Abstract
Proteases play fundamentally important roles in normal physiology and disease pathology. Methods for detection of active proteolysis may greatly aid in the diagnosis of disease progression, and suggest modes of therapeutic intervention. Most assays for proteolytic potential are limited by a lack of specificity and/or quantification. We have developed a solid-phase activity assay for members of the matrix metalloproteinase (MMP) family that is specific and can be used to quantify active enzyme concentration. The assay has two principal components: a capture antibody that immobilizes the MMP without perturbing the enzyme active site, and a fluorescence resonance energy transfer substrate for monitoring proteolysis at low enzyme concentrations. The assay was standardized for MMP-1, MMP-3, MMP-13, and MMP-14. The efficiency of the assay was found to be critically dependent upon the quality of the antibodies, the use of substrates exhibiting high specific activities for the enzymes, and enzyme samples that are fresh. The assay was applied to studies of constitutive and induced MMP activity in human melanoma cells. Analysis of several melanoma cell lines, and comparison with prior studies, correlated higher constitutive MMP-13 activity with higher levels of the cell surface receptor CD44. Ligands to two different melanoma cell surface receptors (the α2β1 integrin or CD44) were found to induce different proteolytic profiles, suggesting that the extracellular matrix can modulate melanoma invasion. Overall, the solid-phase MMP activity assay was found to be valuable for analysis of protease activity in cellular environments. The solid-phase assay is suitably flexible to allow studies of virtually any proteolytic enzyme for which appropriate substrates and antibodies are available.
Keywords: matrix metalloproteinase, fluorogenic substrate, melanoma, solid-phase assay, ELISA
The metzincin subclan of enzymes—which includes the matrix metalloproteinase (MMP), ADAM (a disintegrin and metalloproteinase), and ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) families—has been associated with many normal physiological functions.1 Pathologies such as arthritis and tumor metastasis also feature prominent roles for the metzincins. Most MMP, ADAM, and ADAMTS members of the metzincin subclan are expressed in inactive form as zymogens. Once activated, their ability to process substrate is regulated by protease inhibitors that may be general (α2-macroglobulin) or specific (tissue inhibitor of metalloproteinase; TIMP). The vast majority of studies on MMP, ADAM, and ADAMTS regulation have focused on modulation of protein production. However, given the numerous pathways by which these enzymes are regulated, it is important to extend such studies to include the measurement of active enzyme. Ultimately, the ability to quantify enzymatic activity is necessary to discern which MMP, ADAM, and ADAMTS family members are associated with a disease process.
Numerous approaches have emerged by which activity of these proteases can be evaluated. Use of substrate-based assays often involves radiolabeling, dyes, or fluorescent tags.2–5 Alternatively, polyacrylamide gel electrophoresis can be used in combination with densitometry or substrate zymography to analyze activity,6–10 while enzyme-linked immunosorbent assay (ELISA) and Western blotting can be utilized to differentiate between active enzyme and zymogen forms.8,11 Substrate hydrolysis can also be monitored by using antibodies to neoepitopes exposed only after cleavage12–15 or release of hydrolysis products from co-polymerized polyacrylamide beads.15,16 A continuous assay method, such as one that utilizes an increase in fluorescence upon hydrolysis, allows for rapid and convenient kinetic evaluation of proteolytic activity. Quenched fluorescent substrates that rely on fluorescence resonance energy transfer (FRET)/intramolecular fluorescence energy transfer (IFET) have been constructed by incorporation of a fluorophore and a quencher on the same peptide chain. Fluorogenic substrates have several advantages over other methods, in that they (a) can be monitored continuously and at reasonably low concentration ranges, (b) can be used to determine individual kinetic parameters, and (c) are readily accommodated by high-throughput analytical techniques.
Several different fluorogenic substrates have been described for MMPs and ADAMs, and the relative merits of these substrates have been discussed previously.17–20 In several cases, fluorogenic substrates that are selective for specific MMPs have been described.21–23 However, it is unrealistic to expect that singly selective substrates can be designed for each member of the MMP family. Selectivity can be obtained with antibodies, and thus the present studies have examined the use of antibodies in combination with fluorogenic substrates to create assays selective for MMP activity.
MATERIALS AND METHODS
Peptides
The synthesis and characterization of the general fluorogenic triple-helical peptide (THP) substrates fTHP-4, fTHP-5, and fTHP-7, and the MMP-3-specific fluorogenic substrate NFF-3 have been described previously.20,21,24,25 The synthesis and characterization of the triple-helical ligands for melanoma α2β1 integrin [C10-(α1(IV)382–393)-NH2 THP, C16-(α1(IV)382–393)-NH2 THP] and CD44 [C16-(α1(IV)1263–1277)-NH2 THP] have also been described.22,26–28
Matrix Metalloproteinases
ProMMP-1 and proMMP-3 were expressed in Eschericia coli and folded from the inclusion bodies as described previously.29 ProMMP-1 was activated by reacting with 1 mM 4-aminophenylmercuric acetate (APMA) and an equimolar amount of MMP-3 at 37°C for 6 h. After activation, MMP-3 was completely removed from MMP-1 by affinity chromatography using an anti-MMP-3 IgG Affi-Gel 10 column. ProMMP-3 was activated by reacting with 5 μg/mL chymotrypsin at 37°C for 2 h. Chymotrypsin was inactivated with 2 mM diisopropylfluorophosphate. ProMMP-13 was a generous gift from Dr. P.G. Mitchell, Pfizer, Inc. (Groton, CT). ProMMP-13 was activated with 1 mM APMA. The amounts of active MMP-1, MMP-2, MMP-3, and MMP-13 were determined by titration with recombinant TIMP-130 over a concentration range of 0.1 to 3 μg/mL. Recombinant MMP-14 with the linker and C-terminal hemopexin-like domains deleted [residues 279–523; designated MMP-14(Δ279–523)] was purchased from Chemicon International (Temecula, CA). MMP-14(Δ279–523) was expressed in the active form with Tyr112 at the N-terminus. MMP-14(Δ279–523)—which, in contrast to MMP-14, does not undergo rapid autoproteolysis—was used in the present studies due to the relatively small differences in MMP-14(Δ279–523) and MMP-14 triple-helical peptidase activities noted previously.31 The amount of active MMP-14 was determined by comparison of activity measurements between several different MMP-14 preparations that had been directly titrated with TIMP-2.23,31
Cell Culture
The human melanoma cell lines M14P, M14#5, and M14#11 were obtained as a generous gift from Dr. Barbara M. Mueller (La Jolla Institute for Molecular Medicine, La Jolla, CA). Tissue culture reagents were obtained from Fisher Scientific (Atlanta, GA) unless otherwise stated. All immunologicals were supplied by Chemicon International (Temecula, CA). M14 cells were routinely cultured with RPMI 1640 and supplemented with 10% fetal bovine serum (BioWhittaker, Walkersville, MD) and the following antibiotics: 0.1 mg/mL gentamicin sulfate, 50 units/mL penicillin, and 0.05 mg/mL streptomycin sulfate. Cells were cultured for up to 8 passages and then replaced by frozen stocks to minimize phenotypic drift. Human aortic endothelial cells (Clonetics/Biowhittaker/Cambrex, Walkersville, MD) were cultured according to the supplier’s recommendations (cultured in EGM-2 and discarded at passage 9). All cells were maintained at 37°C in a humidified incubator containing 5% CO2.
Induction of Melanoma Cells
Pro-Bind 96-well assay plates were conditioned at room temperature overnight prior to initiation of the induction experiment with 10 μM C10-(α1(IV)382–393)-NH2 THP, C16-(α1(IV)382–393)-NH2 THP, or C16-(α1(IV)1263–1277)-NH2 THP. Previous studies had determined that these were optimal ligand concentrations for cell adhesion.22,27,28,32,33 The plates were then blocked by adding 2 mg/mL bovine serum albumin (BSA) in phosphate-buffered saline (PBS), pH 7.4, and incubated overnight at room temperature. M14P cultures used for induction experiments were typically 60–80% confluent before release from growth flasks with PBS containing 5 mM ethylenediaminetetraacetic acid (EDTA) (pH 7.3). Subsequent to release, cells were washed with adhesion media [RPMI-1640 containing 20 mM N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES)] and seeded at approximately 7500 cells/well. Melanoma cells were allowed to adhere to ligands for 60 min at 37°C. Three washes of adhesion media were used to remove nonadherent cells. Aliquots of the melanoma-conditioned media were harvested at regular intervals (T = 0, 6, 12, 18, and 24 h) for later determination of metalloproteinase levels. Samples were stored at −20°C until analyses could be performed. Conditioned media was isolated by withdrawal of the media from growing cells and centrifuging at 1000 × g (to remove any floating cells).
Matrix Metalloproteinase Assay
MMP capture antibodies were from Chemicon and were as follows: MMP-1, monoclonal antibody (mAb) MAB1346; MMP-3, polyclonal antibody (pAb) AB810; MMP-13, mAb MAB3321; and MMP-14, pAb AB815. The general assay protocol was as follows: A 96-well plate was incubated with the appropriate MMP Ab for at least 18 h at 4°C. Nonspecific binding sites were blocked by incubating with PBS containing 0.05% Tween 20 and 2 mg/mL BSA for at least 4 h at 4°C. Either MMP standards or unknown samples were added to each well and the plate gently shaken for at least 18 h at 4°C. Unbound enzyme was removed by washing wells 3 times with enzyme assay buffer (50 mM Tricine, 50 mM NaCl, 10 mM CaCl2, 0.05% brij-35). The appropriate fluorogenic substrate was added to each well, and the plate incubated at 37°C in a humidified atmosphere for 0–18 h. Fluorescence readings were taken at appropriate intervals. More specific details for optimized assay conditions are given in the Results and Discussion section. For calculation purposes, the following MMP molecular weight values were used: MMP-1, 41.0 kDa; MMP-3, 43.0 kDa; MMP-13, 42.0 kDa; and MMP-14(Δ279–523), 20.5 kDa34,35. Assays were performed in at least duplicate.
RESULTS AND DISCUSSION
Standardization of Solid-Phase Assay
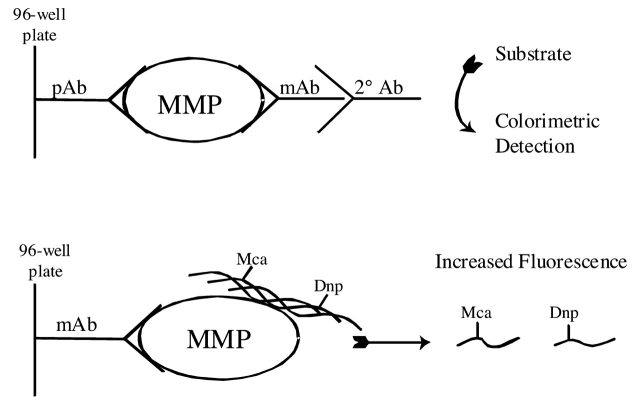
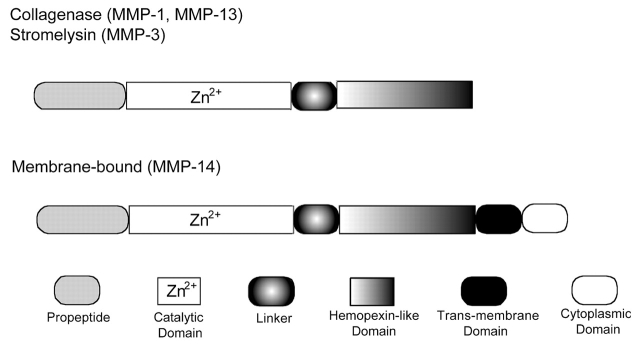
The solid-phase assay was standardized using four MMP family members: MMP-1, MMP-3, MMP-13, and MMP-14. MMP-1, MMP-13, and MMP-14 assays were developed using general triple-helical substrates fTHP-4, fTHP-5, or fTHP-7 (Fig. 1). The MMP-3 assay was developed using the MMP-3 specific substrate NFF-3 (Fig. 1). The general principle of the assay is given in Figure 2. For a traditional solid-phase indirect sandwich ELISA,36 three antibodies are needed: (a) one to immobilize (trap/capture) the MMP, typically a pAb; (b) one to detect the MMP, typically an mAb; and (c) a secondary antibody to detect the mAb, which, in turn, is linked to an enzyme. Colorimetric detection is achieved by conversion of exogenous substrate. Alternatively, a fluorescently labeled secondary antibody can be used. ELISA is typically used to measure protein amounts, but does not discriminate for enzyme activity. For our solid-phase assay, only the immobilization antibody is needed. However, this antibody must bind the MMP selectively and efficiently without significantly inhibiting catalytic activity. MMPs can be divided into four domains: propeptide, catalytic, linker, and hemopexin-like (Fig. 3). The appropriate MMP capture antibodies were thus chosen based on prior application in ELISA and recognizing regions other than the MMP active site within the catalytic domain. After examining a variety of antibodies, the ones specifically selected were as follows: MMP-1 clone 3A9.3, a non-inhibitory mAb raised against human MMP-1; an MMP-3 pAb raised against the human MMP-3 linker region; MMP-13 clone 181–15A12, a non-inhibitory mAb raised against human MMP-13; and an MMP-14 pAb raised against the human MMP-14 linker region.
FIGURE 1.
Sequences of substrates and ligands. The four substrates used in this study are (a) fTHP-4, (b) fTHP-5, (c) fTHP-7, and (d) NFF-3. The three peptide-amphiphile ligands used for induction of melanoma cell MMPs are (e) C10-(α1(IV)382–393)-NH2 THP ligand for the α2β1 integrin, (f) C16-(α1(IV)382–393)-NH2 THP ligand for the α2β1 integrin, and (g) C16-(α1(IV)1263–1277)-NH2 THP ligand for CD44. Sequence numbering for the ligands corresponds to the human α1(IV) gene-derived sequence.57
FIGURE 2.
Schematic diagrams for (top) conventional indirect sandwich ELISA and (bottom) solid-phase MMP activity assay. For both assays, the MMP is captured by an antibody on the solid phase. The indirect sandwich ELISA then requires a detection antibody and an enzyme-labeled antibody. The sandwich ELISA can also be performed directly, in which case the detection antibody is also enzyme-labeled. In the solid-phase MMP activity assay, the fluorogenic substrate serves as the detection system. Dnp, 2,4-dinitrophenyl; Mca, (7-methoxycoumarin-4-yl)acetyl.
FIGURE 3.
Domain structures of MMP-1, MMP-3, MMP-13, and MMP-14.
MMP hydrolysis of the fluorogenic substrates used herein had previously been shown to follow Michaelis-Menton kinetics.21,37 KM values for MMP-1 hydrolysis of fTHP-4, fTHP-5, and fTHP-7, MMP-3 hydrolysis of NFF-3, MMP-13 hydrolysis of fTHP-4, and MMP-14(Δ279–523) hydrolysis of fTHP-4 have previously been determined and fall in the range of 11 to 48 μM.20,21,23–25 Assays were performed using an excess of substrate compared with enzyme but with substrate concentrations typically below enzyme KM values, approximating first-order kinetic conditions. Under these conditions, one can directly correlate enzyme activity to enzyme concentration by comparison with known concentrations of active enzyme.
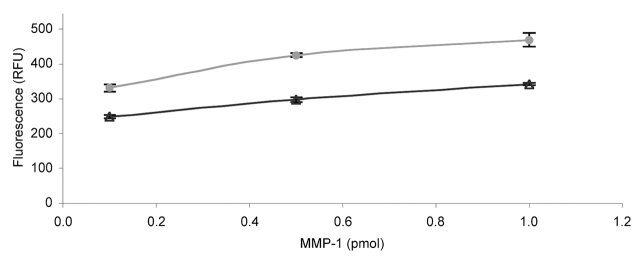
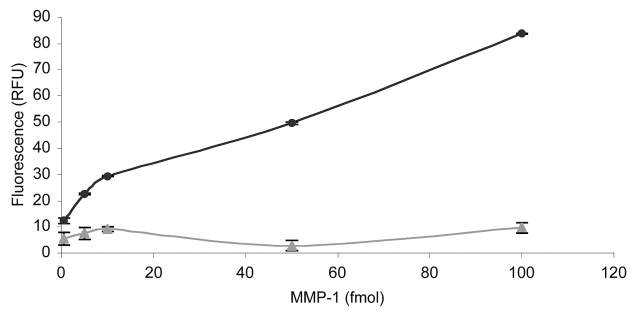
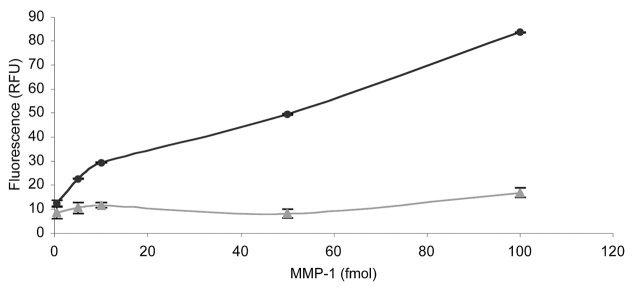
Initial studies were performed with MMP-1 and fTHP-7. First, the appropriate capture mAb concentration was determined by comparing 0.125, 0.25, 0.5, 1.0, 2.0, and 5.0 μg/mL of MMP-1 mAb over a range of MMP-1 concentrations. The capture mAb concentrations were chosen based on literature precedents for MMP ELISA.38,39 The 2.0 μg/mL and 5.0 μg/mL mAb concentrations were effective, but the highest concentration of mAb captured the higher concentrations of MMP-1 slightly more efficiently (Fig. 4). If the anticipated enzyme production is below approximately 50 ng/mL, 2.5 μg/mL capture mAb is likely sufficient for analysis. The limit of detection was then evaluated using substrate fTHP-5, in which the fluorophore (Amp) has a higher quantum yield than the fluorophore of fTHP-7 (Adp).20 The sensitivity of the assay allowed for detection of 1 × 10−15 moles of active MMP-1 in a 200 μL volume (5.0 pM) (data not shown). The specificity of the assay was examined using an irrelevant mAb (one against the β1 integrin subunit) or mouse IgG. MMP-1 activity was not observed over background when the anti-β1 mAb was used as the capture antibody (Fig. 5). Subsequent use of a mouse IgG instead of the anti-β1 mAb as a negative control produced a similar result (data not shown). The cross-reactivity of the MMP-1 capture mAb was examined by comparing its ability to recognize MMP-1 with its ability to recognize MMP-13. MMP-13 activity was not observed over background, demonstrating that the mAb was not cross-reactive and did specifically immobilize active MMP-1 (Fig. 6). Thus, the solid-phase assay was found to specifically quantify MMP-1 activity with good sensitivity over an MMP-1 range of 1.0 fmol to 1.0 pmol.
FIGURE 4.
Efficiency of capture antibody for solid-phase MMP activity assay. The change in fluorescence upon MMP-1 hydrolysis of fTHP-7 was compared after 18 h using 2.5 μg/mL (triangles) or 5 μg/mL (circles) MMP-1 capture mAb. The amounts of MMP-1 added ranged from 0.1–1.0 pmol. Methods were as described in Materials and Methods.
FIGURE 5.
Specificity of the solid-phase MMP activity assay based on the nature of the antibody. The change in fluorescence upon MMP-1 hydrolysis of fTHP-7 was compared after 18 h using 2.5 μg/mL MMP-1 mAb (circles) or 2.5 μg/mL of an anti-β1 integrin subunit mAb (triangles). The amounts of MMP-1 added ranged from 0.5 to 100 fmol. Methods were as described in Results and Discussion.
FIGURE 6.
Specificity of the solid-phase MMP activity assay based on the nature of the MMP. The change in fluorescence upon MMP-1 (circles) or MMP-13 (triangles) hydrolysis of fTHP-7 was compared after 18 h using 2.5 μg/mL MMP-1 mAb. The amounts of MMP-1 added ranged from 0.5 to 100 fmoles. Methods were as described in Results and Discussion.
Based on the MMP-1 results, standard conditions were developed for the solid-phase MMP activity assay. The 96-well plate was incubated with the 100 μL of 5 μg/mL of the appropriate MMP capture Ab in PBS for at least 18 h at 4° C with mixing. Nonspecific binding sites were blocked by incubating with 100 μ L PBS containing 0.05% Tween 20 and 2 mg/mL BSA for at least 4 h at 4 °C with mixing, and the plate washed 3 times with enzyme assay buffer. One-hundred-fifty microliters of either MMP standards or unknown samples were added to each well, followed by 50μL PBS containing 0.2% Tween 20 and 8 mg/mL BSA (to create final concentrations of 0.05% Tween 20 and 2 mg/mL BSA), and the plate was mixed for at least 18 h at 4°C. All liquid was removed and 200 μL enzyme assay buffer or 2 mM APMA (where applicable) was added to each well. This was followed by a 2 h incubation at 37°C. The wells were washed 3 times with enzyme assay buffer and 200μL of the appropriate fluorogenic substrate (5–10μM of fTHP-4, fTHP-5, or fTHP-7; 2.5–5μM of NFF-3) was added to each well. The plate was incubated at 37°C in a humidified atmosphere for 0 to 18 h. Fluorescence readings (λexcitation = 324 nm and λ emission = 393 nm for NFF-3, fTHP-4, and fTHP-5; λ excitation = 348 nm and λ emission = 436 nm for fTHP-7) were taken at appropriate intervals and a standard curve was created by plotting the increase in fluorescence versus concentration of active enzyme. If enzyme activity is anticipated to be high, data collection is best optimized by monitoring activity over 1 to 4 h. The samples analyzed herein (see later discussion) have relatively low levels of active enzyme. Thus, fluorescence was monitored over an extended period of time (18–24 h).
The efficiency of enzyme capture by the MMP antibodies was evaluated by comparing the solid-phase assay with our previously described solution assay.20–22,24 It is assumed that the change in fluorescence obtained from the solution assay represents 100% of the active enzyme, where active enzyme concentration is determined initially by TIMP-1 titration (see Materials and Methods). Change in fluorescence was thus compared directly between the solution and solid-phase assays. The capture efficiency was 63, 37, and 40% for MMP-1, MMP-3, and MMP-13, respectively, over an MMP concentration range of 10 to 20 ng/mL (data not shown). Below this range, capture efficiency approached quantitative values. The capture efficiency for MMP-14 could not be accurately evaluated due to limitations in commercially available MMP-14 reagents. The MMP-14 standard curve (see below) was generated in the presence of the MMP-14 capture pAb. The modulation of activity due to pAb binding was thus taken into consideration, while capture efficiency was not.
Standard curves were subsequently obtained for MMP-1, MMP-3, MMP-13, and MMP-14(Δ279–523) (Fig. 7). In all cases, reasonable changes in fluorescence were observed with increasing MMP concentrations. The curves were not linear, as capture efficiencies change at higher MMP concentrations (see above). Therefore, standard curves should be optimized based on the levels of active enzymes in a given set of samples. If the sample activity is unknown, a standard curve covering a wide range of known concentrations should be constructed and outlying concentrations can be adjusted upon completion of the experiment. The most sensitive assay observed herein was for MMP-3, where active enzyme could be readily detected at less than 1 ng/mL (less than 23 pM). The high kcat/KM value for MMP-3 hydrolysis of NFF-321 resulted in substantial changes in fluorescence at even very low MMP-3 concentrations. The next most sensitive assay was the one for MMP-1, where active enzyme could be readily detected at 1–2 ng/mL (23–47 pM), followed by conservative limits of 3–5 ng/mL for MMP-13 (71–119 pM) and MMP-14(Δ279–523) (146–244 pM).
FIGURE 7.

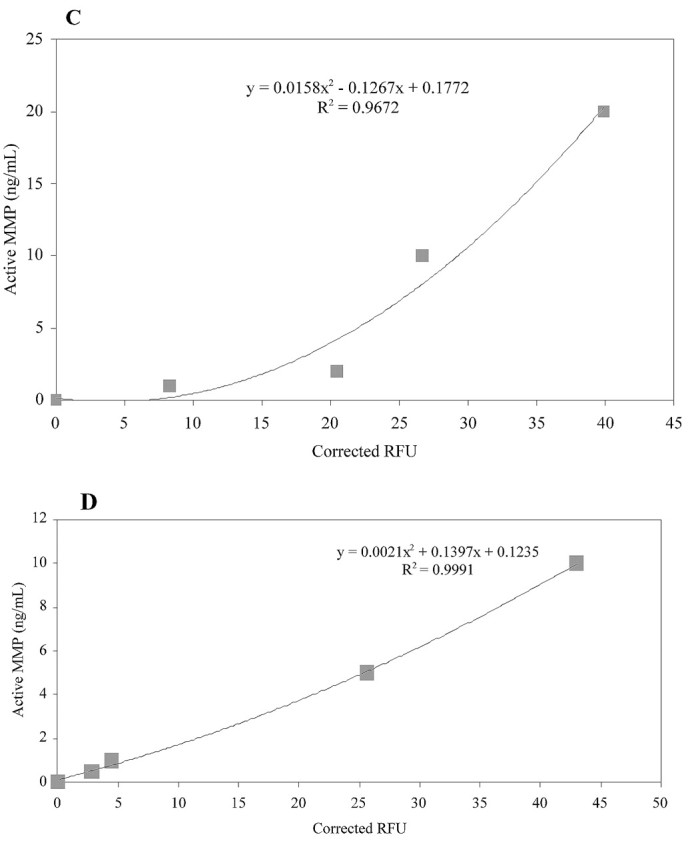
Standard curves for solid-phase MMP activity assay. Active MMP concentration was correlated to a change in substrate (fTHP-4 or fTHP-5) fluorescence using (A) MMP-1, (B) MMP-3, (C) MMP-13, and (D) MMP-14(Δ279–523). The amounts of MMP-1, MMP-3, and MMP-13 added ranged from 1.0 to 20 ng/mL, while a range of 0.5 to 10 ng/mL of MMP-14(Δ279–523) was used. Methods were as described in Results and Discussion. Corrected RFU refers to the relative fluorescence units minus background (no enzyme).
These results were somewhat surprising, considering that both MMP-13 and MMP-14(Δ279–523) hydrolyze fTHPs more efficiently than MMP-1.23,24,31 This suggests that during an extended capture and assay process, MMP-13 and MMP-14 lose some of their activity. Loss of activity was probably due to limitations in antibody recognition, or circumstances where the capture antibody obscures the MMP active site and/or destabilizes enzyme tertiary structure. Alternatively, activity loss could result from MMP autoproteolysis and/or dissociation of structural metals.34,35,40,41 Since MMP-14 capture efficiency could not be calculated, the apparently low MMP-14 sensitivity may reflect inefficient capture. With standard curves generated for MMP-1, MMP-3, MMP-13, and MMP-14, the solid-phase MMP assay was utilized to examine constitutive and inducible MMP activity in melanoma cells.
Constitutive Production of Soluble, Active MMP-3, MMP-13, and MMP-14 by Melanoma Cells
MMP-1, -2, -3, -8, and -13 are expressed in numerous human melanoma cell lines, but not melanocytes, with MMP-2 and MMP-8 expression at higher constitutive levels than other MMPs.42,43 Conversely, MMP-14 is expressed in both melanocytes and melanoma.42 MMP-14 activity has been correlated with the invasion of numerous tumor types, including melanoma.44–46 MMP-14 activity may be membrane-bound or soluble, where nonautocatalytic shedding of MMP-14 is responsible for generation of soluble activity.35 The solid-phase assay was utilized to examine constitutive MMP-1, MMP-3, MMP-13, and soluble MMP-14 (sMMP-14) activity in human melanoma cells as well as in human aortic endothelial cells, which serve as a normal control cell line. MMP activity was quantified for M14P, M14#5, and M14#11 melanoma and aortic endothelial cells by assaying conditioned media. The melanoma cells differ in their surface expression of certain receptors. M14#5 cells have higher levels of CD44 than M14P, while M14#11 cells have lower levels of CD44 than M14P.27 In contrast, the levels of the β1 integrin subunit are similar for M14P, M14#5, and M14#11.27 Finally, M14#5 expresses CD44 but does not express MPG/MCSP—another cell surface proteoglycan receptor.47 It is of interest to learn if receptor distribution has a role in active MMP production.
All three melanoma cell lines produced equivalent, low amounts of active MMP-1 and MMP-3, and levels were overall slightly higher than for endothelial cells (Table 1). M14P melanoma cells had the lowest levels of active MMP-13 and sMMP-14, followed by M14#11 and then M14#5 (Table 1). The accuracy of enzyme quantification was confirmed by spiking melanoma cell conditioned media with known concentrations of MMPs (data not shown). It is interesting to note that the cell line with the highest levels of CD44 (M14#5) had the highest levels of MMP-13 and sMMP-14 activity, since CD44 and MMP-14 are known to associate at the tumor cell migration front46,48 and MMP-14 can serve as an activator of proMMP-13.49 It will be interesting to learn from future studies if increased surface-bound MMP-14 activity also correlates with CD44 and/or MMP-13 levels. Endothelial cells exhibited constitutive MMP-13 activity that was lower than the M14#5 melanoma cell line, further suggesting that the level of this enzyme in M14#5 melanoma cells is functionally significant.
TABLE 1.
Analysis of Constitutive MMP Activity in Melanoma and Endothelial Cell Media
| Cell line | MMP-1 (ng/mL) | MMP-3 (ng/mL) | MMP-13 (ng/mL) | sMMP-14 (ng/mL) |
| M14P melanoma | 1.37 | 1.47 | 1.71 | 0.89 |
| M14#5 melanoma | 0.89 | 1.69 | 27.1 | 2.19 |
| M14#11 melanoma | 0.72 | 1.49 | 14.3 | 1.73 |
| Aortic endothelial | 0.79 | 0.59 | 7.09 | 2.17 |
Induction of Active MMP-1 by Engagement of Melanoma Cell Receptors
Engagement of the α2β1 integrin and CD44 by extracellular matrix ligands results in intracellular signaling.50,51 However, the precise nature of this signaling within melanoma cells has not been documented. The solid-phase assay was further utilized to examine induction of proteolysis upon melanoma cell α2β1 integrin and CD44 binding to “peptide-amphiphiles.” Peptide-amphiphiles consist of peptide sequences covalently linked to hydrocarbon chains, and are designed to house discrete binding sites (such as those found in various extracellular matrix proteins). These small biomolecules assume stable “mini-protein” structures (α-helical or triple-helical) at physiological temperatures, allowing for the study of cellular behaviors in response to discreet binding sites that maintain the three-dimensional structure of the parent protein. For the present studies, melanoma cells were bound to triple-helical peptide-amphiphile ligands that incorporated either an α2β1 integrin or a CD44 binding site from type IV collagen.22,27,28,33
Induction of active MMP-1 was quantified by the solid-phase assay using fTHP-4. Active enzyme induction was compared for three ligands: C10-(α1(IV)382–393)-NH2 THP, C16-(α1(IV)382–393)-NH2 THP, and C16-(α1(IV)1263–1277)-NH2 THP. The first two ligands are specific for the α2β1 integrin, while the third ligand binds CD44. The solid-phase assay showed more activity induced by the α2β1 integrin than by CD44 (4.0–7.0 versus 2.5 ng/mL) (Fig. 8). This result may be related to increased MMP-3, seen in response to the α2β1 integrin ligand.28 MMP-3 is an activator of proMMP-1.34 Prior studies had shown that binding of the α2β1 integrin to collagen results in increased production of MMP-150,52.
FIGURE 8.
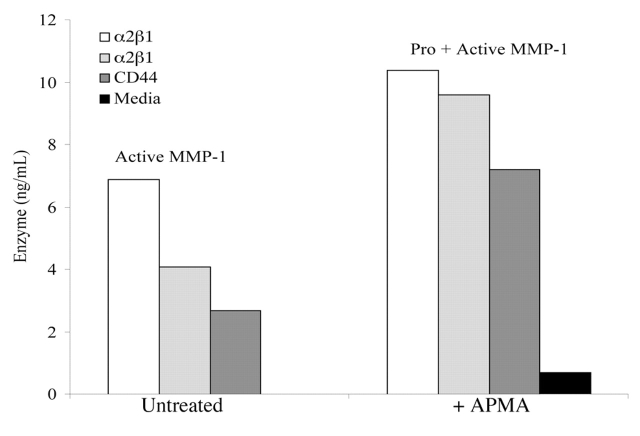
MMP-1 activity levels in conditioned media following melanoma cell adhesion to the α2β1 integrin specific C10-(α1(IV)382–393)-NH2 THP (white bar) and C16-(α1(IV)382–393)-NH2 THP (light grey bar) and the CD44/CSPG specific C16-(α1(IV)1263–1277)-NH2 THP (dark grey bar). Cell supernatants were added to 96 well plates containing 2.5 μg/mL MMP-1 mAb. Samples were either untreated (left panels) or activated with 2 mM APMA (right panels). Three washes of enzyme assay buffer were used to remove unbound proteins. The fTHP-4 substrate was added, and hydrolysis was monitored at 18 h. The black bar at the far right was media alone. Methods are as described in Materials and Methods. Results are the mean of triplicate assays.
Between the two α2β1 integrin ligands, the one with the C10 modification induced higher levels of MMP-1 activity than did the C16 modified ligand (7.0 versus 4.0 ng/mL) (Fig. 8). Differing MMP-1 induction based on the length and subsequent clustering of the α2β1 integrin ligand is reminiscent of prior studies showing different melanoma cell responses (adhesion, spreading, signaling) based on the length or density and thus accessibility of ligand.53–56 Melanoma responses to ligand are clearly dependent upon how much of the ligand is accessible for cell binding.
Treatment of samples with an activator of proMMPs (APMA) resulted in a further increase in MMP-1 activity on the order of 3.5 to 5.5 ng/mL (Fig. 8). Thus, engagement of either the α2β1 integrin or CD44 induces production of both MMP-1 and proMMP-1.
CONCLUSIONS
The solid-phase MMP activity assay has numerous advantages over prior methods for analyzing MMP activity, including combining (a) high sensitivity, (b) enzyme selectivity, and (c) high-throughput analysis. The efficiency of the assay was critically dependent upon the quality of the antibodies and enzyme standards, the use of a substrate with high specific activity, and enzyme samples that were fresh (immediately stored at −20°C and analyzed within one month). The assay did require the use of pure enzyme as a control to ensure proper quantification, although comparisons between samples can readily be made without a standard if the overall goal is to assess relative activities. Antibodies to MMP linker regions, while avoiding the enzyme active site, may be problematic if MMP autolysis occurs within the linker but the MMP retains substantial activity (i.e., MMP-3).
The solid-phase MMP activity assay was utilized to demonstrate differences in constitutive and inducible levels of active MMP production by melanoma cells. Of particular interest was the low constitutive level of MMP-1 and MMP-3 activity by all cell types while one melanoma cell line had significant MMP-13 constitutive activity. These initial studies warrant further investigation of the interactive role of CD44, MMP-13, and MMP-14 in melanoma progression and comparison between surface-bound and soluble active MMP-14 levels. Future analysis of surface-bound MMP-14 will be especially valuable, as current methods cannot quantify MMP-14 levels in cellular environments. Analysis of surface-bound MMP-14 will probably require cell lysis and inhibition of nonmetalloproteases to minimize processing of capture antibodies.
Differences in MMP-1 induction by engagement of the α2β1 integrin compared with CD44 suggested that extracellular matrix components such as type IV collagen can modulate melanoma invasion. The solid-phase MMP activity assay appears to be useful for studies on microenvironmental regulation of cellular proteolytic potential.
Acknowledgments
We gratefully acknowledge support of this work from the National Institutes of Health (AR 39189 to H.N., CA 77402, EB 00289, and CA 98799 to G.B.F.), the Wellcome Trust (reference number 057508 to H.N.), and the FAU Center of Excellence in Biomedical and Marine Biotechnology (contribution #P200410).
The solid-phase MMP activity assay was initially presented at Proteases, Extracellular Matrix and Cancer: American Association for Cancer Research Special Conference in Cancer Research, Hilton Head Island, SC, October 9–13, 2002. Solid-phase MMP activity assay kits are currently commercially available for MMP-1, MMP-8, and MMP-13 (Fluorokine E kits, R&D Systems, Minneapolis, MN).
REFERENCES
- 1.Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes, 2nd ed, vol. 1. Amsterdam: Elsevier/Academic Press; 2004.
- 2.Komsa-Penkova RS, Rashap RK, Yomtova VM. Advantages of orange-labelled collagen and gelatine as substrates for rapid collagenase activity measurement. J Biochem Biophys Methods 1997;34:237–249. [DOI] [PubMed] [Google Scholar]
- 3.Cawston TE, Koshy P, Rowan AD. Assay of matrix metalloproteinases against matrix substrates. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:389–397. [DOI] [PubMed]
- 4.Hembry RM. Detection of focal proteolysis using Texas-Red-gelatin. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Human Press, 2001:417–424. [DOI] [PubMed]
- 5.Billington CJ. Cartilage proteoglycan release assay. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:451–456. [DOI] [PubMed]
- 6.Zucker S, Mancuso P, DiMassimo B, Lysik RM, Conner C, Wu C-L. Comparison of techniques for measurement of gelatinases/type IV collagenases: Enzyme-linked immunoassays versus substrate degradation assays. Clin Exp Metastasis 1994;12:13–23. [DOI] [PubMed] [Google Scholar]
- 7.Gogly B, Groult N, Hornebeck W, Godeau G, Pellat B. Collagen zymography as a sensitive and specific technique for the determination of subpicogram levels of interstitial collagenase. Anal Biochem 1998;255:211–216. [DOI] [PubMed] [Google Scholar]
- 8.Knauper V, Murphy G. Methods for studying activation of matrix metalloproteinases. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:377–387. [DOI] [PubMed]
- 9.Hawkes SP, Li H, Taniguchi GT. Zymography and reverse zymography for detecting MMPs and TIMPs. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:399–410. [DOI] [PubMed]
- 10.George SJ, Johnson JL. In situ zymography. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:411–415. [DOI] [PubMed]
- 11.Fujimoto N, Iwata K. Use of EIA to measure MMPs and TIMPs. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:p 347–358. [DOI] [PubMed]
- 12.Fosang AJ, Last K, Jackson DC, Brown L. Antibodies to MMP-cleaved aggrecan. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:425–449. [DOI] [PubMed]
- 13.Billinghurst RC, Ionescu M, Poole AR. Immunoassay for collagenase-mediated cleavage of types I and II collagens. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:457–472. [DOI] [PubMed]
- 14.Miller JA, Liu R-Q, Davis GL, Pratta MA, Trzaskos JM, Copeland RA. A microplate assay specific for the enzyme aggrecanase. Anal Biochem 2003;314:260–265. [DOI] [PubMed] [Google Scholar]
- 15.Kashiwagi M, Enghild JJ, Gendron C, et al. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J Biol Chem 2004;279:10109–10119. [DOI] [PubMed] [Google Scholar]
- 16.Nagase H, Woessner JF, Jr. An improved assay for proteases and polysaccharides employing a cartilage proteoglycan substrate entrapped in polyacrylamide particles. Anal Biochem 1980;107:385–392. [DOI] [PubMed] [Google Scholar]
- 17.Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers 1996;40:399–416. [DOI] [PubMed] [Google Scholar]
- 18.Amour A, Slocombe PM, Webster A, et al. TNF-α converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett 1998;435:39–44. [DOI] [PubMed] [Google Scholar]
- 19.Fields GB. Using fluorogenic peptide substrates to assay matrix metalloproteinases. In: Clark IM (ed). Methods in Molecular Biology 151: Matrix Metalloproteinase Protocols. Totowa, NJ: Humana Press, 2001:495–518. [DOI] [PMC free article] [PubMed]
- 20.Lauer-Fields JL, Kele P, Sui G, Nagase H, Leblanc RM, Fields GB. Analysis of matrix metalloproteinase activity using triple-helical substrates incorporating fluorogenic l- or d-amino acids. Anal Biochem 2003;321:105–115. [DOI] [PubMed] [Google Scholar]
- 21.Nagase H, Fields CG, Fields GB. Design and characterization of a fluorogenic substrate selectively hydrolyzed by stromelysin 1 (matrix metalloproteinase-3). J Biol Chem 1994;269:20952–20957. [PubMed] [Google Scholar]
- 22.Lauer-Fields JL, Sritharan T, Stack MS, Nagase H, Fields GB. Selective hydrolysis of triple-helical substrates by matrix metalloproteinase-2 and -9. J Biol Chem 2003; 278:18140–18145. [DOI] [PubMed] [Google Scholar]
- 23.Minond D, Lauer-Fields JL, Nagase H, Fields GB. Matrix metalloproteinase triple-helical peptidase activities are differentially regulated by substrate stability. Biochemistry 2004;43:11474–11481. [DOI] [PubMed] [Google Scholar]
- 24.Lauer-Fields JL, Broder T, Sritharan T, Nagase H, Fields GB. Kinetic analysis of matrix metalloproteinase triple-helicase activity using fluorogenic substrates. Biochemistry 2001;40:5795–5803. [DOI] [PubMed] [Google Scholar]
- 25.Lauer-Fields JL, Fields GB. Triple-helical peptide analysis of collagenolytic protease activity. Biol Chem 2002;383:1095–1105. [DOI] [PubMed] [Google Scholar]
- 26.Malkar NB, Lauer-Fields JL, Borgia JA, Fields GB. Modulation of triple-helical stability and subsequent melanoma cellular responses by single-site substitution of fluoroproline derivatives. Biochemistry 2002;41: 6054–6064. [DOI] [PubMed] [Google Scholar]
- 27.Lauer-Fields JL, Malkar NB, Richet G, Drauz K, Fields GB. Melanoma cell CD44 interaction with the a1(IV)1263–1277 region from basement membrane collagen is modulated by ligand glycoslyation. J Biol Chem 2003;278:14321–14330. [DOI] [PubMed] [Google Scholar]
- 28.Baronas-Lowell D, Lauer-Fields JL, Borgia JA, et al. Differential modulation of human melanoma cell metalloproteinase expression by α2β1 integrin and CD44 triple-helical ligands derived from type IV collagen. J Biol Chem 2004;279:43503–43513. [DOI] [PubMed] [Google Scholar]
- 29.Chung L, Shimokawa K, Dinakarpandian D, Grams F, Fields GB, Nagase H. Identification of the RWTNNFREY(183–191) region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J Biol Chem 2000;275:29610–29617. [DOI] [PubMed] [Google Scholar]
- 30.Huang W, Suzuki K, Nagase H, Arumugam S, Van Doren S, Brew K. Folding and characterization of the amino-terminal domain of human tissue inhibitor of metalloproteinases-1 (TIMP-1) expressed at high yield in E. coli. FEBS Lett 1996;384:155–161. [DOI] [PubMed] [Google Scholar]
- 31.Hurst DR, Schwartz MA, Ghaffari MA, et al. Catalytic- and ecto-domains of membrane type 1-matrix metalloproteinase have similar inhibition profiles but distinct endopeptidase activities. Biochem J 2004;377:775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malkar NB, Lauer-Fields JL, Juska D, Fields GB. Characterization of peptide-amphiphiles possessing cellular activation sequences. Biomacromolecules 2003;4:518–528. [DOI] [PubMed] [Google Scholar]
- 33.Baronas-Lowell D, Lauer-Fields JL, Fields GB. Induction of endothelial cell activation by a triple-helical α2b1 integrin ligand derived from type I collagen α1(I)496–507. J Biol Chem 2004;279:952–962. [DOI] [PubMed] [Google Scholar]
- 34.Woessner JF, Nagase H. Matrix Metalloproteinases and TIMPs. Oxford: Oxford University Press, 2000.
- 35.Toth M, Hernandez-Barrantes S, Osenkowski P, et al. Complex pattern of membrane type I matrix metalloproteinase shedding. J Biol Chem 2002;277:26340–26350. [DOI] [PubMed] [Google Scholar]
- 36.Crowther JR. ELISA: Theory and Practice. Totowa, NJ: Humana Press, 1995.
- 37.Lauer-Fields JL, Nagase H, Fields GB. Use of Edman degradation sequence analysis and matrix-assisted laser desorption/ionization mass spectrometry in designing substrates for matrix metalloproteinases. J Chromatogr A. 2000;890:117–125. [DOI] [PubMed] [Google Scholar]
- 38.Tamei H, Azumano I, Iwata K, et al. One-step sandwich enzyme immunoassays for human matrix metalloproteinase 13 (collagenase-3) using monoclonal antibodies. Connect Tissue 1998;30:15–22. [Google Scholar]
- 39.Maliszewska M, Mader M, Scholl U, et al. Development of an ultrasensitive enzyme immunoassay for the determination of matrix metalloproteinase-9 (MMP-9) levels in normal human cerebrospinal fluid. J Neuroimmunol 2001;116:233–237. [DOI] [PubMed] [Google Scholar]
- 40.Okada Y, Nagase H, Harris ED, Jr. A metalloproteinase from human rheumatoid synovial fibroblasts that digests connective tissue matrix components. J Biol Chem 1986;261:14245–14255. [PubMed] [Google Scholar]
- 41.Wilhelm SM, Shao ZH, Housley TJ, et al. Matrix metalloproteinase-3 (stromelysin-1). J Biol Chem 1993;268: 21906–21913. [PubMed] [Google Scholar]
- 42.Giambernardi TA, Sakaguchi AY, Gluhak J, et al. Neutrophil collagenase (MMP-8) is expressed during early development in neural crest cells as well as in adult melanoma cells. Matrix Biol 2001;20:577–587. [DOI] [PubMed] [Google Scholar]
- 43.Ntayi C, Lorimier S, Berthier-Vergnes O, Hornebeck W, Bernard P. Cumulative influence of matrix metalloproteinase-1 and -2 in the migration of melanoma cells within three-dimensional type I collagen lattices. Exp Cell Res 2001;270:110–118. [DOI] [PubMed] [Google Scholar]
- 44.Ellerbroek SM, Stack MS. Membrane associated matrix metalloproteinases in metastasis. BioEssays 1999;21: 940–949. [DOI] [PubMed] [Google Scholar]
- 45.Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J Cell Biol 2000; 149:1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seiki M. Membrane-type 1 matrix metalloproteinase: A key enzyme for tumor invasion. Cancer Lett 2003; 194:1–11. [DOI] [PubMed] [Google Scholar]
- 47.Knutson JR, Iida J, Fields GB, McCarthy JB. CD44/chondroitin sulfate proteoglycan and α2b1 integrin mediate human melanoma cell migration on type IV collagen and invasion of basement membranes. Mol Biol Cell 1996;7:383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorne RF, Legg JW, Isacke CM. The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J Cell Sci 2004;117:373–380. [DOI] [PubMed] [Google Scholar]
- 49.Knauper V, Will H, Lopez-Otin C, et al. Cellular mechanisms for human procollagenase-3 (MMP-13) activation: Evidence that MT1-MMP (MMP-14) and gelatinase A (MMP-2) are able to generate active enzyme. J Biol Chem 1996;271:17124–17131. [DOI] [PubMed] [Google Scholar]
- 50.Vogel WF. Collagen-receptor signaling in health and disease. Eur J Dermatol 2001;11:506–514. [PubMed] [Google Scholar]
- 51.Turley EA, Noble PW, Bourguignon LYW. Signaling properties of hyaluronan receptors. J Biol Chem 2002;277:4589–4592. [DOI] [PubMed] [Google Scholar]
- 52.Heino J. The collagen receptor integrins have distinct ligand recognition and signaling functions. Matrix Biol 2000;19:319–323. [DOI] [PubMed] [Google Scholar]
- 53.Yu Y-C, Pakalns T, Dori Y, McCarthy JB, Tirrell M, Fields GB. Construction of biologically active protein molecular architecture using self-assembling peptide-amphiphiles. Meth Enzymol 1997;289:571–587. [DOI] [PubMed] [Google Scholar]
- 54.Lauer JL, Gendron CM, Fields GB. Effect of ligand conformation on melanoma cell α3b1 integrin-mediated signal transduction events: Implications for a collagen structural modulation mechanism of tumor cell invasion. Biochemistry 1998;37:5279–5287. [DOI] [PubMed] [Google Scholar]
- 55.Fields GB, Lauer JL, Dori Y, Forns P, Yu Y-C, Tirrell M. Proteinlike molecular architecture: Biomaterial applications for inducing cellular receptor binding and signal transduction. Biopolymers 1998;47:143–151. [DOI] [PubMed] [Google Scholar]
- 56.Dori Y, Bianco-Peled H, Satija SK, Fields GB, McCarthy JB, Tirrell M. Ligand accessibility as a means to control cell response to bioactive bilayer membranes. J Biomed Mater Res 2000;50:75–81. [DOI] [PubMed] [Google Scholar]
- 57.Hostikka SL, Tryggvason K. The complete primary structure of the α2 chain of human type IV collagen and comparison with the α1(IV) chain. J Biol Chem 1988;263:19488–19493. [PubMed] [Google Scholar]