Abstract
Plasma from different species is the most accessible and valuable source for biomarker discovery in clinical and animal samples. However, due to the high abundance of some proteins such as albumin and immunoglobulins, low-abundant proteins are often undetectable in proteomic analysis of plasma. We have established a plasma depletion scheme using chicken antibodies against various abundant proteins. This immunoaffinity purification procedure is able to deplete albumin across multiple species. The high binding capacity and specificity of the chicken antibody enables the efficient capture of its ligand from microliter volumes of plasma sample. The resulting two-dimensional gel analyses of the depleted and captured samples show significant enhancement of the low-abundant proteins and specific capture of the abundant ligand. By utilizing this sample preparation scheme, it is now possible to analyze the plasma proteome from multiple species in a potentially rapid and large-scale capacity for biomarker discovery, drug target discovery, and toxicology studies.
Keywords: 2-DE, 2-dimensional gel electrophoresis, HSA, human serum albumin; IgG, immunoglobulin
Serum and plasma are a rich and accessible source for the detection of diagnostic markers and therapeutic targets in many human diseases, such as hemophilia, osteoarthritis, cancer, and cardiovascular diseases.1–5 Many disease processes are correlated with quantitative and physiological changes of proteins in body fluids.6,7 Proteins from damaged tissues can enter the bloodstream and serve as leakage markers.8–10 In general, plasma can contain transport proteins, enzymes, cytokines, receptors, hormones, blood clotting factors, and other soluble proteins. In one study using sequence homology and similarity searches after mass spectrometry (MS), 325 distinct proteins were identified from among the 3700 distinct plasma protein spots.11 But much more work is needed to understand these posttranslational modifications. Currently, only a handful of plasma proteins are routinely used in the clinical diagnosis of diseases. This is largely due to our lack of understanding of many low-abundant plasma proteins—specifically their identification, posttranslational modifications, and relevant physiological functions.
Recent advances in matrix-assisted laser desorption/ionization (MALDI) MS, tandem MS, and two-dimensional electrophoresis (2DE) have contributed greatly to our understanding of protein composition in serum and plasma.12–14 Additionally, other new technologies have surfaced for proteomic study including fluorescence difference gel electrophoresis, isotope-coded affinity tag labeling, protein microarray,15–17 shotgun protein sequencing,12 multidimensional chromatography,18 narrow-range immobilized pH gradient,19 prefractionation procedures,20 and microfluidic devices.8 Oftentimes, high-resolution protein separation by 2DE and MS are followed by bioinformatic database analysis and statistical analysis to advance data accuracy.21
Plasma proteins can express in a dynamic range of milligrams per milliliter (e.g., albumin) to picograms per milliliter (for some interleukins). Such a dynamic range of protein expression represents an enormous problem for detection and quantitation in a single assay. The unusually high abundance of albumin and immunoglobulin G (IgG) in serum can significantly affect the resolution and sensitivity of many techniques. For instance, plasma proteins resolved on 2DE become crowded from 45 to 80 kDa and pI range of 4.5 to 6. The removal of highly abundant proteins such as albumin from serum and plasma is absolutely necessary for the detection of low-abundant proteins by 2DE, isotope-coded affinity tag labeling, or multidimensional protein identification technology proteomic methods.22 The search for novel biomarkers is also heavily dependent on the analysis of the plasma proteome from mammalian model organisms. The removal of albumin from nonhuman mammalian serum/plasma has largely relied on the binding of albumin to Cibacron blue, such as with the Montage Albumin Depletion Kit (Millipore, Bedford, MA, USA). This method lacks specificity and removes potentially important proteins from the sample. The use of monoclonal antibodies against human serum albumin (HSA) as the immunoaffinity depletion reagent is far more efficient and specific than the dye-based resins.22 The use of affinity-purified polyclonal antibodies to specifically remove albumin or IgG is also successful in enriching low-abundant proteins from human plasma samples.23 However, antigen-specific cross-species plasma depletion remains unsolved.
In this report, a novel method for the specific removal of albumin from both human and rat serum/plasma is described. Chicken IgY antibodies (Genway Biotech, San Diego, CA, USA) raised against HSA, which are covalently linked to UltraLink hydrazide beads (Pierce, Rockford, IL), are coupled with high-throughput liquid chromatography for depletion of albumin. A major advantage of chicken IgY antibodies over mammalian IgG antibodies results from the evolutionary distance between chickens and mammals, which allows for greater immunogenicity against conserved mammalian proteins such as albumin. With the platform described herein, very specific and high-throughput depletion of albumin from serum/plasma of multiple mammalian species is achieved using a high-throughput liquid chromatography system, the Applied Biosystems Vision protein workstation (Foster City, CA).
MATERIALS AND METHODS
Albumin Depletion
Human and rat serum was supplied by Bioreclamation, Inc. (Hicksville, NY, USA). Serum was centrifuged at 21,000 × g for 10 min at 4°C to remove insoluble material. Immunodepletion of serum was performed on the Applied Biosystems Vision Workstation liquid chromatography system. Serum was diluted 1:10 in Tris-buffered saline (TBS) and injected at 0.1 mL/min onto a column containing anti-HSA IgY antibodies linked to UltraLink hydrazide beads equilibrated in TBS. The flow-through (depleted) fraction was then collected, and the column eluted in 100 mM glycine-HCl, pH 2.5 at 1 mL/min for albumin retrieval. The column was then washed with 200 mM Tris, pH 7.5, and then re-equilibrated in TBS prior to application of subsequent samples. Depletion using the Montage kit was performed as described in the manufacture’s protocol.
One-Dimensional Electrophoresis
Following depletion, samples were concentrated on Amicon Ultra 4 (Millipore, Milford MA) 10-Kd cutoff ultracentrifugation columns. The protein concentration of the concentrated sample was determined by the Bradford assay. Four micrograms of the concentrated samples was reduced with 50 mM dithiothreitol and electrophoresed on 4% to 12% Bis-Tris SDS NuPage (Invitrogen, Carlsbad CA) gels according to the manufacture’s protocol.
Two-Dimensional Electrophoresis
Prior to isoelectric focusing (IEF), samples were acetone precipitated and solubilized in 40 mM Tris, 7 M urea, 2 M thiourea, and 2% CHAPS, reduced with tri-butylphosphine, and alkylated with 10 mM acrylamide for 90 min at room temperature. Following a second round of acetone precipitation, the pellet was solubilized in 7 M urea, 2 M thiourea, and 2% CHAPS and subjected to IEF on 11-cm, pH 3–10 immobilized pH gradient (IPG) strips (Proteome Systems, Sydney, NSW, Australia). Following IEF, IPG strips were equilibrated in 6 M urea, 2% SDS, 50 mM Tris-acetate buffer (pH 7.0), 0.01% bromophenol blue, and subjected to SDS polyacrylamide electrophoresis on 6%–15% Gel Chips (Proteome Systems, Sydney, Australia). All gels were stained in Sypro Ruby (Molecular Probes, Eugene, Oregon, USA) and imaged by CCD camera on a fluorescent imager (Alpha Innotech, San Leandro, CA, USA).
RESULTS
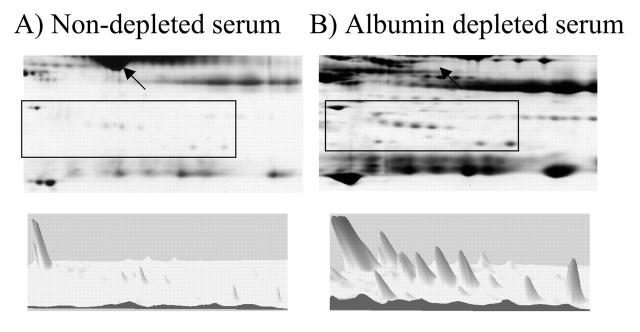
To determine the efficacy and specificity of the IgY antibodies for depletion of abundant proteins from serum or plasma, anti-HSA IgY antibodies linked to UltraLink hydrazide beads were used to deplete albumin from human serum. Nondepleted serum, albumin-depleted serum, and the albumin-associated fractions were subjected to 2DE on pH 3–10 IPG strips and 6% to 15% SDS-PAGE (Fig. 1). The lack of albumin in the albumin-depleted fraction (Fig. 1B) compared with the nondepleted serum (Fig. 1A) indicates the efficacy of IgY antibodies for the depletion of abundant proteins. The specificity of the anti-HSA IgY antibody for albumin is revealed by the lack of abundant nonalbumin proteins detectable in the albumin fraction (Fig. 1C). The appearance of low-abundant proteins in the albumin-depleted fraction (Fig. 2B) that were not detectable in the nondepleted serum (Fig. 2A) demonstrates the usefulness of such depletion paradigms. The three-dimensional view of a representative region of the gel (boxed) elucidates the enrichment of lower abundance proteins in the albumin-depleted sample.
FIGURE 1.
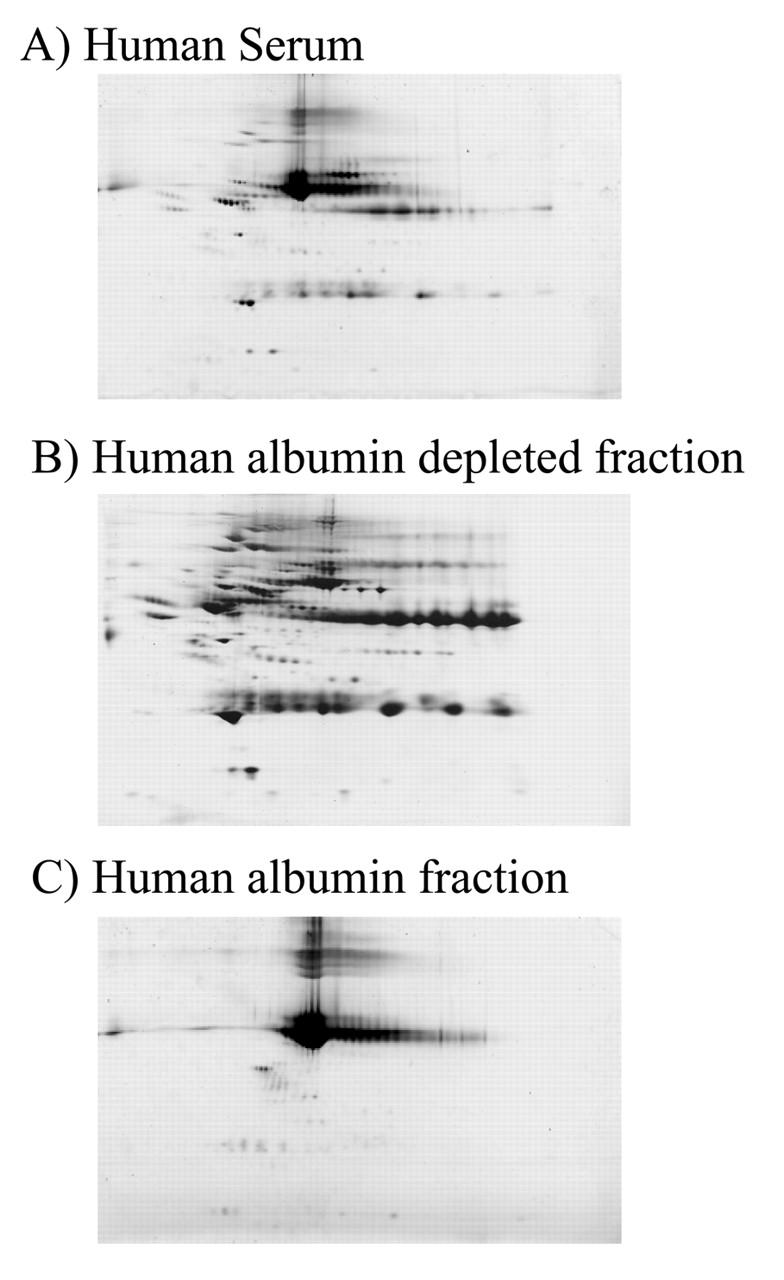
Anti-HAS IgY-mediated depletion of human serum albumin (HSA). Seventy-five micrograms of fractionated human serum (A), anti-HSA IgY-depleted serum (B), and the albumin fraction (C) are subjected to 2DE on a pH 3–10 immobilized pH gradient strip and a 6%–15% SDS-containing polyacrylamide gel.
FIGURE 2.
Enhanced spot detection resulting from human serum albumin depletion. Three-dimensional view of the boxed region of representative 2D gels containing nondepleted human serum (A) and albumin-depleted human serum (B). An increase in the number and intensity of resolved spots in the depleted sample is revealed. The region of the gel containing albumin in A and the lack of albumin in B are denoted by the arrows.
The evolutionary distance between avian species and mammals suggests that antibodies raised to conserved mammalian proteins, such as serum albumin, may cross-react with many mammalian species. This was confirmed by determining the ability of anti-HSA IgY antibodies to specifically deplete albumin from rat serum. Following the same conditions as used with human serum, albumin was depleted from rat serum. The lack of albumin in the albumin-depleted fraction (Fig. 3B) compared with the nondepleted serum (Fig. 3A) indicates the efficacy of IgY antibodies for the depletion of abundant proteins from multiple mammalian species. The specificity of the anti-HSA IgY antibody for rat albumin is revealed by the lack of abundant nonalbumin proteins detectable in the albumin fraction (Fig. 3C).
FIGURE 3.
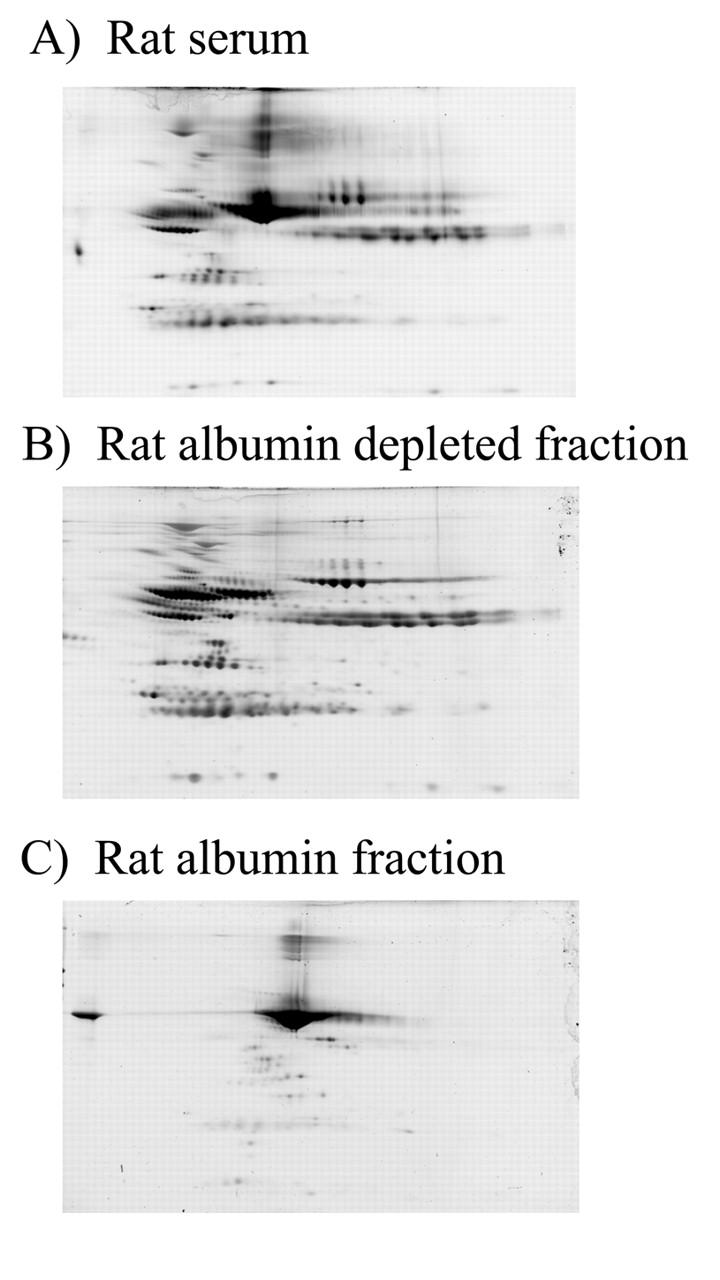
Anti-HSA IgY-mediated depletion of rat serum albumin. Seventy-five micrograms of fractionated rat serum (A), anti-HSA IgY-depleted serum (B), and the albumin fraction (C) are subjected to 2DE on a pH 3–10 immobilized pH gradient strip and a 6%–15% SDS-containing polyacrylamide gel.
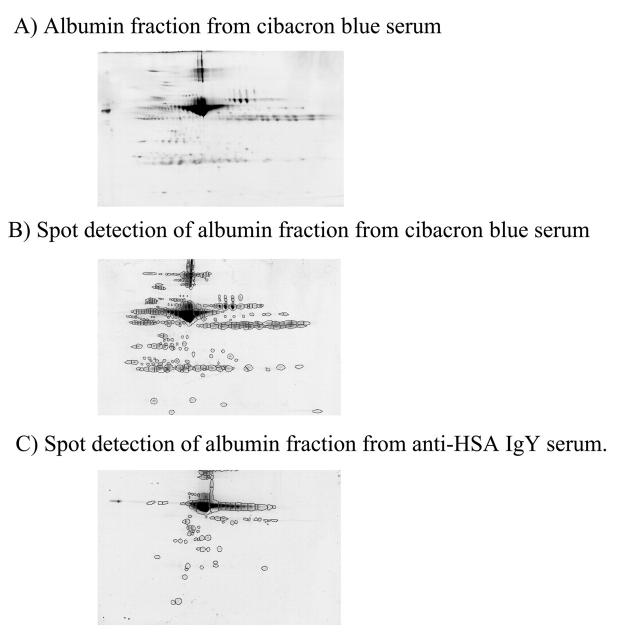
Currently, the depletion of albumin from nonhuman mammals depends upon the binding of albumin to Cibacron blue (Montage kit). To evaluate the potential difference in specificity for albumin inherent in the Montage kit and the anti-HSA IgY antibody, the albumin-associated fraction from rat serum subjected to albumin depletion on the Montage kit was assessed. The albumin-associated fraction from the Montage kit was subjected to 2DE (Fig. 4). The prevalence of abundant nonalbumin proteins reveals the reduced specificity of the Montage kit for albumin compared with the anti-HSA IgY antibody (Fig. 4C).
FIGURE 4.
Comparison of specificity of anti-HSA IgY antibodies and Cibacron blue for rat serum albumin. Seventy-five micrograms of Cibacron blue (albumin)-associated protein was subjected to 2DE on a pH 3–10 immobilized pH gradient strip and a 6%–15% SDS-containing polyacrylamide gel (A). The total number of spots in the albumin-associated fraction from both Cibacron blue (B, 304 spots) and anti-HSA IgY (C, 98 spots) were determined using Progenesis Discovery spot detection.
To quantitate the difference in specificity of the Montage kit and the anti-HSA IgY antibody for rat albumin, the total numbers of spots present in the albumin fractions were compared. Spots were detected using Progenesis Discovery (Nonlinear Dynamics, Manchester, UK) (Fig. 4). There were 304 protein spots in the albumin-associated fraction from the Montage kit and 98 protein spots in the albumin-associated fraction from the anti-HSA IgY depletion, indicating an increase in albumin specificity in the latter case.
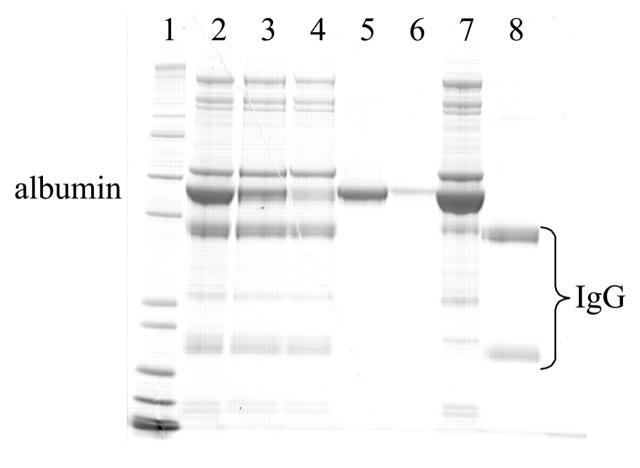
In order to ensure capability for high-throughput analysis of plasma samples, the Applied Biosystems Vision Workstation was deployed. The workstation allows in-series depletion of multiple antigens from plasma or serum. This is achieved by assembling individual columns containing affinity reagents targeting a specific antigen of the plasma, such as anti-serum albumin or protein G resin. It is thus possible to elute the bound protein from individual columns for further analysis, e.g., albumin-associated proteins or IgG-associated proteins. In Figure 5, rat serum was subjected to the depletion and elution procedures of the Vision system. Upon just one pass through the column, most of the serum albumin was depleted from the sample (lane 3). With an additional round of depletion, most of the remaining albumin was removed (lane 4). By eluting the IgY anti-HSA and Poros-G columns at low pH, both albumin (lanes 5 and 6, first and second rounds of depletion, respectively) and the heavy chain and light chain of IgGs are visible (lane 8). Thus, other low-abundant proteins that associate with these abundant proteins may be identified.
FIGURE 5.
SDS-PAGE analysis of anti-HSA IgY and Applied Biosystems Poros-G-mediated depletion of rat serum albumin and IgG using two rounds of depletion. Lane 1 contains Mark 12 molecular weight marker. Albumin and IgG are denoted. Fractions contain nondepleted serum (lane 2), serum depleted for albumin once (lane 3) and twice (lane 4), albumin associated from the first round (lane 5) and the second round (lane 6), serum depleted for IgG (lane 7), and the heavy chain and light chain of IgGs (lane 8).
DISCUSSION
The study of the human and other mammalian species serum/plasma proteome is becoming increasingly important. Serum/plasma are believed to contain many yet uncharacterized posttranslationally modified forms of proteins. These modified forms potentially reflect the physiological state under normal and diseased conditions and thus may be important diagnostic markers, prognostic markers, or drug targets. Furthermore, plasma is readily accessible from human or animal subjects, allowing careful studies in the area of patient monitoring or animal model testing. Traditionally, it is the most abundant tissue being stored for subsequent analysis.
However, the plasma proteome is difficult to study because of the presence of abundant proteins. Albumin and IgG account for more than 70% of the total protein species. Without proper depletion methodologies to specifically remove these proteins, many low-abundant proteins are not detectable with even the most sensitive mass spectrometer. This challenge is further complicated by the huge dynamic range of plasma protein concentrations, between 1010 and 1012. Current instrumentation, such as liquid chromatography MS or 2D gels, can only examine proteins spanning the range of 103 to 104 proteins. The removal of abundant proteins from plasma can significantly increase the relative protein concentration of low-abundant proteins being studied and thus enable their detection and quantitation.
Our study illustrates a novel method for the depletion of albumin from serum/plasma of both human and rat species. The advantage of this depletion scheme versus other methods is that the chicken anti-HSA IgY antibodies have a high affinity and specificity for albumin from multiple mammalian species (Figs. 1–3). Preliminary data have also shown cross-species capability of the anti-albumin antibody for additional species, such as mouse, goat, and pig (data not shown). The affinity purified polyclonal chicken antibodies provide an advantage over other available immunoaffinity methods that use monoclonal antibodies, as the latter rely on single epitopes for protein capture. In disease conditions, different posttranslational modifications can occur and alter the binding epitope of the antigen. By using polyclonal antibodies, one can circumvent this technical difficulty. Other immunoaffinity-based approaches that are dependent on rabbit- or mouse-produced antibodies lack reactivity across multiple mammalian species and thus are typically limited to human samples. It can be costly and time consuming to produce antibodies against albumin and other abundant proteins from every species.
Alternative technologies using immobilized textile dyes, such as Cibacron blue-based kits, lack specificity and thus may remove important proteins from the analysis (Fig. 4). Recent MS identification of proteins that bind to Cibacron blue but not specifically to albumin resulted in 60 unique protein species.24 Oftentimes, the albumin-associated fractions are excluded from further studies. In our study (data not shown) and those done by other groups,25 albumin-associated protein complexes under both normal or diseased conditions contain many uncharacterized proteins.
The antibody conjugation scheme, which was employed with the chicken antibodies, orients the antibody with the antigen-binding site facing outward, with the constant portion of the antibodies covalently coupled to a solid phase material. This allows maximum efficiency of antigen binding. In a column format, the reagent can be regenerated many times to deplete the same sample or multiple samples after proper washing of the column. The high reusability of the column enables multiple rounds of depletion of the same sample to achieve complete removal of undesirable proteins. The liquid chromatography column format is also amenable to automation in a high-throughput scale. To analyze clinical samples, speed and reproducibility are critical for the accumulation of statistically reliable information.
With our current high-throughput Vision liquid chromatography system, it is possible to include additional chicken IgY antibodies for the removal of other abundant proteins from plasma. These may include alpha-1-antitrypsin, IgA, IgG, IgM, transferrin, haptoglobin, and fibrinogen among others. Normally, serum is collected from plasma through clotting and the removal of fibrinogen. With an anti-fibrinogen antibody, plasma can be analyzed directly without activation of many proteolytic enzymes that occur during clotting. By further depleting other abundant proteins from plasma, one can advance the current understanding of low-abundant plasma proteins in disease progression and intervention utilizing chicken IgY antibodies.
Acknowledgments
We are grateful for the technical advice given to us by colleagues from GenWay Biotech. Dr. Jerry Feitelson was excellent in coordinating and delivering the necessary IgY reagents for our study. Mr. Gary Smejkal of PSI Woburn was very helpful by sharing his expertise on 2DE. Thanks are also given to Dr. Jim Jersey for his guidance and support on this project.
REFERENCES
- 1.Sinz A, Bantscheff M, Mikkat S, et al. Mass spectrometric proteome analyses of synovial fluids and plasmas from patients suffering from rheumatoid arthritis and comparison to reactive arthritis or osteoarthritis. Electrophoresis 2002;23:3445–3456. [DOI] [PubMed] [Google Scholar]
- 2.Imam-Sghiouar N, Laude-Lemaire I, Labas V, et al. Subproteomics analysis of phosphorylated proteins: Application to the study of B-lymphoblasts from a patient with Scott syndrome. Proteomics 2002;2:828–838. [DOI] [PubMed] [Google Scholar]
- 3.Vejda S, Posovszky C, Zelzer S, et al. Plasma from cancer patients featuring a characteristic protein composition mediates protection against apoptosis. Mol Cell Proteomics 2002;1:387–393. [DOI] [PubMed] [Google Scholar]
- 4.Qin LX, Tang ZY. The prognostic molecular markers in hepatocellular carcinoma. World J Gastroenterol 2002;8: 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson PJ. A framework for the molecular classification of circulating tumor markers. Ann NY Acad Sci 2001;945:8–21. [DOI] [PubMed] [Google Scholar]
- 6.Tissot JD, Vu DH, Aubert V, et al. The immunoglobulinopathies: From physiopathology to diagnosis. Proteomics 2002;2:813824. [DOI] [PubMed] [Google Scholar]
- 7.Hutter G, Sinha P. Proteomics for studying cancer cells and the development of chemoresistance. Proteomics 2001;1:1233–1248. [DOI] [PubMed] [Google Scholar]
- 8.Li J, LeRiche T, Tremblay TL, et al. Application of microfluidic devices to proteomics research: Identification of trace-level protein digests and affinity capture of target peptides. Mol Cell Proteomics 2002;1:157–168. [DOI] [PubMed] [Google Scholar]
- 9.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics 2002;1(11):845–867. [DOI] [PubMed] [Google Scholar]
- 10.Ruepp SU, Tonge RP, Shaw J, Wallis N, Pognan F. Genomics and proteomics analysis of acetaminophen toxicity in mouse liver. Toxicol Sci 2002;65:135–150. [DOI] [PubMed] [Google Scholar]
- 11.Pieper R, Gatlin CL, Makusky AJ, et al. The human serum proteome: Display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics 2003;3:1345–1364. [DOI] [PubMed] [Google Scholar]
- 12.Wu SL, Choudhary G, Ramstrom M, Bergquist J, Hancock WS. Evaluation of shotgun sequencing for proteomic analysis of human plasma using HPLC coupled with either ion trap or Fourier transform mass spectrometry. J Proteome Res 2003;2:383–393. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol 2003;21:660–666. [DOI] [PubMed] [Google Scholar]
- 14.Choudhary G, Wu SL, Shieh P, Hancock WS. Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J Proteome Res 2003; 2:59–67. [DOI] [PubMed] [Google Scholar]
- 15.Tam SW, Wiese R, Lee S, Gilmore J, Kumble KD. Simultaneous analysis of eight human Th1/Th2 cytokines using microarrays. J Immunol Methods 2002;261:157–165. [DOI] [PubMed] [Google Scholar]
- 16.Sloane AJ, Duff JL, Wilson NL, et al. High throughput peptide mass fingerprinting and protein macroarray analysis using chemical printing strategies. Mol Cell Proteomics 2002;1:490–499. [DOI] [PubMed] [Google Scholar]
- 17.Huang JX, Mehrens D, Wiese R, et al. High-throughput genomic and proteomic analysis using microarray technology. Clin Chem 2001;47:1912–1916. [PubMed] [Google Scholar]
- 18.Wu SL, Amato H, Biringer R, Choudhary G, Shieh P, Hancock WS. Targeted proteomics of low-level proteins in human plasma by LC/MSn: Using human growth hormone as a model system. J Proteome Res 2002;1:459–465. [DOI] [PubMed] [Google Scholar]
- 19.Starita-Geribaldi M, Roux F, Garin J, Chevallier D, Fenichel P, Pointis G. Development of narrow immobilized pH gradients covering one pH unit for human seminal plasma proteomic analysis. Proteomics 2003;3: 1611–1619. [DOI] [PubMed] [Google Scholar]
- 20.Herbert B, Righetti PG. A turning point in proteome analysis: Sample prefractionation via multicompartment electrolyzers with isoelectric membranes. Electrophoresis 2000;21:3639–3648. [DOI] [PubMed] [Google Scholar]
- 21.Muller M, Gras R, Binz PA, Hochstrasser DF, Appel RD. Molecular scanner experiment with human plasma: Improving protein identification by using intensity distributions of matching peptide masses. Proteomics 2002;2:1413–1425. [DOI] [PubMed] [Google Scholar]
- 22.Steel LF, Trotter MG, Nakajima PB, Mattu TS, Gonye G, Block T. Efficient and specific removal of albumin from human serum samples. Mol Cell Proteomics 2003;2: 262–270. [DOI] [PubMed] [Google Scholar]
- 23.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Multi-component immunoaffinity subtraction chromatography: An innovative step towards a comprehensive survey of the human plasma proteome. Proteomics 2003;3:422–432. [DOI] [PubMed] [Google Scholar]
- 24.Bailey J, Zhang K, Zolotarjova N, Nicol G, Szafranski C. Removing high-abundance proteins from serum. Genet Eng News 2003;23(19):32–36. [Google Scholar]
- 25.Conray et al. Presented at Human Proteome Organisation meeting, 2003, Montreal.