Abstract
Tagging proteins with mono- or poly-ubiquitin is now recognized as a multifaceted and universal means of regulating cell growth and physiology. It does so by controlling the cellular lifetime of nearly all eukaryotic proteins and the cellular localization of many critical proteins. Enzymes of the ubiquitin pathway add (ligases) or remove (deubiquitinases [DUBs]) ubiquitin tags to or from their target proteins in a selective fashion. Similarly to the kinases and their corresponding phosphatases, ubiquitin ligases and DUBs have become actively studied molecular oncology targets for drug discovery. Approximately 79 functional DUBs exist in the human proteome, suggesting that selective intervention is a reasonable therapeutic objective, with the goal of downregulating or ablating oncogene products or, alternatively, upregulating or sparing tumor suppressors. In the following review, this fascinating class of regulatory enzymes will be described, and specific examples of DUBs that are viable targets for anticancer therapy will be considered.
Keywords: associated molecule with the SH3-domain of STAM, cylindromatosis gene, deubiquitinating enzymes, isopeptidase, proteasomal degradation, ubiquitin, ubiquitin-specific protease 2a, ubiquitin-specific protease 7, ubiquitin-specific protease 20
Ubiquitin & ubiquitin-like proteins
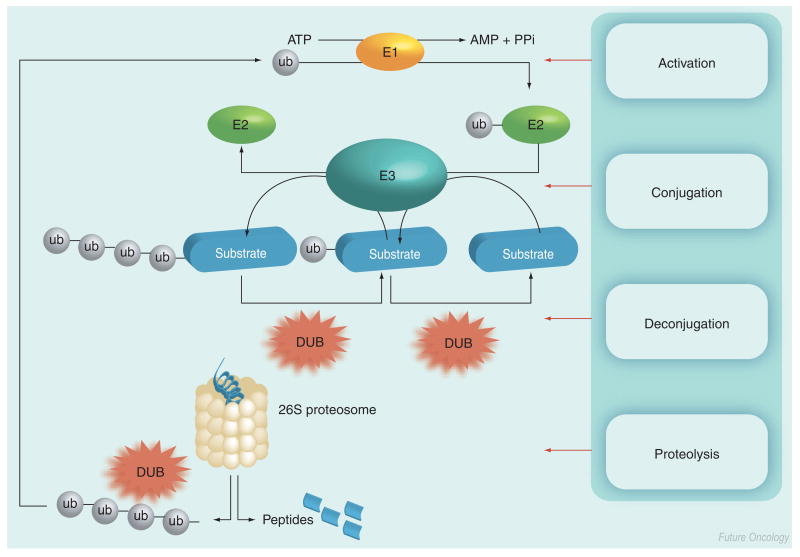
The content of most proteins in the cell is regulated by the ubiquitin–proteasomal pathway [1]. Ubiquitin and ubiquitin-like proteins (UBLs), such as SUMO, NEDD8, ISG15 and FAT10, regulate proteins via additional mechanisms, for example, intracellular compartmentation, signal transduction and the regulation of some E3 ligases [2]. Degradation of a targeted protein by the ubiquitin system involves the activation of ubiquitin by the enzyme E1, which links the ubiquitin C-terminus to a cysteine side chain of the enzyme in an ATP-dependent manner [1]. Activated ubiquitin is transferred as a thioester to enzyme E2, which catalyzes ubiquitin transfer to the ε-amino group of a lysine residue of a target polypeptide that is bound to the third enzyme in the sequence, E3, commonly called ubiquitin ligase (Figure 1) [1]. Subsequently, additional ubiquitin moieties can be conjugated to the ubiquitin to form linear or branch-chained poly-ubiquitinated proteins. Typically, poly-ubiquitinated polypeptides are delivered to the proteasome complex, which hydrolyzes the polypeptide into short oligopeptides and releases free ubiquitin, which is then recycled. The process is reversible; ubiquitin, as well as other UBLs, can be deconjugated by proteases, referred to generically as isopeptidases. In this review, the term isopeptidase will be used to refer to proteases that specifically cleave ubiquitin or UBL after the terminal carboxyl group of ubiquitin (Gly76) or UBL (Figure 1) [3]. Isopeptidases that cleave at the carboxy terminus of ubiquitin are termed deubiquitinases (DUBs) and have been divided into five distinct groups. Four of the five subfamilies identified to date are cysteine proteases: ubiquitin C-terminal hydrolases (UCH); ubiquitin-specific proteases (UBP/USP); Machado-Joseph Domain (MJD) and ovarian tumor related (OTU) [3,4]. By contrast, the JAMM motif DUBs are Zn2+-containing metalloproteases. Collectively, direct experimental and bioinformatic approaches have identified 90 putative DUBs, of which 79 are postulated to be functional [4]. In addition to the DUBs, there are multiple families of UBL-specific isopeptidases, such as SENP1 and P2, which cleave SUMO, DEN1 (SENP8), which cleaves Nedd8, and UBP43, which has been reported to cleave ISG15 in vitro [5–7].
Figure 1. Ubiquitin pathway showing ubiquitin conjugation (E1, E2 and E3) and DUB activities.
In some cases, ubiquitin chain assembly factor (E4) has been shown to enhance the conjugation of ub to certain substrates (not shown).
AMP: Adenosine monophosphate; DUB: Deubiquitinase; PPi: Phosphate; ub: Ubiquitin.
Importance of DUBs: roles in cancer
The approval of the proteasome inhibitor bortezomib (Velcade®) for the treatment of multiple myeloma that had failed one prior treatment schedule validated the targeting of the proteasome for the treatment of cancer [8]. Unfortunately, extended treatment with bortezomib is associated with toxicity and drug resistance, limiting its efficacy [9]. By contrast, therapeutic strategies that target specific aspects of the ubiquitin–proteasome pathway upstream of the proteasome, including DUBs, are predicted to be better tolerated.
The crystal structures of a number of DUBs in the USP/UBP class have been resolved, including USP7(herpesvirus-associated ubiquitin-specific protease [HAUSP]) and USP2, providing the basis for molecular recognition studies of these proteases in their active (ubiquitin or ubiquitin aldehyde-complexed) states [10,11]. These structural studies demonstrated that the mechanism for ubiquitin recognition is the same for the two DUBs, which are homologous only within their catalytic site regions, and it was hypothesized that this recognition mechanism is common to all DUBs of the USP/UBP class (which is by far the most numerous).
Genomics has identified at least 530 human genes that putatively encode enzymes involved in the conjugation and deconjugation of ubiquitin. Of these, at least 79 are thought to encode functional DUBs, some of which have multiple isoforms [4,12]. Considerable progress has been made in the study of ubiquitin conjugation, however, the study of DUBs, is still in its nascent stages. Early research has been promising, implicating a number of DUBs, such as USP4 (UNP), USP6 (Tre-2), USP8 (UBPY), USP28 and UCHL5 (UCH37) in neoplastic disease. However, in the following, we will restrict our discussion to six relatively well studied DUBs representing targets for anticancer therapy. The first five act as oncoproteins, thus, inhibitors of these DUBs would be appropriate therapeutic agents. The sixth DUB functions as a tumor suppressor; drug discovery efforts using this, and similar targets must be directed at activating them or sparing them from degradation.
Oncogene products
USP7
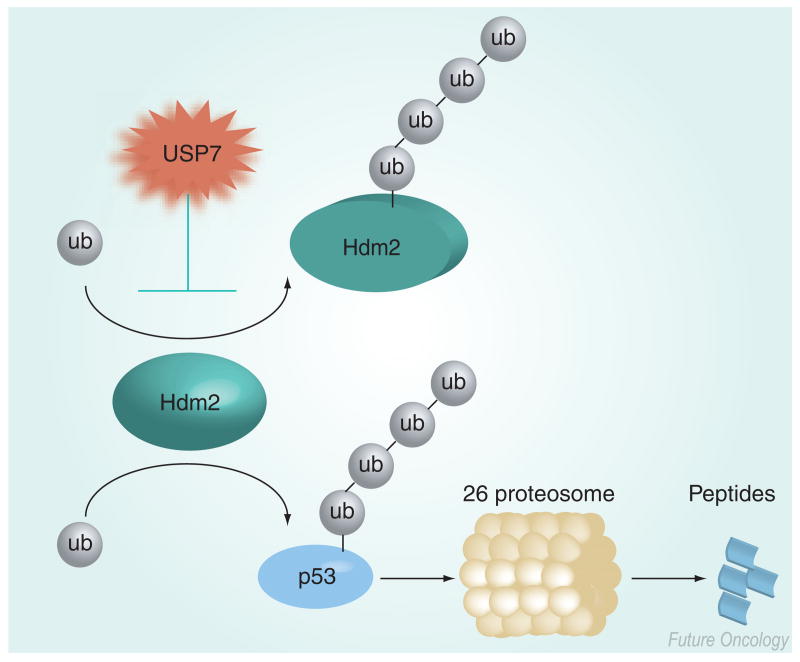
USP7 (HAUSP) was originally identified by its binding activity to a herpes viral protein, ICP0 [13]. USP7 plays a key role in regulating the ubiquitination of the RING-finger E3 ligase Mdm2 (and its human homolog Hdm2) [14,15]. Hdm2 binds the tumor suppressor p53 and facilitates its degradation by the proteasome as a result of polyubiquitinating p53 [16,17]. Similar to many other RING-finger E3 ligases, Hdm2 is capable of autoubiquitination, thereby promoting its own proteolytic degradation [18]. Nevertheless, under normal conditions Hdm2 ubiquitinates p53, resulting in the degradation of p53 via the proteasome (Figure 2). USP7 was originally thought to primarily function in the deubiquitination of p53, thus increasing the level of p53 [19]. However, more recent genetic and biochemical studies have found that, with respect to p53 and Hdm2, the primary target of USP7 is Hdm2 [14,15]. Structural biology studies corroborated these data by revealing that Hdm2 and p53 recognized the tumor necrosis factor-receptor associated factor (TRAF) domain of USP7 in a mutually exclusive manner and, furthermore, that Hdm2 bound to the TRAF domain with a higher affinity than p53 [20]. Space limitations preclude a complete description of the regulation of Hdm2 and p53, and the consequences of the regulation of Hdm2 by USP7 are discussed in further detail below.
Figure 2. USP7 preferentially deubiquitinates Hdm2.
Relative to p53, USP7 has a higher affinity for Hdm2, thus USP7 preferentially deubiquitinates Hdm2, preventing Hdm2 from inducing its own degradation due to autoubiquitination. Subsequently, Hdm2 ubiquitinates p53, which is degraded by the proteasome. An inhibitor of USP7 is predicted to promote the degradation of Hdm2 and therefore abrogate the degradation of p53 by the ubiquitin proteosome system.
ub: Ubiquitin; USP: Ubiquitin-specific protease.
In contrast to the majority of human tumors, mutations or deletions of p53 in common hematological malignancies such as multiple myeloma (MM), acute myeloid leukemia and chronic lymphocytic leukemia (CLL) are relatively rare at initial diagnosis, and the activation of p53 may offer a therapeutic benefit [21,22]. Importantly, a number of synthetic Hdm2 inhibitors have now been developed, including nutlin-3 (Roche), and these have been demonstrated to induce both p53 and apoptosis in a number of p53 wild-type tumors [23,24]. Nutlin-3a was found to activate p53 and induce apoptosis in p53 wild type, but not in p53 mutant MM and CLL cells [25,26]. Furthermore, nutlin-3a induced p53 and cell death in the majority of the primary human MMs, even in the presence of bone marrow stromal cells [25]. These observations suggest that promoting Hdm2 degradation by inhibiting USP7 and, thus, activating the tumor suppressor p53 will offer a therapeutic benefit for treating a range of p53 wild-type cancers.
An additional target of USP7 is the forkhead transcription factor, forkhead box class O (FOXO)4. FOXO transcription factors are post-translationally regulated by phosphorylation, acetylation and ubiquitination [27,28]. Over-expression of FOXO4 induces growth suppression in cell lines, including a Ras transformed cell line, by transcriptionally activating the cyclin-dependent kinase inhibitor p27 [29]. One mechanism of FOXO4 activation is mono-ubiquitination and subsequent translocation to the nucleus following an increase in oxidative stress [28]. Subsequently, USP7 deubiquitinates FOXO4 in a p53-independent manner, resulting in nuclear export and providing a mechanism for inactivating FOXO4 [28]. Hence, an additional benefit of inhibiting USP7 may be to prevent the abrogation of FOXO4 activity.
USP2a
The two isoforms of USP2, USP2a (USP2–69; UBP-t2) and USP2b (USP2–45; UBP-t1), were originally detected by a cloning strategy designed to identify USP enzymes in rat testes [30]. As products of alternative splicing at the 5′ end of the gene, both isoforms share a common catalytic core domain at the 3′ end of the gene [30–32]. An additional variant of USP2, UBP41, has been reported, however, subsequent studies described the lack of an appropriate initiation codon and suggested that this isoform may be a sequencing artifact representing the catalytic core domain of USP2 and a short N-terminal extension [32,33]. In vitro studies suggested that the amino terminal extensions of USP2a and 2b inhibited the isopeptidase activity of the catalytic core domain, yet the catalytic core and USP2a were both able to deubiquitinate high molecular weight proteins from testis extracts with high efficiency [31].
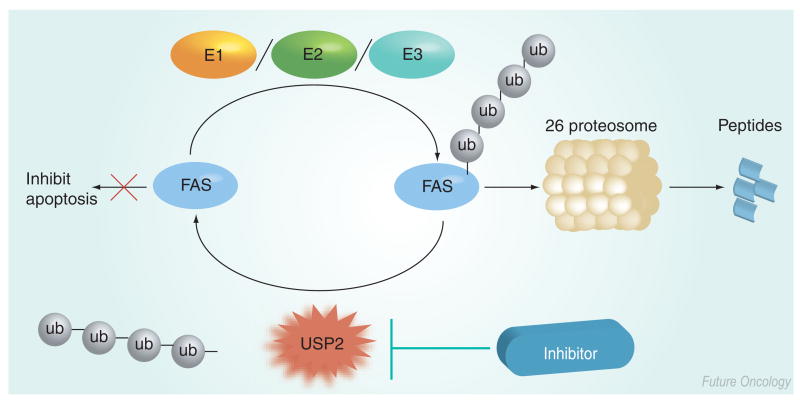
Overexpression of the catalytic core domain of USP2 (identified as UBP41 in the paper) induced apoptosis in human 293T cells [34]. The authors were unable to identify any specific substrates of UBP41, but observed a large decrease in high molecular weight ubiquitin conjugates and, thus, concluded that overexpression of UBP41 interfered with the ubiquitin proteasome system. By contrast, USP2a has been reported to have an anti-apoptotic function in prostate cancer. USP2a is androgen-regulated and overexpressed in prostate tumors [35]. The functional consequence of USP2a overexpression is the stabilization of fatty acid synthase (FAS) (Figure 3) [35,36]. Recently, FAS has been recognized as an emerging oncology target. For example, FAS is overexpressed in many tumors, including prostate cancer, and FAS expression correlates significantly with the tumor grade in human prostate cancer [37]. In addition, FAS inhibition in estrogen receptor (ER)-positive breast and endometrial cancer modulates ER expression, ER-dependent gene transcription and tumor cell death by promoting apoptosis [38]. Inhibition of FAS by 4-methylene-2-octyl-5-oxo-tetrahydro-furan-3-carboxylic acid (C75) or cerulenin results in a reduction in cell proliferation and induction of apoptosis in some tumor cell lines [39,40]. Specific knockdown of USP2a using small interfering (si)RNA increases polyubiquitinated FAS levels (in the presence of the proteasome inhibitor MG132), demonstrating that USP2a deubiquitinates FAS in LNCaP cells [35]. In addition, USP2a siRNA reduces FAS protein levels and induces apoptosis in LNCaP cells [35]. Further proof of the oncogenic role of USP2a was provided by the observation that ectopic expression of USP2a in nontransformed cells promotes oncogenic behavior in vitro and in vivo [36]. Notably, Priolo and colleagues also demonstrated that, in addition to FAS, Hdm2 was deubiquitinated by USP2a, further highlighting the importance of USP2a as a therapeutic target.
Figure 3. Model for inhibition of USP2a.
An inhibitor of USP2a is predicted to promote the degradation of FAS, thus abrogating the antiapoptotic activity of FAS.
FAS: Fatty acid synthase; ub: Ubiquitin; USP: Ubiquitin-specific protease.
Associated molecule with the SH3-domain of signal-transducing adaptor molecule
Recently, Urbe and colleagues described a JAMM domain-containing protein that is linked with the signal transduction associated with the endosomal sorting (trafficking between the membrane and endosomal/lysosomal compartments) of the epidermal growth factor receptor (EGFR). This protein has been named Associated Molecule with the SH3-domain of signal-transducing adaptor molecule (STAM; AMSH), a protein that regulates receptor endosomal sorting [41]. The EGFR regulates numerous cellular functions by initiating signal transduction cascades [42]. During the cellular lifetime of the EGFR, it is recycled from the membrane to the early (sorting) endosome, before finally being selected for sorting to the late endosome and lysosome, where it is degraded by acid proteases. The EGFR participates in signal transduction both at the membrane and in the early endosome compartment.
Whilst much of the signaling is concerned with the regulation of cell growth and other functions, one component of signal transduction regulates the trafficking of the EGFR itself. The E3 ligase Cbl mediates the ubiquitination of phosphorylated EGFR. Subsequent signaling events result in the degradation of the receptor in late endosomes/lysosomes. According to a recent model, ubiquitinated-EGFR is recognized by the protein Hrs at the endosomal surface, whereupon further interactions with the endosomal-associated complex required for transport result in translocation to internal vesicles of the multivesicular body, committing EFGR to protease degradation in the lysosome [41]. McCullough and colleagues demonstrated that recombinant AMSH functions as a DUB in vitro [41]. In addition, ablation of AMSH activity by the incubation of cells with AMSH siRNA enhances the degradation of EGFR [41].
These observations lead to the hypothesis that AMSH inhibitors will decrease the cellular content of EGFR and have activity against EGFR-mediated diseases, the pre-eminent of which is cancer [42]. Space limitations prohibit an adequate discussion of the complete signal transduction scheme that interacts with, and governs, the membrane–endosomal trafficking of EGFR and other receptors. However, given the results obtained with siRNA, the therapeutic hypothesis associated with AMSH exists independently of these details. If a compound can be found to selectively inhibit AMSH, then diseases that are linked to EGFR activity can be addressed by upregulating the natural, physiological destruction of the receptor, rather than by introducing a nonphysiological, potentially toxic mechanism of intervention.
USP20 & USP33
von Hippel-Lindau disease (VHL) is characterized as an autosomal-dominant disease that predisposes individuals to a variety of tumors, including clear cell carcinomas of the kidneys, hemangioblastomas in the CNS and retina and islet cell tumors of the pancreas [43,44]. Whilst most of the tumors are benign, VHL patients have a lifetime risk of more than 70% of developing renal clear cell carcinomas, the principal cause of mortality of patients with VHL disease [44]. VHL disease is a consequence of mutations in the pVHL protein, which is a component of an E3 ligase, consisting of elongin B, elongin C and Cullin 2 [43]. USP33 (VHL-interacting deubiquitinating enzyme 1) was originally identified as a protein that interacts with pVHL by a yeast two-hybrid screen [45]. Subsequent homology searches of the GenBank database identified a related protein, USP20 (VDU2) [46]. USP33 and USP20 proteins have a shared identity of approximately 59%, with strong homology at the amino and carboxy termini. Further analysis reveals that USP20 and USP33 have a higher homology with each other than with other members of the USP family. Both USP20 and USP33 bind competitively to the substrate recognition β-domain of pVHL, a region commonly mutated in patients with VHL disease [45,46]. Furthermore, USP20 and USP33 have been demonstrated to undergo ubiquitination by the pVHL E3 ligase complex in vitro and in transiently transfected transformed African green monkey kidney fibroblast cells (COS-7) cells [45,46].
An additional substrate of the pVHL E3 ligase complex is the α-subunit of the hypoxia-inducible factor (HIF)1. HIF1 is a heterodimeric transcription factor consisting of an α-subunit and a constitutively expressed β subunit, and regulates genes involved in angiogenesis, metastasis, cell survival and glucose metabolism, and it is overexpressed in many human cancers [47]. In contrast to HIF1β, HIF1α, protein levels are highly regulated. Under conditions of normoxia, prolyl hydroxylases modify Pro402 and 564 of HIF1α resulting in the binding of the hydroxylated HIF1α to the pVHL E3 ligase complex and subsequent ubiquitination and degradation by the proteasome. By contrast, during hypoxia, the concentration of oxygen becomes rate-limiting for the prolyl hydroxylases and there is an accumulation of HIF1α protein and an increase in HIF1 activity [47]. Interestingly, Li and colleagues demonstrated that USP20, but not USP33, binds to HIF1α in vitro and deubiquitinates HIF1α in transfected human epithelial kidney (HEK)293 cells [48]. Furthermore, the ectopic expression of USP20 increases steady-state levels of HIF1α in HEK293 cells under normoxic conditions. In addition, overexpression of USP20 significantly increases hypoxia-inducible element gene transcription in a reporter assay. The tight regulation of USP20 and USP33 protein levels by the tumor suppressor, pVHL, suggests that USP20 and USP33 proteins play a role in carcinogenesis. Intriguingly, a recent study found that USP33 was overexpressed in B-cell acute lymphoblastic leukemia (ALL) relative to the T-cell ALL [49]. Furthermore, given the ability of HIF1 to activate genes that promote carcinogenesis, USP20 may be an important therapeutic target for the treatment of a number of different types of cancer.
Tumor suppressor
Cylindromatosis gene
The tumor suppressor role of the protein encoded by the cylindromatosis gene (CYLD), which is mutated in familial cylindromatosis, is well established. Mice in which CYLD is knocked out are prone to chemical tumorigenesis [50], and CYLD levels are downregulated in diverse tumor types [51,52]. In addition, it has recently been established that the CYLD gene product is a DUB that removes lysine 63-linked ubiquitin chains from TRAF2, blocking its ability to activate the IκB kinase, IKK, thereby inhibiting activation of the transcription factor nuclear factor (NF)-κB, which would otherwise lead to anti-apoptotic gene expression and a survival phenotype [53]. In addition, CYLD removes a single ubiquitin from the oncoprotein Bcl3, preventing its entry into the nucleus and the promotion of proliferation-linked transcription by interacting with nuclear factor (NF)-κB subunits [50]. Thus, CYLD potentially blocks tumor formation and maintenance by both antiproliferative and proapoptotic mechanisms. The hypothesis that a loss of CYLD function can be relieved through treatment with aspirin derivatives that inhibit NF-κB activity was validated by the observation that treatment of cylindromatosis patients with a topical preparation of salicyclic acid leads to some complete remissions [53,54]. Similar therapeutic effects may be possible using drugs that activate CYLD.
Conclusions
In this review we have described six different DUBs and their roles in neoplastic disease. In some cases, the DUB acts directly on an oncogene; in the case of USP7 it acts on the regulator of a tumor suppressor and, in the case of CYLD, the DUB acts on the regulator of a prosurvival transcription factor. In all cases, however, modulation of DUB activity has the potential to offer therapeutic benefit for the treatment of cancer.
Future perspective
Originally, the notion that a therapy as toxic as a proteasome inhibitor could be efficacious for the treatment of cancer was viewed with a great deal of skepticism. However, the clinical trials and subsequent approval of the proteasome inhibitor bortezomib have demonstrated that functional blockade of the proteasome is a viable therapy for MM [8]. Still, however, prolonged exposure to bortezomib is associated with toxicity [9]. To address this observation, a more targeted approach is required. Considerable effort has been made to study ubiquitin conjugation with some preclinical success, but this is yet to translate to the clinic. We believe that the direct targeting of isopeptidases offers an alternative strategy for a targeted therapy. As with all enzymes, it should be noted that modulating aberrant DUB activity in cancer may result in unwanted side effects. However, it is hoped that these side effects would be minimal relative to those experienced by patients undergoing conventional cytotoxic chemotherapies or treatment with a proteasome inhibitor such as Bortezomib.
No DUB inhibitors (or activators) have successfully entered the clinic, however, an understanding of the mechanism of action of CYLD in cylindromatosis allowed Oosterkamp and colleagues to perform the first proof-of-concept study in humans [53,54]. These data further validate DUBs as viable therapeutic targets for cancer treatment.
Additionally, owing to space limitations, the foregoing did not address the potential for targeting UBL isopeptidases. UBLs have been shown to be critical for the regulation of numerous processes, such as cellular localization, transcriptional regulation, signal transduction and, perhaps most pertinently, the regulation of some E3 ligases [2]. The scientific community is only beginning to explore the role of isopeptidases in tumorigenesis, and we predict rapid progress over the next decade, culminating in the clinical trials of one or more isopeptidase inhibitors for the treatment of cancer.
Resolving the compound discovery bottleneck
One of the primary hurdles in the discovery and development of compounds that modulate DUB activity is the lack of a high fidelity, robust, high-throughput assay for the screening for inhibitors/activators of USPs. Progenra [101] has developed such an assay and is now screening for small molecules that act to modulate DUB activity [55]. One of the key advantages of the Progenra assay is its ability to rapidly screen compounds against multiple UBL isopeptidases. As a consequence of our, and other companies', screening campaigns, we anticipate that multiple DUB inhibitors will be entering the clinic in the next decade.
Executive summary.
Ubiquitin & ubiquitin-like proteins
The cellular content of the majority of proteins is regulated by the ubiquitin–proteasome system.
Ubiquitin is conjugated to proteins by the sequential action of E1, E2 and E3 enzymes and deconjugated by deubiquitinating enzymes (DUBs).
Ubiquitin and ubiquitin-like proteins (UBLs) also regulate protein function by post-translational modifications.
Importance of DUBs: roles in cancer
The approval of bortezomib validated targeting the proteasome as a treatment for cancer, but was associated with toxicities. The targeted therapeutic approach of directly inhibiting DUBs is predicted to be better tolerated.
Genomics has identified at least 530 human genes that are thought to encode enzymes involved in the addition or removal of ubiquitin; at least 79 are thought to encode functional DUBs. Considerable study has been invested in the study of ubiquitin conjugation; less attention has been given to DUBs.
USP7
USP7 deubiquitinates the E3 ligase Hdm2, resulting in increased levels of Hdm2 and decreased levels of the tumor suppressor p53.
Inhibitors of USP7 are predicted to offer therapeutic benefit in p53 wild-type tumors, such as hematological malignancies, by activating p53.
Inhibitors of USP7 are also postulated to prevent abrogation of FOXO4 activity, and thus promote growth suppression.
USP2a
Ectopic expression of USP2a promotes oncogenic behavior in vitro and in vivo.
USP2a deubiquitinates the anti-apoptotic enzyme, fatty acid synthase (FAS), in prostate cancer cells. Therefore, inhibitors of USP2a are predicted to promote FAS degradation and apoptosis.
USP2a is also reported to deubiquitinate Hdm2.
Inhibition of FAS is also thought to increase apoptosis in estrogen-receptor-positive breast and endometrial cancer, potentially increasing the indications for inhibitors of USP2a.
Associated molecule with the SH3-domain of signal-transducing adaptor molecule
Regulates epithelial growth factor receptor (EGFR) degradation by removing ubiquitin from EGFR, thus rescuing EGFR from degradation by the lysosome.
Inhibition of the associated molecule with the SH3-domain of signal-transducing adaptor molecule (STAM; AMSH) by small interfering (si)RNA enhanced the degradation of EGFR, thus, inhibitors of AMSH are predicted to downregulate the cellular content of EGFR, an important cancer target.
USP20 & USP33
USP20 and USP33 bind competitively to the β-domain of von Hippel-Lindau (VHL) disease, the substrate-binding component of an E3 ligase complex, and are subsequently ubiquitinated.
The tight regulation of USP20 and USP33 protein levels by the tumor suppressor pVHL suggests that USP20 and USP33 proteins play a role in carcinogenesis.
Cylindromatosis
In contrast to the other DUBs discussed above, cylindromatosis (CYLD) is a tumor suppressor which is mutated in familial cylindromatosis and downregulated in diverse tumor types.
CYLD deubiquitinates tumor necrosis factor receptor-associated factor-2, preventing it from activating nuclear factor (NF)-κB, via activation of the IκB kinase, IKK. Therefore, activation of CYLD is predicted to abrogate the activity of the prosurvival transcription factor, NF-κB.
CYLD also deubiquitinates the oncogenic transcription factor Bcl3, preventing it from localizing to the nucleus.
Inhibition of NF-κB activation by aspirin resulted in complete remission for some patients with familial cylindromatosis.
Future perspective
To date, no DUB inhibitors or activators have been tested in the clinic, however, functional ablation of NF-κB activation in cylindromatosis patients provides a proof-of-concept for modulating DUB activity in cancer.
Isopeptidases that cleave other UBLs are also relevant to the progression of neoplastic disease and are only just beginning to be explored by the scientific community.
Novel high-throughput, high-fidelity assays, such as the one developed by Progenra, will enable the rapid determination of selective modulators of DUBs and speed the progress of lead compounds to the clinic.
Acknowledgments
We gratefully acknowledge Keith Wilkinson (Emory University, USA), Arthur Haas (Louisiana State University, USA), Mark Hochstrasser (Yale University, USA) and Rohan Baker (Australian National University, Canberra, Australia) for valuable discussions, as well as support from grant 1R43CA115205 awarded to Michael R Mattern by the NCI/NIH.
Contributor Information
Benjamin Nicholson, Progenra, Inc., 271A Great Valley Parkway, Malvern, PA 19355, USA, Tel.: +1 610 644 6974; Fax: +1 610 644 8616; nicholson@progenra.com.
Jeffrey G Marblestone, Progenra, Inc., 271A Great Valley Parkway, Malvern, PA 19355, USA, Tel.: +1 610 644 6974; Fax: +1 610 644 8616; marblestone@progenra.com.
Tauseef R Butt, Progenra, Inc., 271A Great Valley Parkway, Malvern, PA 19355, USA, Tel.: +1 610 644 6974; Fax: +1 610 644 8616; butt@progenra.com.
Michael R Mattern, Progenra, Inc., 271A Great Valley Parkway, Malvern, PA 19355, USA, Tel.: +1 610 644 6974; Fax: +1 610 644 8616; mattern@progenra.com.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]; •• Complete overview of the ubiquitin conjugation and deconjugation system.
- 2.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 3.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]; • Comprehensive review of deubiquitnases (DUBs).
- 4.Nijman SM, Luna-Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]; • Review of DUBs including a genomic perspective.
- 5.Cheng J, Bawa T, Lee P, Gong L, Yeh ET. Role of desumoylation in the development of prostate cancer. Neoplasia. 2006;8:667–676. doi: 10.1593/neo.06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan-Erdene T, Nagamalleswari K, Yin L, Wu K, Pan ZQ, Wilkinson KD. Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem. 2003;278:28892–28900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 7.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 8.Kane RC, Farrell AT, Sridhara R, Pazdur R. United States Food and Drug Administration approval summary: bortezomib for the treatment of progressive multiple myeloma after one prior therapy. Clin Cancer Res. 2006;12:2955–2960. doi: 10.1158/1078-0432.CCR-06-0170. [DOI] [PubMed] [Google Scholar]; • US FDA approval summary for bortezomib.
- 9.Chauhan D, Catley L, Li G, et al. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from bortezomib. Cancer Cell. 2005;8:407–419. doi: 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Saridakis V, Sheng Y, Sarkari F, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein–Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 11.Renatus M, Parrado SG, D'Arcy A, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Outlines the structure of the core catalytic domain of USP2a in complex with ubiquitin.
- 12.Wong BR, Parlati F, Qu K, et al. Drug discovery in the ubiquitin regulatory pathway. Drug Discov Today. 2003;8:746–754. doi: 10.1016/s1359-6446(03)02780-6. [DOI] [PubMed] [Google Scholar]
- 13.Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53–MDM2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 15.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:486. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 16.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. MDM2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Fang S, Jensen JP, Ludwig RL, Vousdenm KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Chen D, Shiloh A, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 20.Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53–MDM2 pathway. PLoS Biol. 2006;4:E27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Structural validation of the observation that USP7 preferentially deubiquitinates Hdm2.
- 21.Cheon KW, Baek KH. HAUSP as a therapeutic target for hematopoietic tumors (review) Int J Oncol. 2006;28:1209–1215. [PubMed] [Google Scholar]
- 22.Peller S, Rotter V. TP53 in hematological cancer: low incidence of mutations with significant clinical relevance. Hum Mutat. 2003;21:277–284. doi: 10.1002/humu.10190. [DOI] [PubMed] [Google Scholar]
- 23.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]; • Description of a novel class of Hdm2 inhibitors.
- 24.Tovar C, Rosinski J, Filipovic Z, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuhmer T, Chatterjee M, Hildebrandt M, et al. Nongenotoxic activation of the p53 pathway as a therapeutic strategy for multiple myeloma. Blood. 2005;106:3609–3617. doi: 10.1182/blood-2005-04-1489. [DOI] [PubMed] [Google Scholar]
- 26.Coll-Mulet L, Iglesias-Serret D, Santidrian AF, et al. MDM2 antagonists activate p53 and synergize with genotoxic drugs in B-cell chronic lymphocytic leukemia cells. Blood. 2006;107:4109–4114. doi: 10.1182/blood-2005-08-3273. [DOI] [PubMed] [Google Scholar]
- 27.Burgering BM, Kops GJ. Cell cycle and death control: long live forkheads. Trends Biochem Sci. 2002;27:352–360. doi: 10.1016/s0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- 28.van der Horst A, de Vries-Smits AM, Brenkman AB, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 29.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 30.Lin H, Keriel A, Morales CR, et al. Divergent N-terminal sequences target an inducible testis deubiquitinating enzyme to distinct subcellular structures. Mol Cell Biol. 2000;20:6568–6578. doi: 10.1128/mcb.20.17.6568-6578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin H, Yin L, Reid J, Wilkinson KD, Wing SS. Divergent N-terminal sequences of a deubiquitinating enzyme modulate substrate specificity. J Biol Chem. 2001;276:20357–20363. doi: 10.1074/jbc.M008761200. [DOI] [PubMed] [Google Scholar]; • Functional dissection of the multiple isoforms of USP2.
- 32.Gousseva N, Baker RT. Gene structure, alternate splicing, tissue distribution, cellular localization, and developmental expression pattern of mouse deubiquitinating enzyme isoforms USP2–45 and USP2–69. Gene Expr. 2003;11:163–179. doi: 10.3727/000000003108749053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek SH, Choi KS, Yoo YJ, et al. Molecular cloning of a novel ubiquitin-specific protease, UBP41, with isopeptidase activity in chick skeletal muscle. J Biol Chem. 1997;272:25560–25565. doi: 10.1074/jbc.272.41.25560. [DOI] [PubMed] [Google Scholar]
- 34.Gewies A, Grimm S. UBP41 is a proapoptotic ubiquitin-specific protease. Cancer Res. 2003;63:682–688. [PubMed] [Google Scholar]
- 35.Graner E, Tang D, Rossi S, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]; •• Comprehensive description of the ability of USP2a to regulate fatty acid synthase in prostate cancer.
- 36.Priolo C, Tang D, Brahamandan M, et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Res. 2006;66:8625–8632. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- 37.Baron A, Migita T, Tang D, Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem. 2004;91:47–53. doi: 10.1002/jcb.10708. [DOI] [PubMed] [Google Scholar]
- 38.Lupu R, Menendez JA. Targeting fatty acid synthase in breast and endometrial cancer: an alternative to selective estrogen receptor modulators? Endocrinology. 2006;147:4056–4066. doi: 10.1210/en.2006-0486. [DOI] [PubMed] [Google Scholar]
- 39.Pizer ES, Thupari J, Han WF, et al. Malonyl-coenzyme-A is a potential mediator of cytotoxicity induced by fatty-acid synthase inhibition in human breast cancer cells and xenografts. Cancer Res. 2000;60:213–218. [PubMed] [Google Scholar]
- 40.Menendez JA, Vellon L, Mehmi I, et al. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (ERBB-2) oncogene overexpression in cancer cells. Proc Natl Acad Sci USA. 2004;101:10715–10720. doi: 10.1073/pnas.0403390101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Description of the ability of associated molecule with the SH3-domain of signal-transducing adaptor molecule to deubiquitinate epidermal growth factor receptor (EGFR) and, thus, rescue EGFR from degradation.
- 42.Yarden Y. The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37 4:S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 43.Shuin T, Yamasaki I, Tamura K, Okuda H, Furihata M, Ashida S. von Hippel-Lindau disease: molecular pathological basis, clinical criteria, genetic testing, clinical features of tumors and treatment. Jpn J Clin Oncol. 2006;36:337–343. doi: 10.1093/jjco/hyl052. [DOI] [PubMed] [Google Scholar]
- 44.Maher ER, Kaelin WG., Jr von Hippel-Lindau disease. Medicine (Baltimore) 1997;76:381–391. doi: 10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 45.Li Z, Wang D, Na X, Schoen SR, Messing EM, Wu G. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2002;277:4656–4662. doi: 10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Wang D, Na X, Schoen SR, Messing EM, Wu G. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem Biophys Res Commun. 2002;294:700–709. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- 47.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1α. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the ability of USP20, but not USP33 to deubiquitinate hypoxia-inducible factor 1α.
- 49.De Pitta C, Tombolan L, Campo Dell'Orto M, et al. A leukemia-enriched cDNA microarray platform identifies new transcripts with relevance to the biology of pediatric acute lymphoblastic leukemia. Haematologica. 2005;90:890–898. [PubMed] [Google Scholar]
- 50.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 51.Strobel P, Zettl A, Ren Z, et al. Spiradenocylindroma of the kidney: clinical and genetic findings suggesting a role of somatic mutation of the CYLD1 gene in the oncogenesis of an unusual renal neoplasm. Am J Surg Pathol. 2002;26:119–124. doi: 10.1097/00000478-200201000-00016. [DOI] [PubMed] [Google Scholar]
- 52.Hirai Y, Kawamata Y, Takeshima N, et al. Conventional and array-based comparative genomic hybridization analyses of novel cell lines harboring HPV18 from glassy cell carcinoma of the uterine cervix. Int J Oncol. 2004;24:977–986. [PubMed] [Google Scholar]
- 53.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]; • Description of a functional small interfering RNA screen for DUBs and the discovery that CYLD inhibits the activation of nuclear factor (NF)-κB.
- 54.Oosterkamp HM, Neering H, Nijman SM, et al. An evaluation of the efficacy of topical application of salicylic acid for the treatment of familial cylindromatosis. Br J Dermatol. 2006;155:182–185. doi: 10.1111/j.1365-2133.2006.07224.x. [DOI] [PubMed] [Google Scholar]; •• First proof-of-concept clinical study demonstrating that functional ablation of hyperactivated NF-κB results in some complete remissions in cylindromatosis patients.
- 55.Nicholson B, Leach CA, Kodrasov MP, et al. A novel deubiquitinase assay platform for drug discovery and diagnosis. Presented at: SBS 12th Annual Conference and Exhibition; Seattle, WA, USA. September 2006.pp. 17–21. [Google Scholar]
- 101.Progenera website www.progenra.com