In this personal perspective, we recall the history of bioinformatics and computational biology in China, review current research and education, and discuss future prospects and challenges. The field of bioinformatics in China has grown significantly in the past decade despite a delayed and patchy start at the end of the 1980s by a few scientists from other disciplines, most noticeably physics and mathematics, where China's traditional strength has been. In the late 1990s and early 2000s, rapid expansion of the field was fueled by the Internet boom and genomics boom worldwide and in China. Today bioinformatics research in China is characterized by a great variety of biological questions addressed and the close collaborative efforts between computational scientists and biologists, with a full spectrum of focuses ranging from database building and algorithm development to hypothesis generation and biological discoveries. Although challenges remain, the future of bioinformatics in China is promising thanks to advances in both computing infrastructure and experimental biology research, a steady increase of governmental funding, and most importantly a critical mass of bioinformatics scientists consisting of not only converts from other disciplines but also formally trained overseas returnees and a new generation of domestically trained bioinformatics Ph.D.s.
Introduction
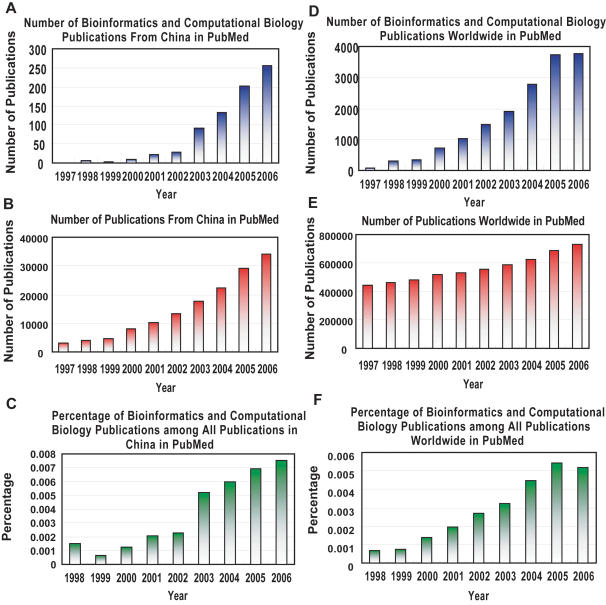
The field of bioinformatics has enjoyed significant growth in China. A rough yet useful indicator of the field is the number of bioinformatics and computational biology publications from China indexed in PubMed at NCBI. As shown in Figure 1A, this number has been increasing significantly over the past decade. The number of all publications from China indexed in PubMed has also been increasing (Figure 1B), but if we plot the percentage of bioinformatics publications from China among all PubMed publications from China, we observe that the percentage itself has been increasing rapidly (Figure 1C), indicating that the contribution of bioinformatics research is becoming more significant within the life sciences in China. Comparing it with the situation worldwide, we observe that the number of bioinformatics publications from China is growing faster than the number of bioinformatics publications worldwide (Figure 1A versus 1D). Additionally, the number of PubMed publications from China has also been growing faster than the total number of PubMed publications (Figure 1B versus 1E). Furthermore, we observe that, very interestingly, for each year starting from 2003, the percentage of bioinformatics publications among all PubMed publications is greater in China than worldwide (Figure 1C versus 1F), indicating that bioinformatics as a field within the life sciences is enjoying a faster growth rate in China than elsewhere in the world.
Figure 1. The number and percentage of bioinformatics publications from China and in all of PubMed in the past decade.
(A) The number of bioinformatics and computational biology publications from China in PubMed, retrieved from NCBI by the Entrez query “China[affiliation] AND (bioinformatics OR computational biology)”. MESH has a heading “Computational Biology” of which “Bioinformatics” is an Entry Term. The numbers provide a rough indication of growth of the field. They tend to be underestimated because some bioinformatics publications cannot be retrieved by keywords “bioinformatics OR computational biology”. However adding more keywords would increase the false positive rate significantly. (B) The number of all publications from China in PubMed, retrieved from NCBI by the Entrez query “China[affiliation]”. (C) The percentage of bioinformatics publications in China among all publications in China, in PubMed. (D) The number of bioinformatics and computational biology publications in all of PubMed, retrieved from NCBI by the Entrez query “bioinformatics OR computational biology”. (E) The number of all publications in PubMed. (F) The percentage of bioinformatics publications among all publications, in PubMed.
These numbers and trends, although only rough estimates, are fascinating and deserve a closer look. In this article, we (see Box 1 for Authors' Biographies) take a historical, as well as a horizontal, survey of bioinformatics in China. Due to the nature of personal perspectives and space limitations, what we present is by no means exhaustive, but merely suggestive.
Box 1. Authors' Biographies
Liping Wei is Professor and Director of the Center for Bioinformatics at Peking University and Associate Director of the National Laboratory of Protein Engineering and Plant Genetic Engineering in China. She received her undergraduate training in Electrical Engineering and Information Sciences from the University of Science and Technology of China. She holds a Master's degree in Applied Mathematics from Brown University and a PhD in Medical Informatics (now called Biomedical Informatics) from Stanford University. She worked in the biotech industry for four years, first at Exelixis, Inc., and then at Nexus Genomics, Inc., while continuing to serve as a Consulting Assistant Professor of bioinformatics in the Department of Medicine at Stanford University. She moved back to China in early 2004 to resume an academic career in bioinformatics at Peking University. Her current research interests include the regulation by, and of, noncoding RNAs and antisense transcripts and the signaling and regulatory networks underlying neurobiological disorders such as autism and addiction.
Jun Yu is a professor in the Beijing Institute of Genomics, Chinese Academy of Sciences (CAS). Dr. Yu obtained a B.S. degree in biochemistry from Jilin University in 1983 and a PhD in biomedical sciences from New York University School of Medicine in 1990. He joined the University of Washington Genome Center in 1993 and attuned his primary research interests toward genomics and bioinformatics. He started to work in China in 1998 and has led many major genome projects there, such as the International Human Genome Project (the Chinese effort), the Superhybrid Rice Genome Project, and the Silkworm Genome Project. His current research interests include comparative genome analysis, transcriptome modeling, human genetic diversity, and model organisms for phenotypic plasticity. He has published more than 120 scientific papers and won numerous academic awards, such as Scientific Leader of the Year, 2002 (Scientific American), 100-Talent Plan (CAS, 2002–2005), and China–US Biology Examination and Application (CUSBEA, 1983).
The Early Years
China had a late and rather sluggish start in bioinformatics, largely due to lack of institutional support for bioinformatics and governmental funding during the early years. It was not until 1996 that the first Center for Bioinformatics in China was established within the College of Life Sciences at Peking University and the first government funding dedicated to bioinformatics was established as part of the National High Technology Development 863 Program by the Ministry of Science and Technology (MOST). Despite the difficulties, starting from the end of the 1980s bioinformatics research was pioneered by a few scientists from other disciplines, most noticeably physics and mathematics where China's traditional strength has been, applying theoretical frameworks and analytical tools from their original specialty to study biological questions.
A great example of these early scientists was Bailin Hao, who, trained in the former Soviet Union, was at the time already an accomplished theoretical physicist. Fascinated by computable and predictable characteristics of biological systems, he first studied protein structures and then devoted himself to the analysis of DNA and protein sequences. In visualizing very long DNA sequences, including the complete genomes of several bacteria and yeast and segments of human genome, his group observed fractal-like patterns [1]; subsequently they proposed the description of a genome using statistics of K-strings (by counting a series of K-tuples in a linear sequence of symbols), enabling a new paradigm for phylogenetic analysis without sequence alignments [2]. Chunting Zhang, another established physicist, also started his bioinformatics research at the end of the 1980s working on protein structure prediction [3]. His group later created the concept of Z-curve to display DNA composition dynamics in geometric spaces [4],[5]. Z-curve has been successfully applied to genome sequence analyses such as prediction of coding genes in yeast genome [6], isochores in human genome [7], and replication origins in archaeal genomes [8]. Runsheng Chen, with a background in biophysics, was involved in the early phases of Chinese genomics research and later pioneered small RNA research in China [9]–[17]. His group identified hundreds of novel small noncoding RNAs in the nematode C. elegans and characterized their sequence features and expression patterns [11],[13]. A few other early scientists with important contributions include Yanda Li [18]–[24], Luhua Lai [25]–[34], Yunyu Shi [35], Liaofu Luo [36]–[40], Dafu Ding [41]–[43], and Zhirong Sun [44]–[48].
In the early to mid-1990s, many Chinese biologists were still not familiar with international biological databases, and some in remote cities did not even have reliable Internet connections to overseas Web sites. To promote bioinformatics as well as biology research, since 1995 Jingchu Luo and Xiaocheng Gu at the Center for Bioinformatics, Peking University, dedicated themselves to setting up official mirror sites of major biological databases, providing bioinformatics services, and organizing bioinformatics training workshops. From them, many Chinese scientists received their first exposure to biological data and analysis tools. Through close collaboration with the NCBI, EBI, and EMBnet, Luo's group continues to maintain the largest online bioinformatics resource in China at http://www.cbi.pku.edu.cn.
The Internet Boom and the Genomic Boom
Two significant developments in the second half of the 1990s proved critical to the expansion of bioinformatics in China. They were the Internet boom and the genomic boom. It was not until 1994 that full TCP/IP Internet connection between China and the rest of the world was established. At that time, the only established connection was an existing link, a dial-up X.25 connection, between the Institute of High-Energy Physics (IHEP) in Beijing and the Stanford Linear Accelerator Center (SLAC) at Stanford University. On May 17, 1994, the official connection to FIX-West was announced, and the U.S.-based Energy Sciences Network (ESnet) agreed to carry China IP traffic. Despite the late start, China caught up quickly with the worldwide Internet boom and established fast and broad Internet connections which were critical for biological data exchange.
At about the same time, the genomic boom in the late 1990s to early 2000s exemplified by the Human Genome Project [49] started to generate huge and exponentially growing amounts of data. After earlier efforts such as sequencing of full-length cDNAs [50], Chinese scientists officially participated in the Human Genome Project in 1999 and sequenced 1% of the human genome [9],[49]. Since then the Chinese genome centers have independently sequenced several large genomes including those of rice (Oryza sativa L. ssp. indica) [51], silkworm (Bombyx mori) [52], and numerous microbial genomes such as that of Thermoanaerobacter tengcongensis, a rod-shaped, gram-negative, anaerobic eubacterium isolated from a freshwater hot spring in Tengchong, China [53].
Data is a major driving force for bioinformatics—it demands new methods for data storage and analyses and motivated the computational discovery of biological patterns. In all genomic projects, bioinformatics teams played significant roles in data analysis. Databases were created to store the genomic data, for instance BGI-RIS [54] and SilkDB [55] for rice and silkworm genomic sequence data and related biological information, and ChickVD [56] for chicken genomic sequence variations. Methods and software packages were developed to analyze the huge amount of genomic data, including for instance BGF, an ab initio gene prediction method developed for the rice genome based on Hidden Markov Model (HMM) and dynamic programming [57], a nonsupervised gene prediction algorithm for bacterial and archaeal genomes [58], and a method for genome comparison using Gene Ontology (GO) with statistical testing [59].
Present Day: A Diversified Landscape
Since the end of the 1990s, bioinformatics in China has experienced significant growth fueled by not only the aforementioned Internet and genomic boom but also by official institutional support, steadily increasing government funding, and very importantly, a growing number of Chinese scientists returning to China after formal overseas training and research experience in bioinformatics and genomics. Today the field is characterized by a great variety of biological questions addressed and the close collaborations between computational scientists and bench biologists, with a full spectrum of focuses ranging from providing resources and services, developing databases and algorithms, to generating hypotheses and making biological discoveries.
Providing biological data resources and services is still an important part of bioinformatics. A good example is the Shanghai Center for Bioinformation Technology (http://www.scbit.org/), led by Yixue Li and founded in 2002. It has served as the central repository for biological data generated throughout Shanghai and neighboring areas. It has also provided extensive public online bioinformatics resources. Furthermore, many new primary databases with raw data and value-added secondary databases were created by Chinese scientists (Table 1), and many locally developed methods were implemented and incorporated into Web servers (Table 2). These databases and tools address a broad range of biological questions and are open to the entire international scientific community, and are enjoyed by a large worldwide user base.
Table 1. Examples of biological databases developed and maintained in China.
| Category | Name | Main content | URL | Reference |
| Genome resources | BGI-RIS | An integrated information resource and comparative analysis workbench for rice genomics | http://rise.genomics.org.cn/ | [54] |
| SilkDB | A knowledgebase for silkworm biology and genomics | http://silkworm.genomics.org.cn | [55] | |
| ChickVD | A sequence variation database for the chicken genome | http://chicken.genomics.org.cn | [56] | |
| The Z curve database | A graphic representation of genome sequences | http://tubic.tju.edu.cn/zcurve/ | [148] | |
| MED-Start | Predicted translation initiation sites in microbial genomes | http://ctb.pku.edu.cn/main/SheGroup/MED_Start.htm | [100] | |
| ProTISA | A comprehensive resource for translation initiation site annotation in prokaryotic genomes | http://mech.ctb.pku.edu.cn/protisa | [149] | |
| DoriC | A database of oriC regions in bacterial genomes | http://tubic.tju.edu.cn/doric/ | [150] | |
| Transcription regulation resources | DRTF | A database of rice transcription factors | http://drtf.cbi.pku.edu.cn/ | [97] |
| DATF | A database of Arabidopsis transcription factors | http://datf.cbi.pku.edu.cn | [98] | |
| NATsDB | A database of natural antisense transcripts identified in ten genomes | http://natsdb.cbi.pku.edu.cn/ | [151] | |
| NONCODE | An integrated knowledge database of noncoding RNAs | http://noncode.bioinfo.org.cn | [13],[152] | |
| NPInter | A database of noncoding RNA and protein interaction. | http://bioinfo.ibp.ac.cn/NPInter | [14] | |
| ATID | A collection of publicly available alternative translational initiation events | http://bioinfo.au.tsinghua.edu.cn/atie/ | [153] | |
| dbRES | A database for annotated RNA editing sites | http://bioinfo.au.tsinghua.edu.cn/dbRES/ | [154] | |
| PASDB | A collection of genes reported to be alternatively spliced in plants, spanning 44 plant species | http://pasdb.genomics.org.cn | [155] | |
| Protein resources | MPSS | An integrated database of protein annotations | http://www.scbit.org/mpss/ | [156] |
| SPD | A secreted protein database | http://spd.cbi.pku.edu.cn | [157] | |
| SynDB | A synapse protein database based on a synapse ontology | http://syndb.cbi.pku.edu.cn | [158] | |
| DBSub-Loc | Database of protein subcellular localizations | http://www.bioinfo.tsinghua.edu.cn/dbsubloc.html | [45] | |
| dbNEI | A database for neuro-endocrine-immune interaction. | http://bioinfo.au.tsinghua.edu.cn/dbNEIweb/ | [159] | |
| SPIDer | Saccharomyces protein–protein interaction database | http://cmb.bnu.edu.cn/SPIDer/index.html | [130] | |
| InterDom | A database of putative interacting protein domains for validating predicted protein interactions and complexes | http://InterDom.lit.org.sg | [160] |
Table 2. Examples of Web servers and software packages developed and maintained in China.
| Category | Name | Main Function | URL | Reference |
| Genome analysis tools | BGF | Prediction of genes in the rice genome | http://tlife.fudan.edu.cn/bgf/ | [57] |
| CVTree | A phylogenetic tree reconstruction tool based on whole genome sequences without alignment | http://cvtree.cbi.pku.edu.cn | [161] | |
| ZCURVE | A system for recognizing protein coding genes in bacterial and archaeal genomes | http://tubic.tju.edu.cn/Zcurve_B/ | [5] | |
| Zplotter online | A program to draw and manipulate the Z curve online based on a user's input DNA sequence | http://tubic.tju.edu.cn/zcurve/ | [148] | |
| GS-Finder | A program to find bacterial gene start sites with a self-training method | http://tubic.tju.edu.cn/GS-Finder/ | [162] | |
| GC-Profile | A Web-based tool for visualizing and analyzing the variation of GC content in genomic sequences | http://tubic.tju.edu.cn/GC-Profile/ | [163] | |
| FGF | A Web tool for Fishing Gene Family in a whole genome database | http://fgf.genomics.org.cn/ | [164] | |
| BPhyOG | An interactive server for genome-wide inference of bacterial phylogenies based on overlapping genes | http://cmb.bnu.edu.cn/BPhyOG/ | [165] | |
| SAPRED | Using new structural and sequence attributes and Support Vector Machine to predict possible disease association of single amino acid polymorphism | http://sapred.cbi.pku.edu.cn/ | [82] | |
| Expression regulation analysis tools | GBA | EST-based digital gene expression profiling | http://gba.cbi.pku.edu.cn | [166] |
| CEAS | An online server to analyze transcription factor binding sites based on ChIP-chip data | http://ceas.cbi.pku.edu.cn | [99] | |
| OTFBS | Over-represented Transcription Factor Binding Site Prediction Tool | http://www.bioinfo.tsinghua.edu.cn/7Ezhengjsh/OTFBS/index.html | [48] | |
| SVAP | Identification and expression analysis of alternatively spliced isoforms | http://svap.cbi.pku.edu.cn | ||
| CPC | Prediction of the protein-coding potential of transcripts using sequence features and support vector machine | http://cpc.cbi.pku.edu.cn | [107] | |
| RDfolder | A Web server for prediction of RNA secondary structure | http://rna.cbi.pku.edu.cn | [108] | |
| miRAS | A data processing system for miRNA expression profiling study | http://e-science.tsinghua.edu.cn/miras/ | [106] | |
| MiPred | Classification of real and pseudo microRNA precursors using random forest prediction model with combined features | http://www.bioinf.seu.edu.cn/miRNA/ | [103] | |
| RFRCDB-siRNA | Improved design of siRNAs by random forest regression model coupled with database searching | http://www.bioinf.seu.edu.cn/siRNA/index.htm | [167] | |
| Protein analysis tools | EasyGO | Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species | http://bioinformatics.cau.edu.cn/easygo/ | [168] |
| CTKPred | An SVM-based method for the prediction and classification of the cytokine superfamily | http://www.bioinfo.tsinghua.edu.cn/7Ehn/CTKPred/index.html | [169] | |
| GNBSL | A new integrative system to predict the subcellular location for Gram-negative bacteria proteins | http://166.111.24.5/webtools/GNBSL/index.htm | [127] | |
| MeMo | A Web tool for prediction of protein methylation modifications | http://www.bioinfo.tsinghua.edu.cn/∼tigerchen/memo/contact.html | [170] | |
| KOBAS | A Web-based platform for pathway identification | http://kobas.cbi.pku.edu.cn | [171] | |
| IntNetDB | An integrated protein–protein interaction network database generated by a probabilistic model | http://hanlab.genetics.ac.cn/IntNetDB.htm | [172] | |
| Platforms | BOD | A customizable bioinformatics on demand system accommodating multiple steps and parallel tasks | http://e-science.tsinghua.edu.cn/bod/index.jsp | [173] |
| ABCGrid | Application for Bioinformatics Computing Grid | http://abcgrid.cbi.pku.edu.cn | [174] |
Independently or in collaboration with bench scientists, bioinformatics scientists have played significant roles in important biological discoveries. In 2003, China and many other countries suffered from the severe acute respiratory syndrome (SARS) epidemic. A team of bioinformaticians quickly joined the Chinese SARS Molecular Epidemiology Consortium, led by Guo-Ping Zhao, to sequence and analyze SARS coronavirus genomic sequences derived from the early, middle, and late phases of the SARS epidemic as well as viral sequences from palm civets. Their work uncovered the molecular evolution of the SARS coronavirus and the viral invasion from animal to human [60],[61]. Another group proposed a mathematical model to estimate the evolution rate of the SARS coronavirus genome (0.16 base/day) and the time of the last common ancestor of the sequenced SARS strains (August or September of 2002) [62]. To identify potential anti-viral drug targets, proteomic and bioinformatic methods were used to investigate key SARS viral proteins including structural proteins [63], spike protein [64], and 3C-like proteinase [30]. Genomic packaging signals, which may be used to design antisense RNA and RNA interfere (RNAi) drugs treating SARS, were predicted by comparative genomics methods [65]. The three-dimensional structure of 3C-like proteinase was constructed by homology modeling, based on which virtual screening of chemical databases was performed to search for potential inhibitors [31]. Chinese SARS patients were statistically studied to elucidate the association of symptom combinations with different outcome and therapeutic effects [66] and the association between mannose-binding lectin gene polymorphisms and susceptibility to SARS virus infection [67].
The large Chinese population provided invaluable resources for genetic studies. Patient and control groups were genotyped to establish associations between genetic variations and susceptibility to diseases such as esophageal squamous cell carcinoma [68], hepatocellular carcinoma in a particularly high-risk region of China [69], hypertension [70], and coronary heart disease [71]. A link between an Asian-enriched SNP in human sialidase and severe adverse reactions to the anti-viral drug Tamiflu (oseltamivir carboxylate) was proposed and confirmed by bioinformatic methods and enzymatic assays [72]. A number of interesting works studied the migration history of Chinese and East Asian populations by the sequencing and phylogentic analysis of Y-chromosomes [73]–[77]. Recently the Chinese HapMap Consortium participated in the international HapMap project to genotype the Chinese Han population [78]. As more and more genetic data were generated, new algorithms continued to be developed, for instance a Java-based method to analyze linkage disequilibrium [79], software for haplotype block partition and htSNPs selection [80], a method to detect human recombination hotspots based on a multiple-hotspot model and an approximate log-likelihood ratio test [81], and an SVM-based method that used new sequence and structure features to predict the possible disease association of nonsynonmous SNPs with increased accuracy [82].
The increasing number of completely and partially sequenced genomes have enabled the study of the evolution of genomes and gene families. Of particular interest to many Chinese scientists is the study of the evolution of plants, largely due to the country's longstanding strength in plant biology and partly due to its dedication to the sequencing of the rice genome. Two groups had studied the collinearity of duplicated genes along rice chromosomes and discovered that the rice genome had undergone an ancient whole-genome duplication 70 million years ago and a recent segmental duplication involving Chromosomes 11 and 12 five million years ago [83],[84]. Their findings settled previous disputes over whether rice was an ancient aneuploid or an ancient polyploidy, bringing the two contradictory sides to agreement in the form of a graciously co-authored Commentary [85]. In addition to whole genomes, the evolution of many important plant gene families was also studied in detail, such as transcription factor families [86] and enzymes [87]. A group studied the origination of new genes in rice and found a surprisingly high number of retrogenes as well as chimeric genes originated by retroposition, suggesting that retroposition is an important mechanism that governs gene evolution in rice and other grass species [88]. Another group designed a theoretical model based on the molecular clock to test the hypotheses of hybridization as an evolutionary mechanism [89]. Several groups had proposed interesting theories about the genetic code (amino acid codons and stop codons) [36],[37],[40],[90],[91].
When and where genes in a genome are transcribed and translated and how this expression of genes and proteins is regulated are important biological questions that had interested numerous scientists for decades. A first step to study this important question is to measure and compare the expression level of mRNAs. Several Chinese groups had developed algorithms for microarray experiments including designing probes [92], processing raw data by an integrated pipeline [93], detecting differentially expressed genes by relative entropy [94], identifying clusters of co-expressed genes by Hidden Markov Models [47], and finding disease-related genes by an ensemble decision approach [95]. Transcription factors bind to upstream regions of genes to regulate their transcription. About 2,000 transcription factors (∼10% of the genomes) were identified in each of the three sequenced plant genomes, Arabidopsis, rice, and populus, by combining similarity and motif-based approaches [96]–[98]. A new method was developed to identify potential transcription factor binding sites in the upstream regions of genes by searching for over-represented cis-elements with Position Weight Matrix–based similarity scores [48]. Another method uncovered the patterns of conservation, genomic distribution, and co-factor binding of transcription factor binding sites given ChIP-chip tiling array data [99]. A new four-component statistical model was proposed to improve accuracy of the identification of translation initiation sites in microbial genomes [100]. Alternative splicing adds great variety to the proteome. A new method, named Splicing Variant Analysis Platform (SVAP, http://svap.cbi.pku.edu.cn), could identify alternatively spliced isoforms from EST data ten times faster than other existing methods. Increased prediction accuracy of splice sites was achieved by using quadratic discriminant analysis with diversity measure or by introducing a competition mechanism of splice sites selection [22],[101]. The impact of very short alternative splicing on protein structures and functions was studied [21]. An interesting work identified 2,695 newly evolved exons in rodents and calculated the new exon origination rate at about 2.71×10−3 per gene per million years; they suggested that most new exons might originate through “exonization” of intronic sequences and appear to be alternative exons that are expressed at low levels [102].
More recently, several Chinese bioinformatics groups had made progress in studying the regulation of transcription and translation by microRNAs, noncoding RNAs, and natural antisense transcripts. New computational methods were developed to identify precursor and mature microRNAs by sequence features, hair-pin structural features, and cross-species conservations [18],[20],[103],[104]. MicroRNAs in Chlamydomonas reinhardtii, a unicellular green alga, were identified in reads obtained by the highly parallel 454 pyrosequencing technology; the results suggested that the miRNA pathway may be an ancient mechanism of gene regulation that evolved prior to the emergence of multicellularity [105]. To study the expression profile of microRNAs, two methods were developed using data from EST sequencing and SAGE-based total RNA clones, respectively [19],[106]. Longer noncoding RNAs were also studied extensively. Hundreds of novel noncoding RNAs were identified in C. elegans first by a cloning strategy and then by a whole-genome tiling array technology [11],[12]. The expression of intron-encoded noncoding RNAs was profiled using a custom-designed microarray combining noncoding RNAs and their host genes, and many noncoding RNAs were found to be independently transcribed with ncRNA-specific promoter elements [10]. To facilitate future studies, new databases were created to catalogue noncoding RNAs and noncoding RNA–protein interactions [13],[14], and new algorithms were developed to accurately predict the protein-coding potential of a given transcript and the secondary structure of an RNA [107],[108]. Many RNAs function through an antisense mechanism. Several Chinese groups have identified cis- and trans-natural antisense transcripts at the whole-genome scale and found that they are highly abundant and have interesting features of function, expression, and evolution [109]–[111].
Research at the protein level is gaining increasing momentum thanks in part to China's recognition of protein sciences as one of the major research areas to promote and support in the next 20 years. Progress continued to be made in the prediction of protein secondary and tertiary structures [46], [112]–[116]. An interesting new method achieved rapid multiple alignment of protein three-dimensional structures by the use of conformational letters, which were defined as discretized states of 3D segmental structural states [117]. Several effective scoring functions and algorithms were developed for the docking of protein with small compounds [28],[29],[33],[34]. In the field of proteomics, China joined the Human Proteome Organization (HUPO) with a special focus on the study of the human liver proteome, led by Fuchu He [118],[119]. The fetal and adult liver proteomes were extensively profiled using mass spectrometry technologies [120]–[122]. Driven by the need to identify peptides and proteins from mass spectrometry data, new algorithms were developed with increased accuracy [15], [16], [123]–[126]. Computational prediction of subcellular localizations of proteins is an important problem in proteomics, and several good methods had been developed in China using a variety of classification methods such as Support Vector Machine [44],[45], fuzzy k-NN method [23], and an integrated meta-approach [127]. Another method predicted the protein submitochondria locations by hybridizing pseudo-amino acid composition with physicochemical features of segmented sequence [24]. Protein–protein interactions were under intense study. They were predicted from sequence features [128], Gene Ontology annotations [129],[130], and integration of 27 heterogeneous genomic, proteomic, and functional annotation datasets [27]. The protein–protein interfaces were studied in detail, such as the potential of mean force used in the ranking of the binding energies of different protein–protein complexes [25] and the hydrogen bond, hydrophobic and vdW interactions used to estimate the individual contribution of each interfacial residue to the binding [26]. A recent paper reported the design of nonnatural protein–protein interaction pairs by key residues grafting [27]. The topology of the protein–protein interaction network in yeast was analyzed using a spectral method derived from graph theory to uncover hidden topological structures such as quasi-cliques and quasi-bipartites [131], and visualized as a clustering tree [17]. Metabolic pathways were identified using the KEGG Othology as an alternative controlled vocabulary [132].
Interesting progress had been made in the systems biology studies of biological networks. The global dynamical properties and stabilities of the networks had been studied. In particular, the yeast cell-cycle network and the Drosophila segment polarity network appeared to be highly robust [133],[134]. The protein interaction networks of Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster were found to have a scale-free and high-degree clustering nature with a small-world property with similar diameter at 4–5 [135]. Key proteins, subgraphs, and modules were identified and analyzed in the signal transduction networks [136], transcriptional regulation networks in yeast [137], and protein–protein interaction networks during fruitfly and human brain aging [138],[139]. Simulation analysis of the energy metabolism network in mammalian myocardia revealed that the systemic states of metabolic networks did not always remain optimal, but might become suboptimal when a transient perturbation occurs [140]. Finally, the dynamic properties of the arachidonic acid metabolic network which includes several targets for anti-inflammatory drugs was analyzed using ordinary differential equations, and the flux balance in the network was found to be important for efficient and safe drug design [141].
The bioinformatics discipline has grown too broad to be reviewed comprehensively in any one article. Many works cannot be covered here because of the space limitations. Just to give two examples of interesting areas of bioinformatics research that we did not adequately review here: first, a small group of researchers in China have made progress in computational neurobiology, including proposing a computational model as a neurodecoder based on synchronous oscillation in the visual cortex [142] and a simulation study on the Ca2+-independent but voltage-dependent exocytosis and endocytosis in dorsal root ganglion neurons [143]; second, many groups have developed databases and methods of imaging, such as the digital image datasets of Chinese Visible Human (a male and a female) [144], a high-resolution anatomical rat atlas [145], and methods for optical molecular imaging [146].
Education: A New Generation of Locally Trained Bioinformaticians
Bioinformatics talent in China is reaching a critical mass. In addition to local bioinformatics scientists who converted from other disciplines and scientists who returned to China after formal training overseas, two new talent pools are forming. First, more and more scientists from other countries, especially European countries, are drawn to work in China by the improved research and funding environment. A good example is the Max Planck–Chinese Academy of Science Partners Institute in Computational Biology in Shanghai which employs a number of European scientists and plays key roles in facilitating international collaborations.
Second, a growing significant talent pool that has emerged in the past few years is the new generation of young bioinformatics scientists trained in local bioinformatics degree programs. Take the Center for Bioinformatics at Peking University as an example. By summer 2007, it had graduated 24 PhDs and four Master's degrees in bioinformatics. Although a slight majority of the graduates (15 out of 28, or 54%) still chose to pursue postdoctoral positions overseas, the remaining had decided to stay in China, with nine working in academia and four in industry. Many other Chinese universities are now offering bioinformatics degree programs at the PhD, Master's, and even Bachelor's levels (see list in Table 3). In addition to formal degree programs, many university courses and ad hoc workshops have been offered to train bench biologists on how to use bioinformatic databases and tools.
Table 3. Examples of bioinformatics training programs in China.
| City | University/Academy | Affiliation | Degree |
| Beijing | Peking University | Center for Bioinformatics, College of Life Sciences; Center for Theoretical Biology | PhD |
| Beijing | Tsinghua University | Department of Biological Sciences and Biotechnology, Institute of Bioinformatics; Department of Automation | PhD |
| Beijing | Chinese Academy of Sciences | Beijing Institute of Genomics; Center of Systems Biology, Institute of Biophysics; Center of Molecular Systems Biology, Institute of Genetics and Developmental Biology | PhD |
| Beijing | China Agricultural University | College of Biological Sciences | PhD, Master's |
| Beijing | China Pharmaceutical University | School of Life Science and Technology | PhD, Master's |
| Beijing | Beijing Normal University | College of Life Sciences, Laboratory of Computational Molecular Biology | Master's |
| Beijing | The Academy of Military Medical Science | Institute of Basic Medical Science | Master's |
| Baoding | Hebei University | College of Life Science | Master's |
| Chengdu | Sichuan University | School of Life Sciences | PhD, Master's |
| Chengdu | University of Electronic Science and Technology of China | School of Life Science and Technology | PhD, Master's |
| Chongqing | Chongqing University of Post and Telecommunications | School of Bioinformatics | Bachelor's |
| Guangzhou | Sun Yat-Sen University | School of Life Sciences | PhD, Master's |
| Hangzhou | Zhejiang University | James D. Watson Institute of Genome Sciences, College of Life Sciences | PhD, Master's |
| Harbin | Harbin Medical University | Department of Bioinformatics | Master's |
| Hefei | University of Science and Technology of China | School of Life Sciences | PhD, Master's |
| Kunming | Yunnan University | School of Life Sciences | PhD, Master's |
| Lanzhou | Lanzhou University | School of Life Sciences | PhD, Master's |
| Nanjing | Nanjing University | School of Life Science | PhD |
| Nanjing | Southeast University | State Key Laboratory of Bioelectronics, School of Biological Science & Medical Engineering | PhD, Master's |
| Shanghai | Fudan University | Institute of Biodiversity Science, School of Life Sciences | PhD, Master's |
| Shanghai | Shanghai Institute for Biological Sciences | Key Laboratory of Systems Biology | PhD |
| Shanghai | Shanghai Jiao Tong University | Department of Biomedical Engineering, College of Life Science and Biotechnology | PhD, Master's |
| Shanghai | Shanghai University | School of Life Sciences | Master's |
| Shanghai | East China Normal University | School of Life Sciences | Master's |
| Shanghai | Tongji University | School of Life Science and Technology | PhD, Master's, Bachelor's |
| Tianjin | Nankai University | College of Life Sciences | PhD, Master's |
| Tianjin | Tianjin University | Tianjin University BioInformatics Centre, School of Science | PhD, Master's |
| Xiamen | Xiamen University | Department of Chemistry | Master's |
| Wuhan | Huazhong University of Science and Technology | School of Life Science and Technology | PhD, Master's |
| Yangling | Northwest A&F university | School of Life Sciences | PhD, Master's |
| Zibo | Shandong University of Technology | School of Life Sciences | Master's |
The Future: Promises and Challenges
Today bioinformatics research in China still lags behind the best in the world. However, it is catching up quickly, and several positive factors suggest a bright future. First, the aforementioned critical mass of bioinformaticians will be a driving force for the future development of bioinformatics in China. Second, life science research at large is gaining momentum in China and progressing quickly. Collaborating closely with talented local and international biologists, bioinformatics scientists will have access to new and numerous biological data to analyze, important biological questions to solve, and bench experimental laboratories capable of validating their predictions and hypotheses. Third, China's total funding for scientific research has been steadily increasing in the past five years, reaching 1.40% of her GDP in 2006 and a total annual R&D expenditure of 300 billion Yuan (or US$38.5 billion) [147]. The recent national goal for R&D growth has been set to an incremental annual increase of 20%. Last but not least, China is investing heavily in its computing infrastructure, including the China Grid initiated by the Ministry of Education and the China National Grid (CNGrid) initiated by Chinese Academy of Sciences, both supported largely by the Ministry of Science and Technology. Bioinformatics scientists benefit as welcomed major users of these computing infrastructures.
Despite the promising future, current challenges remain. Scientists returning from overseas often face a salary reduction, reverse culture shock, and a different funding system and application process. China lacks the equivalent of the Biomedical Information Science and Technology Initiative Consortium (BISTIC) launched at the NIH in 2000. BISTIC consists of senior-level representatives from each of the NIH institutes and centers plus representatives of other federal agencies interested in biocomputing. It plays important roles in coordinating sustained funding and other support for bioinformatics. No similar initiatives exist in China yet. There is also no official professional bioinformatics society in China. More noticeable perhaps is the lack of a Chinese national center for bioinformation, the equivalent of the US National Center for Biotechnology Information (NCBI) that is the central repository and information hub for biological data and tools. As China is generating a large and growing amount of biological data, a Chinese national center with a dedicated budget and staff could more effectively collect and store the local data and exchange it with the international scientific community. Journals and other publications in Chinese could also be collected locally and shared internationally. A Chinese national center could also offer better and faster online bioinformatics service to the large user base in China and neighboring Euro–Asian countries.
Concluding Remarks
Bioinformatics in China has come a long way since the early years. It has benefited from developments in the rest of the world and has made its own distinct contributions. As bioinformatics in China continues to grow, we expect to see an increasing exchange of data, tools, and talent between China and other countries. We believe the future is bright for bioinformatics in China and worldwide.
Acknowledgments
We thank Chuan-Yun Li and Xiaomo Li for help with compiling the references and tables. We thank Professors Chun-Ting Zhang, Ying Xu, Xiaole Liu, Heping Cheng, Manyuan Long, Louis Tao, Xiaocheng Gu, Jingchu Luo, and the editors for insightful suggestions.
References
- 1.Yu ZG, Anh V, Lau KS. Measure representation and multifractal analysis of complete genomes. Phys Rev E Stat Nonlin Soft Matter Phys. 2001;64:031903. doi: 10.1103/PhysRevE.64.031903. [DOI] [PubMed] [Google Scholar]
- 2.Qi J, Wang B, Hao BI. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J Mol Evol. 2004;58:1–11. doi: 10.1007/s00239-003-2493-7. [DOI] [PubMed] [Google Scholar]
- 3.Zhang CT, Chou KC. An optimization approach to predicting protein structural class from amino acid composition. Protein Sci. 1992;1:401–408. doi: 10.1002/pro.5560010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R, Zhang CT. Z curves, an intutive tool for visualizing and analyzing the DNA sequences. J Biomol Struct Dyn. 1994;11:767–782. doi: 10.1080/07391102.1994.10508031. [DOI] [PubMed] [Google Scholar]
- 5.Guo FB, Ou HY, Zhang CT. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 2003;31:1780–1789. doi: 10.1093/nar/gkg254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang CT, Wang J. Recognition of protein coding genes in the yeast genome at better than 95% accuracy based on the Z curve. Nucleic Acids Res. 2000;28:2804–2814. doi: 10.1093/nar/28.14.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang CT, Zhang R. An isochore map of the human genome based on the Z curve method. Gene. 2003;317:127–135. doi: 10.1016/s0378-1119(03)00665-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang R, Zhang CT. Identification of replication origins in archaeal genomes based on the Z-curve method. Archaea. 2005;1:335–346. doi: 10.1155/2005/509646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muzny DM, Scherer SE, Kaul R, Wang J, Yu J, et al. The DNA sequence, annotation and analysis of human chromosome 3. Nature. 2006;440:1194–1198. doi: 10.1038/nature04728. [DOI] [PubMed] [Google Scholar]
- 10.He H, Cai L, Skogerbo G, Deng W, Liu T, et al. Profiling Caenorhabditis elegans non-coding RNA expression with a combined microarray. Nucleic Acids Res. 2006;34:2976–2983. doi: 10.1093/nar/gkl371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng W, Zhu X, Skogerbo G, Zhao Y, Fu Z, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He H, Wang J, Liu T, Liu XS, Li T, et al. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res. 2007;17:1471–1477. doi: 10.1101/gr.6611807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He S, Liu C, Skogerbo G, Zhao H, Wang J, et al. NONCODE v2.0: decoding the non-coding. Nucleic Acids Res. 2008;36:D170–172. doi: 10.1093/nar/gkm1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu T, Wang J, Liu C, Zhang Y, Shi B, et al. NPInter: the noncoding RNAs and protein related biomacromolecules interaction database. Nucleic Acids Res. 2006;34:D150–152. doi: 10.1093/nar/gkj025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C, Lin Y, Sun S, Cai J, Zhang J, et al. An iterative algorithm to quantify factors influencing peptide fragmentation during tandem mass spectrometry. J Bioinform Comput Biol. 2007;5:297–311. doi: 10.1142/s0219720007002643. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Sun S, Zhu X, Chang S, Liu X, et al. A novel scoring schema for peptide identification by searching protein sequence databases using tandem mass spectrometry data. BMC Bioinformatics. 2006;7:222. doi: 10.1186/1471-2105-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H, Zhu X, Liu H, Skogerbo G, Zhang J, et al. The interactome as a tree–an attempt to visualize the protein-protein interaction network in yeast. Nucleic Acids Res. 2004;32:4804–4811. doi: 10.1093/nar/gkh814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu J, Fu H, Zhang X, Li Y. Identifications of conserved 7-mers in 3-UTRs and microRNAs in Drosophila. BMC Bioinformatics. 2007;8:432. doi: 10.1186/1471-2105-8-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu J, He T, Pei Y, Li F, Wang X, et al. Primary transcripts and expressions of mammal intergenic microRNAs detected by mapping ESTs to their flanking sequences. Mamm Genome. 2006;17:1033–1041. doi: 10.1007/s00335-006-0007-9. [DOI] [PubMed] [Google Scholar]
- 20.Xue C, Li F, He T, Liu GP, Li Y, et al. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen F, Li F, Xia H, Lu X, Zhang X, et al. The impact of very short alternative splicing on protein structures and functions in the human genome. Trends Genet. 2004;20:232–236. doi: 10.1016/j.tig.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Xia H, Bi J, Li Y. Identification of alternative 5′/3′ splice sites based on the mechanism of splice site competition. Nucleic Acids Res. 2006;34:6305–6313. doi: 10.1093/nar/gkl900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Li Y. Prediction of protein subcellular locations using fuzzy k-NN method. Bioinformatics. 2004;20:21–28. doi: 10.1093/bioinformatics/btg366. [DOI] [PubMed] [Google Scholar]
- 24.Du P, Li Y. Prediction of protein submitochondria locations by hybridizing pseudo-amino acid composition with various physicochemical features of segmented sequence. BMC Bioinformatics. 2006;7:518. doi: 10.1186/1471-2105-7-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang L, Gao Y, Mao F, Liu Z, Lai L. Potential of mean force for protein-protein interaction studies. Proteins. 2002;46:190–196. doi: 10.1002/prot.10031. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y, Wang R, Lai L. Structure-based method for analyzing protein-protein interfaces. J Mol Model. 2004;10:44–54. doi: 10.1007/s00894-003-0168-3. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Zhu X, Liang H, Cao A, Chang Z, et al. Nonnatural protein-protein interaction-pair design by key residues grafting. Proc Natl Acad Sci U S A. 2007;104:5330–5335. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pei J, Wang Q, Liu Z, Li Q, Yang K, et al. PSI-DOCK: towards highly efficient and accurate flexible ligand docking. Proteins. 2006;62:934–946. doi: 10.1002/prot.20790. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Lai L. Pocket v.2: further developments on receptor-based pharmacophore modeling. J Chem Inf Model. 2006;46:2684–2691. doi: 10.1021/ci600246s. [DOI] [PubMed] [Google Scholar]
- 30.Fan K, Wei P, Feng Q, Chen S, Huang C, et al. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J Biol Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Huang C, Fan K, Wei P, Chen H, et al. Virtual screening of novel noncovalent inhibitors for SARS-CoV 3C-like proteinase. J Chem Inf Model. 2005;45:10–17. doi: 10.1021/ci049809b. [DOI] [PubMed] [Google Scholar]
- 32.Zhou L, Liu Y, Zhang W, Wei P, Huang C, et al. Isatin compounds as noncovalent SARS coronavirus 3C-like protease inhibitors. J Med Chem. 2006;49:3440–3443. doi: 10.1021/jm0602357. [DOI] [PubMed] [Google Scholar]
- 33.Wang R, Liu L, Lai L, Tang Y. SCORE: A New Empirical Method for Estimating the Binding Affinity of a Protein-Ligand Complex. Journal of Molecular Modeling. 1998;4:379–394. [Google Scholar]
- 34.Wang R, Gao Y, Lai L. LigBuilder: A Multi-Purpose Program for Structure-Based Drug Design. Journal of Molecular Modeling. 2004;6:498–516. [Google Scholar]
- 35.Tang YZ, Chen WZ, Wang CX, Shi YY. Constructing the suitable initial configuration of the membrane-protein system in molecular dynamics simulations. Eur Biophys J. 1999;28:478–488. doi: 10.1007/s002490050230. [DOI] [PubMed] [Google Scholar]
- 36.Luo L, Li X. Coding rules for amino acids in the genetic code: the genetic code is a minimal code of mutational deterioration. Orig Life Evol Biosph. 2002;32:23–33. doi: 10.1023/a:1013963505140. [DOI] [PubMed] [Google Scholar]
- 37.Luo L, Li X. Construction of genetic code from evolutionary stability. Biosystems. 2002;65:83–97. doi: 10.1016/s0303-2647(02)00009-6. [DOI] [PubMed] [Google Scholar]
- 38.Liaofu L, Lu T. Fractal dimension of Nucleic acid sequences and its relation to evolutionary level. Chinese Physics Letters. 1988;5:421–424. [Google Scholar]
- 39.Luo L, Lee W, Jia L, Ji F, Tsai L. Statistical correlation of nucleotides in a DNA sequence. Physical Review. 1998;58:861–871. [Google Scholar]
- 40.Luo LF. The degeneracy rule of genetic code. Orig Life Evol Biosph. 1988;18:65–70. doi: 10.1007/BF01808781. [DOI] [PubMed] [Google Scholar]
- 41.Ding DF, Qian J, Feng ZK. A differential geometric treatment of protein structure comparison. Bull Math Biol. 1994;56:923–943. doi: 10.1007/BF02458274. [DOI] [PubMed] [Google Scholar]
- 42.Xie T, Chen J, Ding DF. An Evolutionary Trace Method for Functional Prediction of Genomes. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 1999;31:433–439. [PubMed] [Google Scholar]
- 43.Wang LM, Zhang Q, Zhu W, He C, Lu CL, et al. Identification of the key amino acids of glial cell line-derived neurotrophic factor family receptor alpha1 involved in its biological function. J Biol Chem. 2004;279:109–116. doi: 10.1074/jbc.M306287200. [DOI] [PubMed] [Google Scholar]
- 44.Hua S, Sun Z. Support vector machine approach for protein subcellular localization prediction. Bioinformatics. 2001;17:721–728. doi: 10.1093/bioinformatics/17.8.721. [DOI] [PubMed] [Google Scholar]
- 45.Guo T, Hua S, Ji X, Sun Z. DBSubLoc: database of protein subcellular localization. Nucleic Acids Res. 2004;32:D122–124. doi: 10.1093/nar/gkh109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hua S, Sun Z. A novel method of protein secondary structure prediction with high segment overlap measure: support vector machine approach. J Mol Biol. 2001;308:397–407. doi: 10.1006/jmbi.2001.4580. [DOI] [PubMed] [Google Scholar]
- 47.Ji X, Li-Ling J, Sun Z. Mining gene expression data using a novel approach based on hidden Markov models. FEBS Lett. 2003;542:125–131. doi: 10.1016/s0014-5793(03)00363-6. [DOI] [PubMed] [Google Scholar]
- 48.Zheng J, Wu J, Sun Z. An approach to identify over-represented cis-elements in related sequences. Nucleic Acids Res. 2003;31:1995–2005. doi: 10.1093/nar/gkg287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 50.Qiang B. Human genome research in China. J Mol Med. 2004;82:214–222. doi: 10.1007/s00109-003-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu J, Wong GK, Liu S, Wang J, Yang H. A comprehensive crop genome research project: the Superhybrid Rice Genome Project in China. Philos Trans R Soc Lond B Biol Sci. 2007;362:1023–1034. doi: 10.1098/rstb.2007.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia Q, Zhou Z, Lu C, Cheng D, Dai F, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 53.Bao Q, Tian Y, Li W, Xu Z, Xuan Z, et al. A complete sequence of the T. tengcongensis genome. Genome Res. 2002;12:689–700. doi: 10.1101/gr.219302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao W, Wang J, He X, Huang X, Jiao Y, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Xia Q, He X, Dai M, Ruan J, et al. SilkDB: a knowledgebase for silkworm biology and genomics. Nucleic Acids Res. 2005;33:D399–402. doi: 10.1093/nar/gki116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, He X, Ruan J, Dai M, Chen J, et al. ChickVD: a sequence variation database for the chicken genome. Nucleic Acids Res. 2005;33:D438–441. doi: 10.1093/nar/gki092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Liu J-S, Xu Z, Jin J, Fang L, et al. Test data sets and evaluation of gene prediction programs on the rice genome. Journal of Computer Science and Technology. 2006;20:446–453. [Google Scholar]
- 58.Zhu H, Hu GQ, Yang YF, Wang J, She ZS. MED: a new non-supervised gene prediction algorithm for bacterial and archaeal genomes. BMC Bioinformatics. 2007;8:97. doi: 10.1186/1471-2105-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai Z, Mao X, Li S, Wei L. Genome comparison using Gene Ontology (GO) with statistical testing. BMC Bioinformatics. 2006;7:374. doi: 10.1186/1471-2105-7-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chinese SMEC. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 61.Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu H, Zhao Y, Zhang J, Wang Y, Li W, et al. Date of origin of the SARS coronavirus strains. BMC Infect Dis. 2004;4:3. doi: 10.1186/1471-2334-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ying W, Hao Y, Zhang Y, Peng W, Qin E, et al. Proteomic analysis on structural proteins of Severe Acute Respiratory Syndrome coronavirus. Proteomics. 2004;4:492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu XJ, Luo C, Lin JC, Hao P, He YY, et al. Putative hAPN receptor binding sites in SARS_CoV spike protein. Acta Pharmacol Sin. 2003;24:481–488. [PubMed] [Google Scholar]
- 65.Qin L, Xiong B, Luo C, Guo ZM, Hao P, et al. Identification of probable genomic packaging signal sequence from SARS-CoV genome by bioinformatics analysis. Acta Pharmacol Sin. 2003;24:489–496. [PubMed] [Google Scholar]
- 66.Li S, Wang R, Zhang Y, Zhang X, Layon AJ, et al. Symptom combinations associated with outcome and therapeutic effects in a cohort of cases with SARS. Am J Chin Med. 2006;34:937–947. doi: 10.1142/S0192415X06004417. [DOI] [PubMed] [Google Scholar]
- 67.Zhang H, Zhou G, Zhi L, Yang H, Zhai Y, et al. Association between mannose-binding lectin gene polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus infection. J Infect Dis. 2005;192:1355–1361. doi: 10.1086/491479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hao B, Wang H, Zhou K, Li Y, Chen X, et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64:4378–4384. doi: 10.1158/0008-5472.CAN-04-0372. [DOI] [PubMed] [Google Scholar]
- 69.Chen X, Wang H, Xie W, Liang R, Wei Z, et al. Association of CYP1A2 genetic polymorphisms with hepatocellular carcinoma susceptibility: a case-control study in a high-risk region of China. Pharmacogenet Genomics. 2006;16:219–227. doi: 10.1097/01.fpc.0000194424.20393.c6. [DOI] [PubMed] [Google Scholar]
- 70.Gu D, Su S, Ge D, Chen S, Huang J, et al. Association study with 33 single-nucleotide polymorphisms in 11 candidate genes for hypertension in Chinese. Hypertension. 2006;47:1147–1154. doi: 10.1161/01.HYP.0000219041.66702.45. [DOI] [PubMed] [Google Scholar]
- 71.Su S, Chen S, Zhao J, Huang J, Wang X, et al. Plasminogen activator inhibitor-1 gene: selection of tagging single nucleotide polymorphisms and association with coronary heart disease. Arterioscler Thromb Vasc Biol. 2006;26:948–954. doi: 10.1161/01.ATV.0000204731.17646.f2. [DOI] [PubMed] [Google Scholar]
- 72.Li CY, Yu Q, Ye ZQ, Sun Y, He Q, et al. A nonsynonymous SNP in human cytosolic sialidase in a small Asian population results in reduced enzyme activity: potential link with severe adverse reactions to oseltamivir. Cell Res. 2007;17:357–362. doi: 10.1038/cr.2007.27. [DOI] [PubMed] [Google Scholar]
- 73.Ke Y, Su B, Song X, Lu D, Chen L, et al. African origin of modern humans in East Asia: a tale of 12,000 Y chromosomes. Science. 2001;292:1151–1153. doi: 10.1126/science.1060011. [DOI] [PubMed] [Google Scholar]
- 74.Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, et al. Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet. 2005;77:408–419. doi: 10.1086/444436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qian Y, Qian B, Su B, Yu J, Ke Y, et al. Multiple origins of Tibetan Y chromosomes. Hum Genet. 2000;106:453–454. doi: 10.1007/s004390000259. [DOI] [PubMed] [Google Scholar]
- 76.Zhang F, Su B, Zhang YP, Jin L. Genetic studies of human diversity in East Asia. Philos Trans R Soc Lond B Biol Sci. 2007;362:987–995. doi: 10.1098/rstb.2007.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng W, Shi B, He X, Zhang Z, Xu J, et al. Evolution and migration history of the Chinese population inferred from Chinese Y-chromosome evidence. J Hum Genet. 2004;49:339–348. doi: 10.1007/s10038-004-0154-3. [DOI] [PubMed] [Google Scholar]
- 78.Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks LD, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding K, Zhou K, He F, Shen Y. LDA–a java-based linkage disequilibrium analyzer. Bioinformatics. 2003;19:2147–2148. doi: 10.1093/bioinformatics/btg276. [DOI] [PubMed] [Google Scholar]
- 80.Ding K, Zhang J, Zhou K, Shen Y, Zhang X. htSNPer1.0: software for haplotype block partition and htSNPs selection. BMC Bioinformatics. 2005;6:38. doi: 10.1186/1471-2105-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J, Zhang MQ, Zhang X. A new method for detecting human recombination hotspots and its applications to the HapMap ENCODE data. Am J Hum Genet. 2006;79:628–639. doi: 10.1086/508066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye ZQ, Zhao SQ, Gao G, Liu XQ, Langlois RE, et al. Finding new structural and sequence attributes to predict possible disease association of single amino acid polymorphism (SAP). Bioinformatics. 2007;23:1444–1450. doi: 10.1093/bioinformatics/btm119. [DOI] [PubMed] [Google Scholar]
- 83.Wang X, Shi X, Hao B, Ge S, Luo J. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol. 2005;165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- 84.Yu J, Wang J, Lin W, Li S, Li H, et al. The Genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paterson AH, Bowers JE, Van de Peer Y, Vandepoele K. Ancient duplication of cereal genomes. New Phytol. 2005;165:658–661. doi: 10.1111/j.1469-8137.2005.01347.x. [DOI] [PubMed] [Google Scholar]
- 86.Xiong Y, Liu T, Tian C, Sun S, Li J, et al. Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol. 2005;59:191–203. doi: 10.1007/s11103-005-6503-6. [DOI] [PubMed] [Google Scholar]
- 87.Yang J, Gu H, Yang Z. Likelihood analysis of the chalcone synthase genes suggests the role of positive selection in morning glories (Ipomoea). J Mol Evol. 2004;58:54–63. doi: 10.1007/s00239-003-2525-3. [DOI] [PubMed] [Google Scholar]
- 88.Wang W, Zheng H, Fan C, Li J, Shi J, et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sang T, Zhong Y. Testing hybridization hypotheses based on incongruent gene trees. Syst Biol. 2000;49:422–434. doi: 10.1080/10635159950127321. [DOI] [PubMed] [Google Scholar]
- 90.Zhang C-T, Zhang R. Analysis of distribution of bases in the coding sequences by a digrammatic technique. Nucleic Acids Research. 1991;19:6313–6317. doi: 10.1093/nar/19.22.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun J, Chen M, Xu J, Luo J. Relationships among stop codon usage bias, its context, isochores, and gene expression level in various eukaryotes. J Mol Evol. 2005;61:437–444. doi: 10.1007/s00239-004-0277-3. [DOI] [PubMed] [Google Scholar]
- 92.Li W, Ying X. Mprobe 2.0: computer-aided probe design for oligonucleotide microarray. Appl Bioinformatics. 2006;5:181–186. doi: 10.2165/00822942-200605030-00006. [DOI] [PubMed] [Google Scholar]
- 93.Wang X, He H, Li L, Chen R, Deng XW, et al. NMPP: a user-customized NimbleGen microarray data processing pipeline. Bioinformatics. 2006;22:2955–2957. doi: 10.1093/bioinformatics/btl525. [DOI] [PubMed] [Google Scholar]
- 94.Yan X, Deng M, Fung WK, Qian M. Detecting differentially expressed genes by relative entropy. J Theor Biol. 2005;234:395–402. doi: 10.1016/j.jtbi.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 95.Li X, Rao S, Wang Y, Gong B. Gene mining: a novel and powerful ensemble decision approach to hunting for disease genes using microarray expression profiling. Nucleic Acids Res. 2004;32:2685–2694. doi: 10.1093/nar/gkh563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu QH, Guo AY, Gao G, Zhong YF, Xu M, et al. DPTF: a database of poplar transcription factors. Bioinformatics. 2007;23:1307–1308. doi: 10.1093/bioinformatics/btm113. [DOI] [PubMed] [Google Scholar]
- 97.Gao G, Zhong Y, Guo A, Zhu Q, Tang W, et al. DRTF: a database of rice transcription factors. Bioinformatics. 2006;22:1286–1287. doi: 10.1093/bioinformatics/btl107. [DOI] [PubMed] [Google Scholar]
- 98.Guo A, He K, Liu D, Bai S, Gu X, et al. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 99.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhu HQ, Hu GQ, Ouyang ZQ, Wang J, She ZS. Accuracy improvement for identifying translation initiation sites in microbial genomes. Bioinformatics. 2004;20:3308–3317. doi: 10.1093/bioinformatics/bth390. [DOI] [PubMed] [Google Scholar]
- 101.Zhang L, Luo L. Splice site prediction with quadratic discriminant analysis using diversity measure. Nucleic Acids Res. 2003;31:6214–6220. doi: 10.1093/nar/gkg805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang W, Zheng H, Yang S, Yu H, Li J, et al. Origin and evolution of new exons in rodents. Genome Res. 2005;15:1258–1264. doi: 10.1101/gr.3929705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang P, Wu H, Wang W, Ma W, Sun X, et al. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang X, Zhang J, Li F, Gu J, He T, et al. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- 105.Zhao T, Li G, Mi S, Li S, Hannon GJ, et al. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tian F, Zhang H, Zhang X, Song C, Xia Y, et al. miRAS: a data processing system for miRNA expression profiling study. BMC Bioinformatics. 2007;8:285. doi: 10.1186/1471-2105-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ying X, Luo H, Luo J, Li W. RDfolder: a web server for prediction of RNA secondary structure. Nucleic Acids Res. 2004;32:W150–153. doi: 10.1093/nar/gkh445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li YY, Qin L, Guo ZM, Liu L, Xu H, et al. In silico discovery of human natural antisense transcripts. BMC Bioinformatics. 2006;7:18. doi: 10.1186/1471-2105-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang H, Chua NH, Wang XJ. Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol. 2006;7:R92. doi: 10.1186/gb-2006-7-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang F. Prediction of protein secondary structure with a reliability score estimated by local sequence clustering. Protein Eng. 2003;16:651–657. doi: 10.1093/protein/gzg089. [DOI] [PubMed] [Google Scholar]
- 113.Guo J, Chen H, Sun Z, Lin Y. A novel method for protein secondary structure prediction using dual-layer SVM and profiles. Proteins. 2004;54:738–743. doi: 10.1002/prot.10634. [DOI] [PubMed] [Google Scholar]
- 114.Liu X, Zhang LM, Zheng WM. Prediction of protein secondary structure based on residue pairs. J Bioinform Comput Biol. 2004;2:343–352. doi: 10.1142/s0219720004000594. [DOI] [PubMed] [Google Scholar]
- 115.Liu X, Zheng WM. An amino acid substitution matrix for protein conformation identification. J Bioinform Comput Biol. 2006;4:769–782. doi: 10.1142/s0219720006002156. [DOI] [PubMed] [Google Scholar]
- 116.Jina L, Fang W, Tanga H. Prediction of protein structural classes by a new measure of information discrepancy. Computational Biology and Chemistry. 2002;27:373–380. doi: 10.1016/s1476-9271(02)00087-7. [DOI] [PubMed] [Google Scholar]
- 117.Liu X, Zhao YP, Zheng WM. CLEMAPS: Multiple alignment of protein structures based on conformational letters. Proteins. 2007 doi: 10.1002/prot.21739. [DOI] [PubMed] [Google Scholar]
- 118.He F. Human liver proteome project: plan, progress, and perspectives. Mol Cell Proteomics. 2005;4:1841–1848. doi: 10.1074/mcp.R500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 119.He F. Proteomics in China. Proteomics. 2006;6:397–403. doi: 10.1002/pmic.200690008. [DOI] [PubMed] [Google Scholar]
- 120.Ying W, Jiang Y, Guo L, Hao Y, Zhang Y, et al. A dataset of human fetal liver proteome identified by subcellular fractionation and multiple protein separation and identification technology. Mol Cell Proteomics. 2006;5:1703–1707. doi: 10.1074/mcp.M500344-MCP200. [DOI] [PubMed] [Google Scholar]
- 121.Zhang X, Guo Y, Song Y, Sun W, Yu C, et al. Proteomic analysis of individual variation in normal livers of human beings using difference gel electrophoresis. Proteomics. 2006;6:5260–5268. doi: 10.1002/pmic.200600006. [DOI] [PubMed] [Google Scholar]
- 122.Chen M, Ying W, Song Y, Liu X, Yang B, et al. Analysis of human liver proteome using replicate shotgun strategy. Proteomics. 2007;7:2479–2488. doi: 10.1002/pmic.200600338. [DOI] [PubMed] [Google Scholar]
- 123.Li D, Fu Y, Sun R, Ling CX, Wei Y, et al. pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics. 2005;21:3049–3050. doi: 10.1093/bioinformatics/bti439. [DOI] [PubMed] [Google Scholar]
- 124.Zhang J, Gao W, Cai J, He S, Zeng R, et al. Predicting molecular formulas of fragment ions with isotope patterns in tandem mass spectra. IEEE/ACM Trans Comput Biol Bioinform. 2005;2:217–230. doi: 10.1109/TCBB.2005.43. [DOI] [PubMed] [Google Scholar]
- 125.Xue X, Wu S, Wang Z, Zhu Y, He F. Protein probabilities in shotgun proteomics: evaluating different estimation methods using a semi-random sampling model. Proteomics. 2006;6:6134–6145. doi: 10.1002/pmic.200600070. [DOI] [PubMed] [Google Scholar]
- 126.Zhang J, Li J, Xie H, Zhu Y, He F. A new strategy to filter out false positive identifications of peptides in SEQUEST database search results. Proteomics. 2007;7:4036–4044. doi: 10.1002/pmic.200600929. [DOI] [PubMed] [Google Scholar]
- 127.Guo J, Lin Y, Liu X. GNBSL: a new integrative system to predict the subcellular location for Gram-negative bacteria proteins. Proteomics. 2006;6:5099–5105. doi: 10.1002/pmic.200600064. [DOI] [PubMed] [Google Scholar]
- 128.Shen J, Zhang J, Luo X, Zhu W, Yu K, et al. Predicting protein-protein interactions based only on sequences information. Proc Natl Acad Sci U S A. 2007;104:4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu X, Zhu L, Guo J, Zhang DY, Lin K. Prediction of yeast protein-protein interaction network: insights from the Gene Ontology and annotations. Nucleic Acids Res. 2006;34:2137–2150. doi: 10.1093/nar/gkl219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu X, Zhu L, Guo J, Fu C, Zhou H, et al. SPIDer: Saccharomyces protein-protein interaction database. BMC Bioinformatics. 2006;7(Suppl 5):S16. doi: 10.1186/1471-2105-7-S5-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bu D, Zhao Y, Cai L, Xue H, Zhu X, et al. Topological structure analysis of the protein-protein interaction network in budding yeast. Nucleic Acids Res. 2003;31:2443–2450. doi: 10.1093/nar/gkg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 133.Li F, Long T, Lu Y, Ouyang Q, Tang C. The yeast cell-cycle network is robustly designed. Proc Natl Acad Sci U S A. 2004;101:4781–4786. doi: 10.1073/pnas.0305937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ma W, Lai L, Ouyang Q, Tang C. Robustness and modular design of the Drosophila segment polarity network. Mol Syst Biol. 2006;2:70. doi: 10.1038/msb4100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li D, Li J, Ouyang S, Wang J, Wu S, et al. Protein interaction networks of Saccharomyces cerevisiae, Caenorhabditis elegans and Drosophila melanogaster: large-scale organization and robustness. Proteomics. 2006;6:456–461. doi: 10.1002/pmic.200500228. [DOI] [PubMed] [Google Scholar]
- 136.Liu W, Li D, Zhang J, Zhu Y, He F. SigFlux: a novel network feature to evaluate the importance of proteins in signal transduction networks. BMC Bioinformatics. 2006;7:515. doi: 10.1186/1471-2105-7-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang Z, Liu C, Skogerbo G, Zhu X, Lu H, et al. Dynamic changes in subgraph preference profiles of crucial transcription factors. PLoS Comput Biol. 2006;2:e47. doi: 10.1371/journal.pcbi.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xue H, Xian B, Dong D, Xia K, Zhu S, et al. A modular network model of aging. Mol Syst Biol. 2007;3:147. doi: 10.1038/msb4100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xia K, Xue H, Dong D, Zhu S, Wang J, et al. Identification of the proliferation/differentiation switch in the cellular network of multicellular organisms. PLoS Comput Biol. 2006;2:e145. doi: 10.1371/journal.pcbi.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Luo RY, Liao S, Tao GY, Li YY, Zeng S, et al. Dynamic analysis of optimality in myocardial energy metabolism under normal and ischemic conditions. Mol Syst Biol. 2006;2:2006 0031. doi: 10.1038/msb4100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang K, Ma W, Liang H, Ouyang Q, Tang C, et al. Dynamic simulations on the arachidonic acid metabolic network. PLoS Comput Biol. 2007;3:e55. doi: 10.1371/journal.pcbi.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Songnian Z, Xiaoyun X, Guozheng Y, Zhi F. A computational model as neurodecoder based on synchronous oscillation in the visual cortex. Neural Comput. 2003;15:2399–2418. doi: 10.1162/089976603322362419. [DOI] [PubMed] [Google Scholar]
- 143.Yang H, Zhang C, Zheng H, Xiong W, Zhou Z, et al. A simulation study on the Ca2+-independent but voltage-dependent exocytosis and endocytosis in dorsal root ganglion neurons. Eur Biophys J. 2005;34:1007–1016. doi: 10.1007/s00249-005-0486-8. [DOI] [PubMed] [Google Scholar]
- 144.Zhang SX, Heng PA, Liu ZJ. Chinese visible human project. Clin Anat. 2006;19:204–215. doi: 10.1002/ca.20273. [DOI] [PubMed] [Google Scholar]
- 145.Bai X, Yu L, Liu Q, Zhang J, Li A, et al. A high-resolution anatomical rat atlas. J Anat. 2006;209:707–708. doi: 10.1111/j.1469-7580.2006.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Du W, Wang Y, Luo Q, Liu BF. Optical molecular imaging for systems biology: from molecule to organism. Anal Bioanal Chem. 2006;386:444–457. doi: 10.1007/s00216-006-0541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Funding increase of R&D in China based on official statistics released by MOST: http://www.most.gov.cn/eng/statistics/2006/200703/t20070309_42000.htm; http://www.most.gov.cn/ztzl/qgkjgzhy/2007/2007mtbd/200702/t20070201_40533.htm. [Google Scholar]
- 148.Zhang CT, Zhang R, Ou HY. The Z curve database: a graphic representation of genome sequences. Bioinformatics. 2003;19:593–599. doi: 10.1093/bioinformatics/btg041. [DOI] [PubMed] [Google Scholar]
- 149.Hu GQ, Zheng X, Yang YF, Ortet P, She ZS, et al. ProTISA: a comprehensive resource for translation initiation site annotation in prokaryotic genomes. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao F, Zhang CT. DoriC: a database of oriC regions in bacterial genomes. Bioinformatics. 2007;23:1866–1867. doi: 10.1093/bioinformatics/btm255. [DOI] [PubMed] [Google Scholar]
- 151.Zhang Y, Li J, Kong L, Gao G, Liu QR, et al. NATsDB: Natural Antisense Transcripts DataBase. Nucleic Acids Res. 2007;35:D156–161. doi: 10.1093/nar/gkl782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu C, Bai B, Skogerbo G, Cai L, Deng W, et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cai J, Zhang J, Huang Y, Li Y. ATID: a web-oriented database for collection of publicly available alternative translational initiation events. Bioinformatics. 2005;21:4312–4314. doi: 10.1093/bioinformatics/bti704. [DOI] [PubMed] [Google Scholar]
- 154.He T, Du P, Li Y. dbRES: a web-oriented database for annotated RNA editing sites. Nucleic Acids Res. 2007;35:D141–144. doi: 10.1093/nar/gkl815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhou Y, Zhou C, Ye L, Dong J, Xu H, et al. Database and analyses of known alternatively spliced genes in plants. Genomics. 2003;82:584–595. doi: 10.1016/s0888-7543(03)00204-0. [DOI] [PubMed] [Google Scholar]
- 156.Hao P, He WZ, Huang Y, Ma LX, Xu Y, et al. MPSS: an integrated database system for surveying a set of proteins. Bioinformatics. 2005;21:2142–2143. doi: 10.1093/bioinformatics/bti306. [DOI] [PubMed] [Google Scholar]
- 157.Chen Y, Zhang Y, Yin Y, Gao G, Li S, et al. SPD–a web-based secreted protein database. Nucleic Acids Res. 2005;33:D169–173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang W, Zhang Y, Zheng H, Zhang C, Xiong W, et al. SynDB: a Synapse protein DataBase based on synapse ontology. Nucleic Acids Res. 2007;35:D737–741. doi: 10.1093/nar/gkl876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhuang Y, Li S, Li Y. dbNEI: a specific database for neuro-endocrine-immune interactions. Neuro Endocrinol Lett. 2006;27:53–59. [PubMed] [Google Scholar]
- 160.Ng SK, Zhang Z, Tan SH, Lin K. InterDom: a database of putative interacting protein domains for validating predicted protein interactions and complexes. Nucleic Acids Res. 2003;31:251–254. doi: 10.1093/nar/gkg079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Qi J, Luo H, Hao B. CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 2004;32:W45–47. doi: 10.1093/nar/gkh362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ou HY, Guo FB, Zhang CT. GS-Finder: a program to find bacterial gene start sites with a self-training method. Int J Biochem Cell Biol. 2004;36:535–544. doi: 10.1016/j.biocel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 163.Gao F, Zhang CT. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006;34:W686–691. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zheng H, Shi J, Fang X, Li Y, Vang S, et al. FGF: a web tool for Fishing Gene Family in a whole genome database. Nucleic Acids Res. 2007;35:W121–125. doi: 10.1093/nar/gkm426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Luo Y, Fu C, Zhang DY, Lin K. BPhyOG: an interactive server for genome-wide inference of bacterial phylogenies based on overlapping genes. BMC Bioinformatics. 2007;8:266. doi: 10.1186/1471-2105-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wu X, Walker MG, Luo J, Wei L. GBA server: EST-based digital gene expression profiling. Nucleic Acids Res. 2005;33:W673–676. doi: 10.1093/nar/gki480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jiang P, Wu H, Da Y, Sang F, Wei J, et al. RFRCDB-siRNA: Improved design of siRNAs by random forest regression model coupled with database searching. Comput Methods Programs Biomed. 2007;87:230–238. doi: 10.1016/j.cmpb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 168.Zhou X, Su Z. EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genomics. 2007;8:246. doi: 10.1186/1471-2164-8-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huang N, Chen H, Sun Z. CTKPred: an SVM-based method for the prediction and classification of the cytokine superfamily. Protein Eng Des Sel. 2005;18:365–368. doi: 10.1093/protein/gzi041. [DOI] [PubMed] [Google Scholar]
- 170.Chen H, Xue Y, Huang N, Yao X, Sun Z. MeMo: a web tool for prediction of protein methylation modifications. Nucleic Acids Res. 2006;34:W249–253. doi: 10.1093/nar/gkl233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xia K, Dong D, Han JD. IntNetDB v1.0: an integrated protein-protein interaction network database generated by a probabilistic model. BMC Bioinformatics. 2006;7:508. doi: 10.1186/1471-2105-7-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Qiao LA, Zhu J, Liu Q, Zhu T, Song C, et al. BOD: a customizable bioinformatics on demand system accommodating multiple steps and parallel tasks. Nucleic Acids Res. 2004;32:4175–4181. doi: 10.1093/nar/gkh756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sun Y, Zhao S, Yu H, Gao G, Luo J. ABCGrid: Application for Bioinformatics Computing Grid. Bioinformatics. 2007;23:1175–1177. doi: 10.1093/bioinformatics/btm086. [DOI] [PubMed] [Google Scholar]