays flowering at least in part by reducing CO abundance, appears to exert an effect non-cell autonomously from the mesophyll cells (Endo et al, 2005). This result suggests that a signalling step downstream of phyB exerts an effect non-cell autonomously to trigger degradation of CO protein, although definitive conclusions on the site of action of phytochromes influencing flowering will require a better understanding of the spatial requirement for other phytochromes, such as phyC and phyE. We showed that expression of COP1 in the vascular tissue from the SUC2 promoter complemented the flowering-time phenotype of cop1–4 mutants under LDs and reduced FT mRNA levels. Under SDs, cop1–4 SUC2:COP1 plants still flowered earlier than WT, but this was probably due to a CO-independent process causing early flowering in the cop1–4 mutant, because cop1–4 co-10 plants also flowered earlier than WT under SDs. In WT plants, SUC2:COP1 also delayed flowering under LDs but not under SDs. The day length specificity of this effect suggests that the overexpression in companion cells affects flowering through CO, and indicates that COP1 levels are a limiting factor on CO degradation under these conditions. Taken together, these results suggest that COP1 exerts an effect in companion cells to regulate FT expression. This observation is consistent with our suggestion that COP1 exerts an effect to degrade CO at the end of the day and during the night, but not as part of the phyB pathway, which exerts an effect mainly in the morning or in red light. The temporal patterns of COP1 activity, therefore, support our understanding of the spatial pattern of activity of the pathways responsible for post-translational regulation of CO (Figure 7).
COP1 and CO interact in vitro and in nuclear speckles in vivo
COP1 directly interacts with target proteins and directs them for degradation (Hoecker, 2005; Jiao et al, 2007). CO is composed of three domains, zinc-finger B-boxes, a central domain and the C-terminal CCT domain (Wenkel et al, 2006). CO and COP1 interact directly in vitro as demonstrated by immunoprecipitation experiments. This interaction was almost abolished when the C-terminal part of CO was removed, suggesting that COP1 interacts with the C-terminal region of CO, as was previously observed for interactions between COP1 and COL3 or between CO and SPA1 (Datta et al, 2006; Laubinger et al, 2006). The interaction between COP1 and HY5 occurs through a defined domain that includes adjacent valine and proline residues that are essential for the interaction (Holm et al, 2001). Conserved pairs of valine-proline residues in the CCT domain of COL3 are also important for the interaction with COP1 (Datta et al, 2006). The region of CO that interacts with COP1 contains three VP motifs, but changing all of these to AA did not impair the interaction with COP1 in vitro (data not shown). A similar result was previously observed for the interaction between CO and SPA1 (Laubinger et al, 2006). Therefore, the interaction between CO and COP1 likely involves a different motif than observed for the interactions between COP1 and COL3 or HY5.
Direct interaction between COP1 and CO was further supported by transient expression of COP1 and CO fused to fluorescent proteins in Arabidopsis leaf cells. The proteins colocalized in the nucleus and both occurred in speckles. Previously, COP1 was shown to colocalize with its target proteins HY5, HYH and LAF1 in nuclear speckles in onion epidermal cells (Osterlund et al, 2000; Holm et al, 2002; Seo et al, 2003). COP1 speckles were proposed to represent nuclear sites for proteasome-mediated protein degradation (Al-Sady et al, 2006). The presence of CO and COP1 in nuclear speckles similar to those observed for other targets of COP1-mediated degradation supports the idea that direct interaction between COP1 and CO is required for CO degradation in the nucleus. This degradation presumably requires the SPA proteins, perhaps acting directly in a larger order complex with COP1, as SPA proteins also regulate CO abundance at least at the end of a LD and interact directly with COP1 (Laubinger et al, 2006). The mechanism by which CO is degraded by the SPA–COP1 complex has therefore strong parallels with that of HY5. However, the precise relationship between SPA1 and COP1 activity and whether the proteins exert an effect in a larger order complex that interacts directly with substrates is still not clear (Saijo et al, 2003; Seo et al, 2003; Hoecker, 2005).
COP1 and the external coincidence model controlling flowering of Arabidopsis in response to photoperiod
CO promotes flowering and FT transcription under LDs but not SDs. CO activity is proposed to be restricted to LDs by an external coincidence model in which circadian clock control and light signalling combine to trigger CO activity (Searle and Coupland, 2004; Imaizumi and Kay, 2006). The cop1–4 mutant causes CO mRNA to accumulate earlier under SDs, so that cop1–4 mutations may in part accelerate flowering under SDs by causing CO mRNA to be expressed in the light, as previously shown for toc1-1 mutants (Yanovsky and Kay, 2002). However, the major time of expression of FT under SDs in cop1–4 is later during the night, suggesting that the earlier phase of CO expression in cop1–4 has a small part in the acceleration of flowering under these conditions. Rather the major role of COP1 in this system appears to be to degrade CO protein in the dark, and thereby ensure that no FT transcription occurs under SDs (Figure 7). The importance of this activity is demonstrated by the high abundance of CO protein in the cop1–4 mutant under SDs and the extreme early flowering of cop1–4 mutants under these conditions. During the final revision of this paper, another study described the role of cryptochrome signalling in suppressing COP1-mediated degradation of CO in the dark (Liu et al, 2008). Our data extend the model of photoperiodic flowering in Arabidopsis by providing a molecular explanation for why CO mRNA expression during the night in SDs does not lead to FT transcription and promotion of flowering. The day length-insensitive early-flowering phenotype of cop1–4 mutants and the strong suppression of this phenotype caused by co null alleles demonstrate that degradation of CO under SDs is essential in conferring a photoperiodic flowering response.
Materials and methods
Plant material
WT Arabidopsis thaliana plants and all mutants used in this study were Col-0. The cop1–4 allele was previously characterized (McNellis et al, 1994). The co-10 allele was previously used (Laubinger et al, 2006) and was confirmed to have a T-DNA insertion 342 bp after the ATG. Homozygous cop1–4 co mutant plants were found using PCR-based markers. The cop1–6 phyB-9 and cop1–6 seeds were kindly provided by Dr Jorge Casal (Boccalandro et al, 2004).
Analysis of flowering time
For analysis of flowering time and gene expression, plants were grown on soil in controlled environment rooms under LDs (16 h light–8 h dark) or SDs (8 h light–16 h dark). Flowering time was measured by scoring the number of rosette and cauline leaves on the main stem of at least eight individuals. Data are expressed as average±s.d.
mRNA expression analysis
Arabidopsis RNA was isolated with the Plant RNeasy kit (Qiagen) according to the manufacturer's instructions. RNA was analysed by RT–PCR. Detailed protocols and the origins of the primer sequences are presented in Supplementary data.
Plant transformation
The COP1 full-length cDNA was isolated by RT–PCR and produced as entry clone through BP reaction of Gateway system from Invitrogen. Then, the entry clone was utilized for the construction of destination vectors for plant transformation, FRET experiments and in vitro-binding assay. All plasmids for plant transformation were introduced into Agrobacterium strain GV3101 (pMP90RK) and transformed into WT Columbia, cop1–4 or SUC2:CO (An et al, 2004) plants by the floral dip method (Clough and Bent, 1998).
In vitro-binding assay
For the in vitro expression of GAD:COP1, we produced the vector pJIC39 containing pT7:GAD:GATEWAY cassette and T7 terminator, so that by LR reaction with the COP1 entry clone, the construct expressing GAD∷COP1 is produced. Vector (Wenkel et al, 2006; Turck et al, 2007) pJIC26 is similar but contains only the GAD domain and was used for expressing full open reading frame of CO or parts of the ORF. COΔB-box and COΔCCT (Laubinger et al, 2006) were also tested for the binding with COP1. The detailed method used for the in vitro precipitation experiments is presented in Supplementary data.
Confocal microscopy, CO:COP1 colocalization and FRET analysis
To express CFP:COP1 and YFP:CO in plants, the CO and COP1 genes were cloned into the GATEWAY vectors pENSG:CFP or pENSG:YFP by recombination reaction. In these vectors, CFP:COP1 and YFP:CO are expressed under the control of the constitutive 35S promoter (Laubinger et al, 2006). The method used to analyse FRET is described in detail in Supplementary data.
Immunological techniques
WT Columbia, cop1–4 and spa1–7 were grown in temperature-controlled light cabinets either under LDs (16 h light and 8 h dark) or SDs (8 h light and 16 h dark). Plants were grown on solid germination medium for 2 weeks, harvested at specified zeitgeber time (ZT), frozen in liquid nitrogen and kept at −80°C until further use. For the red light experiments, 35S:CO and cop1–4 plants were grown in LD (16 h light–8 h dark) for 12 days, moved to red light conditions at ZT 0 and maintained for 16 h under red light. Nuclear extracts were prepared from the plants at different ZT times as described previously (Valverde et al, 2004). Nuclear proteins (17 μg) were separated employing 10% bis-Tris NuPAGE gels (Invitrogen), transferred to nitrocellulose membranes and probed with an anti-CO antibody followed by a horseradish peroxidase-conjugated secondary antibody. Immunoreactive proteins were visualized by Pico chemiluminescence substrate system (Pierce). The membrane was subsequently reprobed with an antibody against histone H3a (Abcam) as a loading control.
Supplementary Material
Supplementary Text
Acknowledgments
We thank Ute Höcker, Andreas Bachmair and Alon Samach for valuable discussions and Ute Höcker for the spa1–7 seeds. We are grateful to Jorge Casal for the cop1–6 phyB-9 seeds. SJ received a KOSEF fellowship and VM was funded within the EC Training Grant ADOPT. FV is a researcher of the ‘Ramon y Cajal' Programme of the Spanish Ministry of Education and Science. This study was partially funded by a GIF grant to GC The laboratory of GC is funded by a core grant from the Max Planck Society.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Ni WM, Kircher S, Schafer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccalandro HE, Rossi MC, Saijo Y, Deng XW, Casal JJ (2004) Promotion of photomorphogenesis by COP1. Plant Mol Biol 56: 905–915 [DOI] [PubMed] [Google Scholar]
- Chen HD, Shen YP, Tang XB, Yu L, Wang J, Guo L, Zhang Y, Zhang HY, Feng SH, Strickland E, Zheng N, Deng XW (2006) Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GHCM, Deng XW, Holm M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH (1991) cop1—a regulatory locus involved in light-controlled development and gene-expression in Arabidopsis. Genes Dev 5: 1172–1182 [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G-beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Endo M, Mochizuki N, Suzuki T, Nagatani A (2007) CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17: 1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Whitelam GC (2003) Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol 131: 1913–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U (2005) Regulated proteolysis in light signaling. Curr Opin Plant Biol 8: 469–476 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Quail PH (2001) The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276: 38173–38178 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH (1999) SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499 [DOI] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20: 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Kiba T, Chua NH (2006) The Arabidopsis SPA1 gene is required for circadian clock function and photoperiodic flowering. Plant J 46: 736–746 [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YL, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 [DOI] [PubMed] [Google Scholar]
- Liu L-J, Zhang Y-C, Li Q-H, Sang Y, Mao J, Lian H-L, Wang L, Yang H-Q (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Kuttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mcnellis TW, Von Arnim AG, Araki T, Komeda Y, Misera S, Deng XW (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Alonso JM, Ecker JR, Zhang YL, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15: 1962–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Komeda Y (2004) Flowering of Arabidopsis cop1 mutants in darkness. Plant Cell Physiol 45: 398–406 [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang HY, Yang JP, Shen YP, Rubio V, Ma LG, Hoecker U, Deng XW (2003) The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J (2002) De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol 12: 1462–1472 [DOI] [PubMed] [Google Scholar]
- Searle I, Coupland G (2004) Induction of flowering by seasonal changes in photoperiod. EMBO J 23: 1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: 855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor Cop1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Wang HY, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhang ZM, Dornan D, Arnott D, Deshaies RJ, Dixit VM (2004) Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JP, Lin RC, James S, Hoecker U, Liu BL, Xu L, Deng XW, Wang HY (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text