Abstract
Nanoscale liquid chromatography coupled to electrospray ionization mass spectrometry was used to identify the nature of the ligand that binds noncovalently to siderocalin (lipocalin 2). The folded state siderocalin–ligand complex was separated from free, unfolded siderocalin using reversed phase chromatography, and the molecular weight of the siderocalin ligand was then determined from the deconvoluted molecular weights of the complex and of the free protein. The ligand was identified as dihydroxybenzoyl-serine, a breakdown product of enterobactin, an iron-chelating compound (“siderophore”) synthesized in bacteria. These results demonstrate that, in some cases, electrostatic noncovalent protein complexes can survive the denaturing conditions of reversed phase liquid chromatography and the gas phase transfer occurring during electrospray ionization.
Keywords: Noncovalent complexes, lipocalin, siderocalin, liquid chromatography, electrospray ionization mass spectrometry
The past decade has seen the widespread application of electrospray ionization mass spectrometry (ESI-MS) not only as a tool for the analysis of individual molecules but also, more recently, for the analysis of macromolecular complexes. Although noncovalent complexes typically dissociate when in flight in the mass spectrometer, it was shown that, in some cases, the energy associated with the ionic interactions between small proteins and ligands is higher than the dissociation energy of the complex.1 In such cases, the noncovalent complex survives the transition from solution to the gas phase and can be detected by a mass spectrometer. First results demonstrating the applicability of ESI-MS for studying noncovalent complexes were published between 1991 and 1993.2 As suggested by several reviews, the number of reports has increased significantly in recent years.3–7
Since many noncovalent complexes are highly sensitive to dissociative processes, one of the critical factors for successful ESI-MS analysis is maintaining proper solution conditions for keeping the complex in its native (folded) state. Direct observation on noncovalent complexes also depends on the ionization parameters (voltage, pressure, and temperature) employed for desolvation of the ESI-generated droplets. In most cases, solution conditions as well as desolvation conditions have to be carefully optimized before noncovalent complexes can be detected. However, some noncovalent complexes based on electrostatic interactions seem to be an exception to these rules because they have shown to be extremely stable, even at very high collision energies.8,9
Here we report the use of nanoscale liquid chromatography (nano-LC) coupled to electrospray ionization mass spectrometry (ESI-MS) to identify the nature of the ligand that binds noncovalently to human siderocalin (lipocalin 2, or neutrophil gelatinase-associated lipocalin), a member of the lipocalin family of binding proteins. Lipocalins are functionally diverse proteins that generally bind small, hydrophobic ligands. Human siderocalin is expressed in epithelial cells in response to inflammatory signals and in neutrophil precursors, but until recently the function of this protein remained a mystery. The structure of siderocalin was previously determined by X-ray crystallography10 and NMR,11 and recent evidence12 shows that siderocalin binds a negatively charged siderophore (ferric enterobactin) through noncovalent electrostatic and cation-π interactions, thus acting as a potent bacteriostatic agent by sequestering iron.
MATERIALS AND METHODS
Protein Expression and Purification
Human siderocalin was expressed in Escherichia coli as a fusion protein with glutathione-S-transferase and purified according to a procedure previously described.13 A red-colored siderocalin complex (holo-siderocalin) was isolated from XL1-Blue competent cells (Stratagene, La Jolla, CA) grown under iron-depleting conditions (Fe3+ concentration below 10−7 M). The same protein, expressed in the BL21 strain of E. coli under normal conditions, produces a nearly colorless protein (apo-siderocalin).
Mass Spectrometry
All experiments were performed on an API-US Q-TOF mass spectrometer (Micromass, Manchester, UK) equipped with the CapLC system (Waters Corp., Milford, MA). The stream select module was configured with a 5-mm × 300-μm ID trap column packed with 5 μm C4 particles (LC Packings, San Francisco, CA) connected by a ZU1XC metallic union (Valco, Houston, TX) to an in-house packed 5-cm × 75-μm ID nanoscale analytical column. The latter was a fused-silica column with an integral frit—“PicoFrit” (360 μm OD × 75 μm ID × 40 cm, 15-μm tip) obtained from New Objective (Cambridge, MA) packed with 5 μm C4 particles (Michrom Bioresources, Auburn, CA) using the pressurized bomb method described by Kennedy and Jorgenson.14 Protein samples (1 μL) were injected onto the trap column at 10 μL/min and desalted for 5 min before being back-flushed to the analytical column at 0.7 μL/min using a linear gradient elution step from 10% to 90% solvent B in 5 min, followed by 90% B for 15 min (solvent A = 5% acetonitrile, 0.1% formic acid; solvent B = 95% acetonitrile, 0.1% formic acid).
The Q-TOF operation parameters were as follows: The electrospray potential was set to 3.5 kV (applied to the Valco union), the cone voltage was set to 40 V, the extraction cone was set to 2V, and the source temperature was 100°C. The MS scan was from m/z 500–3000 with an acquisition time of 1 sec and a collision energy of 5 eV.
RESULTS AND DISCUSSION
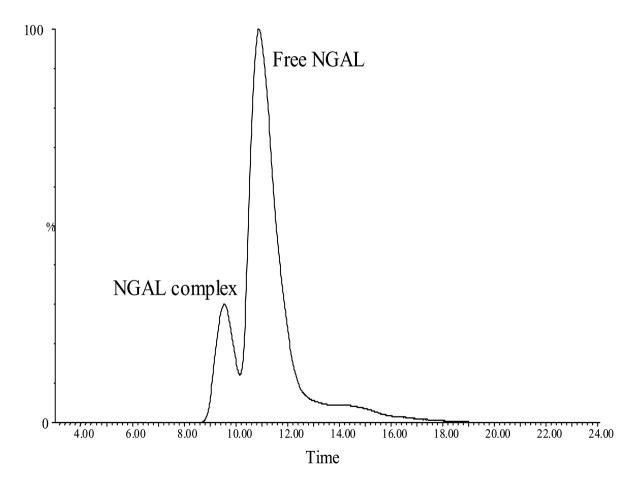
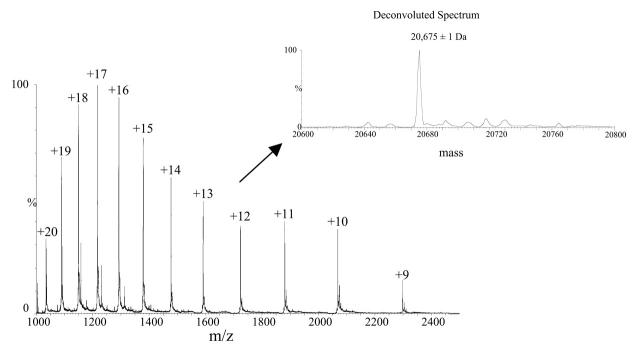
Human siderocalin forms a stable noncovalent complex with a negatively charged ligand when exogenously expressed in XL1-Blue E. coli grown under iron-depleting conditions. The complex exhibits a red color and can be disrupted only after boiling in 8 M urea.12 Native, folded siderocalin complexes can be separated from free, unfolded siderocalin using reversed phase chromatography on a C4 column (see Figure 1). Figure 2 shows the ESI-MS spectrum of free siderocalin. The zero-charge deconvoluted spectrum generated using the MaxEnt1 deconvolution program incorporated in the MassLynx 4.0 software (Micromass) indicated a relative molecular weight of 20,675 ± 1 Da, which matched very well the expected relative molecular weight of siderocalin calculated according to protein’s sequence:
FIGURE 1.
Nano-LC/ESI-MS reconstructed ion chromatogram of the holo-siderocalin sample. The siderocalin complex (1 μL) was injected onto the trap column at 10 μL/min, desalted, and eluted with a 5-min linear gradient from 10 % to 90% solvent B in 5 min (Solvent A = 5% acetonitrile, 0.1% formic acid; solvent B = 95% acetonitrile, 0.1% formic acid). The chromatogram was reconstructed using the signals corresponding to the most abundant charge states present in the ESI-MS spectra of apo-siderocalin (+17) and holo-siderocalin (+10). NGAL, neutrophil gelatinase-associated lipocalin (siderocalin).
FIGURE 2.
ESI-MS spectrum of apo-siderocalin. The mass spectrum was obtained by averaging 50 scans acquired during elution of free siderocalin peak shown in Figure 1. The inset displays the deconvoluted spectrum that resulted upon MaxEnt1 processing of the ESI-MS spectrum. The calculated relative molecular weight of the protein matched the expected molecular weight of 20,675 Da, calculated according to the sequence of the protein.
GSP_QDSTSDLIPAPPLSKVPLQQNFQDNQFQGK WYVVGLAGNAILREDKDPQKMYATIYELKEDKSYN VTSVLFRKKKCDYWIRTFVPGCQPGEFTL GNIKSYPGLTSYLVRVVSTNYNQHAMVFFKKVSQN REYFKITLYGRTKELTSELKENFIRFSKSLGLPENHIVF PVPIDQCID
This sequence contains a three-amino acid N-terminal tag (GSP) left after the glutathione-S-transferase fusion protein was cleaved with thrombin.
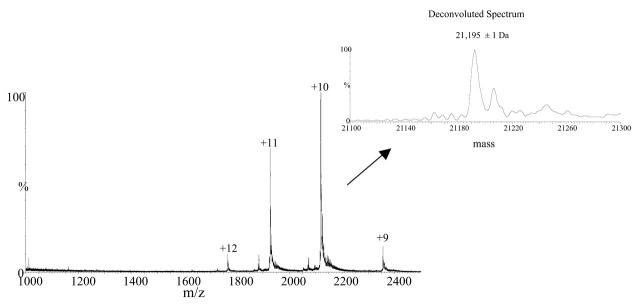
In the case of the siderocalin–ligand complex, only four charge states were observed (+9 to +12) suggesting that the noncovalent complex was preserved in its naturally folded state (Fig. 3).
FIGURE 3.
ESI-MS spectrum of holo-siderocalin. The mass spectrum was obtained by averaging 25 scans acquired during elution of the siderocalin complex peak shown in Figure 1. The inset displays the deconvoluted spectrum resulted upon MaxEnt1 processing of the ESI-MS spectrum. The calculated relative molecular weight of the complex is 21,195 ± 1 Da.
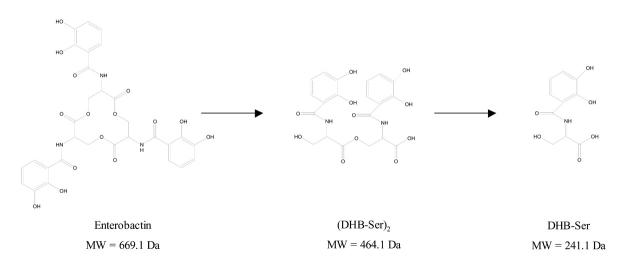
The measured relative molecular weight of the siderocalin–ligand complex was 21,195 ± 1 Da, indicating that the molecular weight of the iron-chelated ligand was 520 Da. The calculated molecular weight of the free ligand of 464 Da corresponded to the molecular weight of (DHB-Ser)2, the linear dimeric ester of dihydroxy-benzoyl-serine (Fig. 4). The expected ligand, enterobactin [cyclo-Tris (2,3-dihydroxy-N-benzoylseryl)] has a molecular weight of 669.1 Da (Fig. 4). In solution, ferric enterobactin rapidly breaks down into dihydroxy-benzoyl-serine (DHB-Ser).15 It is not clear whether the enterobactin decomposition occurred in solution or during the ESI process. Crystallography data also showed ligand degradation,12 which is consistent with the mass spectrometry data provided here.
FIGURE 4.
Degradation products of enterobactin. Chemical formulas of enterobactin, dihydroxy-benzoyl-serine dimer (DHB-Ser)2, and dihydroxy-benzoyl-serine monomer (DHB-Ser).
To solve the iron supply problem caused by the insolubility of Fe3+ (10−9 M), bacteria synthesize iron-chelating compounds called siderophores. Pathogenic bacteria can also obtain iron during infection from heme and iron-binding proteins, such as transferrin, in some cases through a siderophore-mediated mechanism. The immune system wards off bacterial infections by withholding iron from infecting bacteria through such mechanisms as the release of lactoferrin from neutrophil granules. Siderocalin, while not binding iron directly, complements this strategy through a specificity for iron reserved for bacterial use as ferric siderophore complexes. Siderocalin has been shown to be a potent bacteriostatic agent through iron sequestration.12 Our findings help us understand the biological function of siderocalin and should enable future determinations of novel siderocalin ligands.
CONCLUSIONS
In some cases, electrostatic noncovalent protein complexes can survive the denaturing conditions of reversed phase liquid chromatography and the gas phase transfer occurring during ESI. In such cases, mass spectrometry can easily identify the nature of the ligand bound to a protein and help us understand the biological function and recognition specificity of the protein.
Acknowledgments
This work was supported by grants from the National Institutes of Health (AI42200), the FHCRC New Technology Development Fund to R.K.S. and an University of Washington Tools for Transformation Award.
REFERENCES
- 1.Robinson CV, Chung EW, Kragelund BB, et al. Probing the nature of noncovalent interactions by mass spectrometry. A study of Protein-CoA ligand binding and assembly. J Am Chem Soc 1996;118:8646–8653. [Google Scholar]
- 2.Smith RD, Light-Wahl KJ. Perspectives—The observation of noncovalent interactions in solution by electrospray ionization mass spectrometry—Promise, pitfalls, and prognosis. Biol Mass Spectrom 1993:22:493–501. [Google Scholar]
- 3.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev 1997;16:1–23. [DOI] [PubMed] [Google Scholar]
- 4.Loo JA. Electrospray ionization mass spectrometry: A technology for studying noncovalent macromolecular complexes. Int J Mass Spectrom 2000;200:176–186. [Google Scholar]
- 5.Miranker AD. Protein complexes and analysis of their assembly by mass spectrometry. Curr Opin Struct Biol 2000;10:601–606. [DOI] [PubMed] [Google Scholar]
- 6.Sobott F, Robinson CV. Protein complexes gain momentum. Curr Opin Struct Biol 2002;12:729–734. [DOI] [PubMed] [Google Scholar]
- 7.Robinson CV. Protein complexes take flight. Nat Struct Biol 2002;9:505–506. [DOI] [PubMed] [Google Scholar]
- 8.Loo JA, Holler TP, Foltin SK, et al. Application of electrospray ionization mass spectrometry for studying human immunodeficiency virus protein complexes. Proteins 1998;2:28–37 (Suppl.) [DOI] [PubMed] [Google Scholar]
- 9.Sannes-Lowery KA, Hu P, Mack DP, Mei HY, Loo JA. HIV-1 Tat peptide binding to TAR RNA by electrospray ionization mass spectrometry. Anal Chem 1997;69: 5130–5135. [DOI] [PubMed] [Google Scholar]
- 10.Goetz DH, Willie ST, Armen RS, Bratt T, Borregaard N, Strong RK. Ligand preference inferred from the structure of neutrophil gelatinase associated lipocalin. Biochemistry 2000;39:1935–1941. [DOI] [PubMed] [Google Scholar]
- 11.Coles M, Diercks T, Muehlenweg B, et al. The solution structure and dynamics of human neutrophil gelatinase-associated lipocalin. J Mol Biol 1999;289:139–157. [DOI] [PubMed] [Google Scholar]
- 12.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell 2002;10:1033–1043. [DOI] [PubMed] [Google Scholar]
- 13.Bundgaard JR, Sengelov H, Borregaard N, Kjeldsen L. Molecular cloning and expression of a cDNA encoding NGAL: A lipocalin expressed in human neutrophils. Biochem Biophys Res Commun 1994;202:1468–1475. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy RT, Jorgenson JW. Preparation and evaluation of packed capillary liquid chromatography columns with inner diameters from 20 to 50 micrometers. Anal Chem 1989;61:1128–1135. [Google Scholar]
- 15.O’Brien IG, Gibson F. The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim Biophys Acta 1970; 215:393–402. [DOI] [PubMed] [Google Scholar]