Abstract
Anion-exchange chromatography with integrated pulsed amperometric detection (AE-IPAD) separates and directly detects amino acids, carbohydrates, alditols, and glycols in the same injection without pre- or post-column derivatization. These separations use a combination of NaOH and NaOH/sodium acetate eluents. We previously published the successful use of this technique, also known as AAA-Direct, to determine free amino acids in cell culture and fermentation broth media. We showed that retention of carbohydrates varies with eluent NaOH concentration differently than amino acids, and thus separations can be optimized by varying the initial NaOH concentration and its duration. Unfortunately, some amino acids eluting in the acetate gradient portion of the method were not completely resolved from system-related peaks and from unknown peaks in complex cell culture and fermentation media. In this article, we present changes in method that improve amino acid resolution and system ruggedness. The success of these changes and their compatibility with the separations previously designed for fermentation and cell culture are demonstrated with yeast extract-peptone-dextrose broth, M199, Dulbecco’s modified Eagle’s (with F-12), L-15 (Leibovitz), and McCoy’s 5A cell culture media.
Keywords: amino acids, carbohydrates, chromatography, amperometry, media
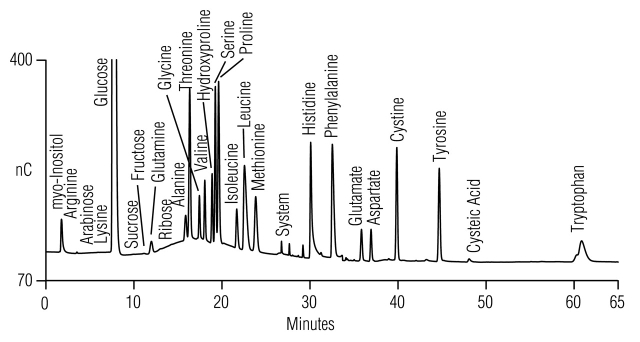
Anion-exchange chromatography with integrated pulsed amperometric detection (AE-IPAD) is an established technique for amino acid analysis.1,2 This technique, also known as AAA-Direct, separates amino acids as anions and directly detects them without pre- or post-column derivatization (Fig. 1). The AminoPac PA10 (Dionex, Sunnyvale, CA) is commonly used for AE-IPAD amino acid analysis. AE-IPAD also detects carbohydrates, glycols, and sugar alcohols. The presence of these compounds, often at high concentrations in cell culture and fermentation broth media, can complicate amino acid determinations. We previously showed that the AminoPac PA10 can separate amino acids from the carbohydrates, glycols, and sugar alcohols in cell media by altering and extending the initial NaOH eluent concentration.1,2 Carbohydrate retention is impacted differently from amino acid retention by a change in NaOH concentration. We used this selectivity difference to design AE-IPAD methods for amino acid determinations of diluted cell culture and fermentation media, including Bacto yeast extract-peptone-dextrose (YPD; yeast culture medium) broth; Luria-Bertani (bacterial culture medium); minimal essential medium; and serum-free, protein-free hybridoma medium (mammalian cell culture media). Since these publications, there have been several modifications of the AE-IPAD method:
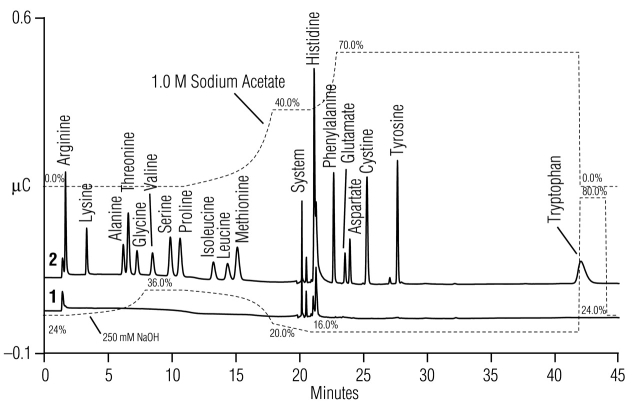
FIGURE 1.
Common amino acids separated using the standard unmodified (Table 1, Method 1) gradient method. Comparison of chromatograms for (1) water blank, and (2) 8 μM amino acid standard mix.
improved long-term stability of the AE-IPAD system,
improved histidine (His) peak shape and resolution,
reduced minor gradient/system peaks, and
reduced carryover of His, aspartate (Asp), and glutamate (Glu).3
By adding NaOH to the water (10 mM, channel A) and the 1-M sodium acetate (25 mM, channel C) eluents, by adding a fourth channel containing 100 mM acetic acid for additional column washing, and by altering the elution program, we were able to combine all the separate modifications into one program. We also modified the gradient method to improve the resolution of phenylalanine (Phe), Asp, Glu, and cystine from the remaining, small coeluting system peaks and from unknown ingredient peaks found in some complex media.
This article shows that the combined changes to the AE-IPAD method improve the performance of our previously published method. The new method successfully determined amino acids in YPD broth, M199, Dulbecco’s modified Eagle’s (with F-12), L-15 (Leibovitz), and McCoy’s 5A cell culture media.
MATERIALS AND METHODS
Amino acids, carbohydrates, alditols, and other compounds investigated in this research were purchased from the National Institute for Standards and Technology (NIST, Standard Reference Material 2389, Gaithersburg, MD), Sigma-Aldrich (St. Louis, MO), Pfanstiehl Laboratories (Waukegan, IL), McNeil Nutritionals (Fort Washington, PA), Eastman Chemical (Rochester, NY), Fisher Scientific (Pittsburgh, PA), J.T. Baker (Phillipsburg, NJ), or EM Science (Gibbstown, NJ).
The cell culture media were as follows: YPD broth (BD Diagnostics, Sparks, MD; cat. no. 242820); Dulbecco’s modified Eagle’s medium F12 (Sigma-Aldrich; cat. no. D6421); medium 199 (Sigma-Aldrich; cat. no. M4530); L-15 medium Leibovitz (Sigma-Aldrich; cat. no. L5520); and McCoy’s 5A medium modified (Sigma-Aldrich; cat. no. M8403).
All solid media (YPD broth) were reconstituted in water to their normal concentrations, centrifuged at 16,000 × g for 10 min, and the supernatant diluted in water. Liquid media or reconstituted media were diluted 10-, 100-, or 1000-fold in water. These were analyzed directly.
The equipment used in this study was as follows. A BioLC system configured for microbore (Dionex, Sunnyvale, CA) consisted of (a) a GP50 gradient pump with vacuum degas option and GM-4 gradient mixer, (b) ED50 electrochemical detector with AAA-Direct certified disposable electrodes (P/N 060082) and combination pH/Ag/AgCl reference electrode (P/N 044198), (c) AS50 autosampler with 25-μL injection loop, (d) AS50 thermal compartment, and (e) E01 eluent organizer, including four 2-L plastic bottles and pressure regulator. Chromeleon chromatography management software (Dionex) was utilized. Helium was 4.5 grade, high purity 99.5%. The filter unit was 0.2 μm nylon (Nalgene 90-mm Media-Plus; Nalge Nunc International, Rochester, NY; P/N 500–118). The vacuum pump was from Gast Manufacturing (Benton Harbor, MI; P/N DOA-P104-AA or equivalent). Vials were 0.5 mL, polypropylene, microinjection, 12–32-mm screw thread cap, and pre-slit Teflon/silicone septum (Dionex; P/N 055428).
Chromatography columns were AminoPac PA10 analytical column, 2 × 250 mm (Dionex; P/N 55406) and AminoPac PA10 guard column, 2 × 50 mm (Dionex; P/N 55407). Chromatography conditions were flow rate 0.25 mL/min, temperature 30°C, injection volume 25 μL (full-loop injection mode), reference electrode mode pH. The waveform for the ED50 detector was as described in refs. 3 and 4. The eluents were as follows: A, 10 mM NaOH; B, 250 mM NaOH; C, 25 mM NaOH in 1 M sodium acetate; and D, 100 mM acetic acid. Deionized water was 18 MΩ-cm resistance or better, free of biological or chemical contamination, prefiltered through 0.2-μm nylon filter to degas. Gradient conditions are described in Table 1.
Table 1.
Gradient Methods
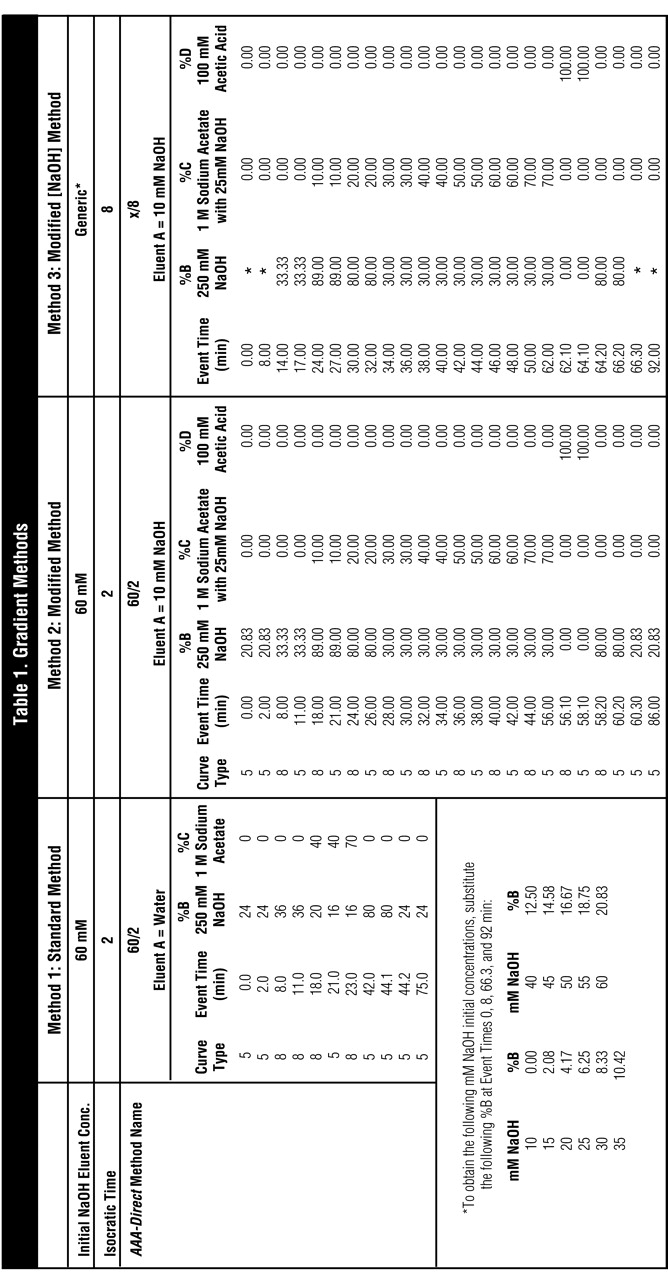
In this article, conditions are described in Table 1, Method 3 as x/y, where x is the initial NaOH eluent concentration in millimolar and y is the isocratic duration for this eluent in minutes. For example, condition “15/8” refers to a method using 15 mM NaOH as the starting eluent concentration which is held for 8 min before the start of the NaOH gradient.
RESULTS AND DISCUSSION
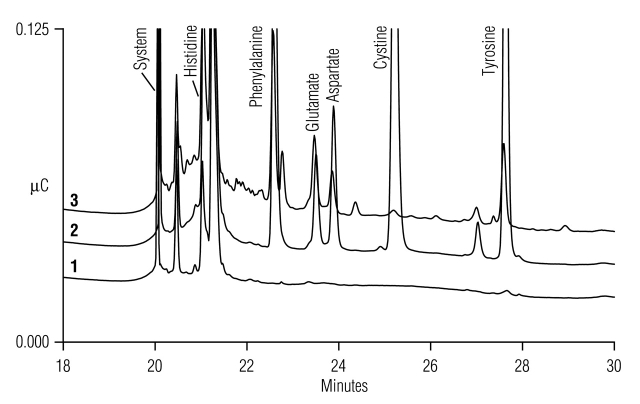
The AE-IPAD system using the standard AAA-Direct gradient method (Table 1, Method 1)1,2 is well suited for most amino acid, carbohydrate, alditol, and glycol separations. However, (a) His coelutes with minor system peaks, (b) there is some His peak distortion, (c) there is 1–4% carryover of His, Asp, Glu, and Tyr from previous injections, (d) there is a tendency for system impurities to increase over time from biological contamination of water and acetate eluent lines, and (e) incomplete resolution of ingredient peaks present in complex culture media eluting during the acetate gradient. These deficiencies are shown in Figure 2.
FIGURE 2.
Acetate gradient region using the standard unmodified (Table 1, Method 1) gradient method for (1) a water blank, (2) amino acid standards, and (3) 1000-fold diluted YPD broth supernatant.
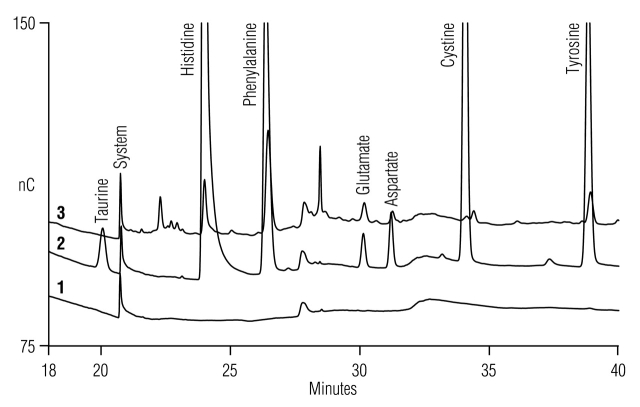
To correct these deficiencies, we developed a new method that included NaOH in both the water (channel A) and 1 M sodium acetate (channel C) eluents to maintain sterility of the eluent lines (Table 1, Method 2). This reduced the appearance of minor system peaks during the acetate gradient, and increased system ruggedness by reducing the need to perform system sanitization. The gradient program was modified to accommodate the NaOH in channels A and C, and to resolve the small system peaks that remain and the unknown cell culture media ingredients from His, Phe, Asp, Glu, and cystine. Finally, the column was washed with 100 mM acetic acid for 2 min to eliminate minor carryover of His, Phe, Asp, Glu, and Tyr, and thus increase the quantitative accuracy for these peaks near their lower limits of detection. Previous approaches to solving some of these deficiencies are described as separate methods.3 Here we present a single method that incorporates all improvements (Fig. 3). The new method reduces system peaks, resolves the remaining minor system peaks from the peaks of interest, increases long-term stability of the AAA-Direct system, eliminates trace carryover from previous injections, and improves resolution of unknown media-related peaks. Good resolution of all amino acids is maintained without frequent system sanitization. Comparing Figures 2 and 3 illustrates the improvements realized with the new method. For example, the unknown peak eluting just after His in the water blank chromatogram using the standard gradient (Fig. 2) is absent in the water blank using the new gradient (Fig. 3). The unknown ingredient peak in YPD broth on the trailing edge of the Phe peak using the standard gradient (Fig. 2) is resolved into about four separate peaks eluting between Phe and Glu using the new method (Fig. 3).
FIGURE 3.
Acetate gradient region using the modified (Table 1, Method 2) gradient method for injections of (1) a water blank, (2) amino acid standards, and (3) 1000-fold diluted YPD broth supernatant.
The improved method (Table 1, Method 2) was tested for compatibility with changes in initial eluent NaOH concentration and change in duration of the initial NaOH concentration. We found that the method changes did not interfere with the selectivity changes achieved by varying initial NaOH concentration and duration of that eluent (Table 1, Method 3). These selectivity changes can be used to optimize separations of amino acids in cell cultures and fermentation broths. Measured retention times of 30 amino acids, and 42 carbohydrates using varying initial eluent NaOH concentrations were published for the previous gradient method,1–2 and can be used to design specific separations; however, most retention times will differ with the new methods.
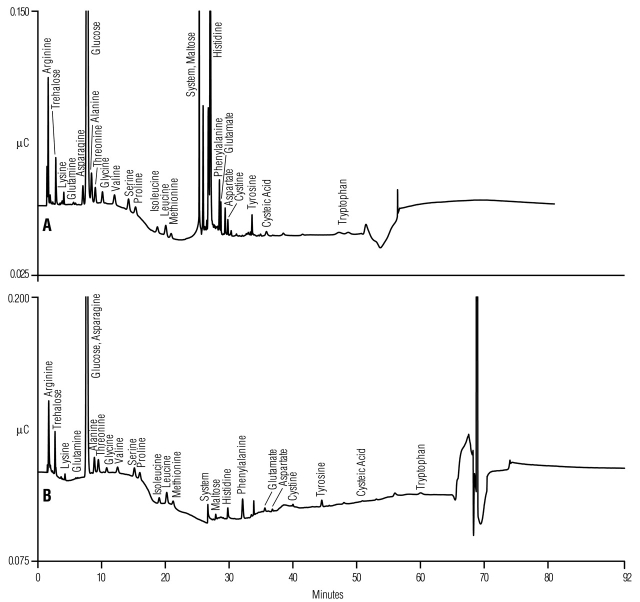
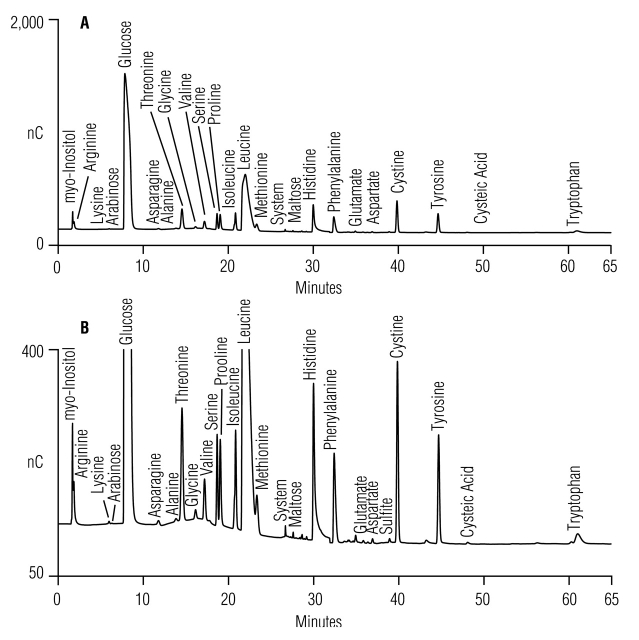
Our improved gradient methods (Table 1, Method 3) were tested for suitability using complex undefined media, such as YPD broth supernatant. Figure 4A shows a separation of YPD broth supernatant using the previously published 40/8 method1,2 where incomplete resolution of some amino acids and unidentified ingredient peaks are observed in the acetate gradient region of the chromatogram. Figure 4B shows the improved resolution of amino acids and unidentified ingredient peaks using the improved 40/8 method (Table 1, Method 3). For example, the improved separation resolves maltose from the main system peak.
FIGURE 4.
Separation of amino acids, carbohydrates, alditols in 1000-fold dilution of YPD broth supernatant (10 μL) using (A) the previously published1,2 AAA-Direct 40/8 gradient method, and (B) modified (Table 1, Method 3) 40/8 gradient method.
In Figure 5, using method 15/8 (Table 1, Method 3) we identified all amino acid and carbohydrate peaks expected to be present in medium 1994 (see Table 2), except Cys, which is converted to cystine under the alkaline conditions used for separation, and deoxyribose, for which standards were unavailable at the time of this work. The presence of phenol red (pH indicator), sodium bicarbonate (buffer), and the many other ingredients (Table 2) did not appear to cause interference at a 10-fold dilution. Trace amounts of fructose and sucrose, both common impurities of dextrose, were detected. Their elution positions are marked, but their peaks are not observed using the scale of Figure 5.
FIGURE 5.
Separation of amino acids in medium 199 (10-fold dilution, 25 μL) using a 15/8 gradient method (Table 1, Method 3).
Table 2.
Media Ingredients
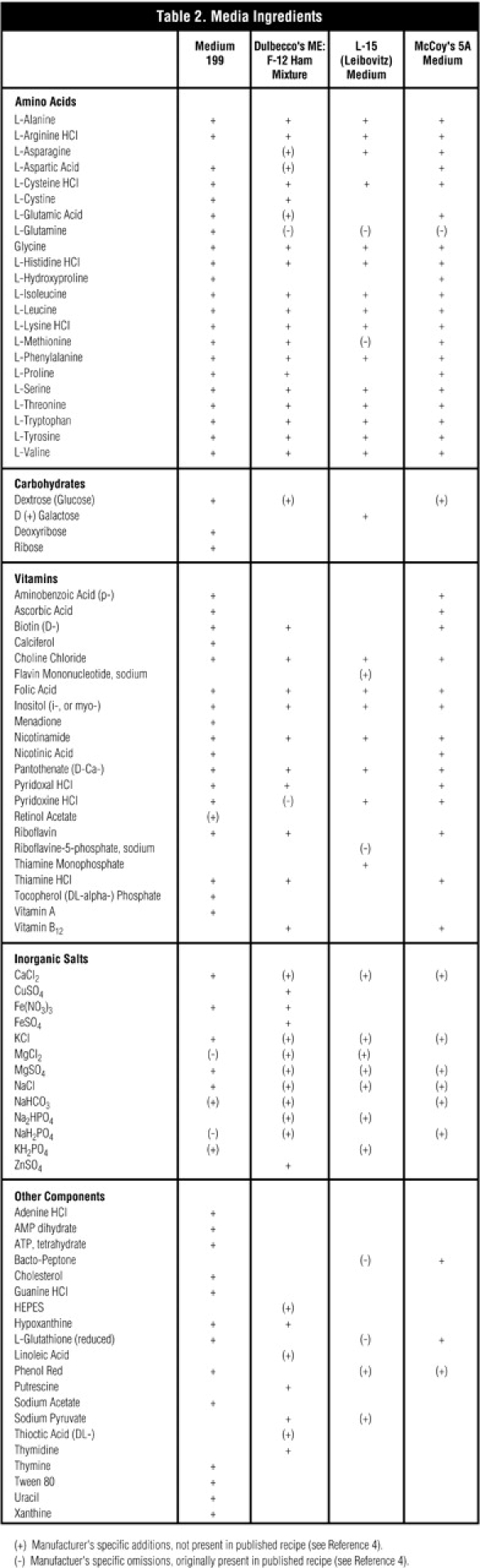
Figure 6 shows that a 15/8 method (Method 3) separates all amino acid and carbohydrate peaks expected to be present in Dulbecco’s modified Eagle’s F-12 Ham mixture (Table 2), except Cys for the reason explained above. Of particular interest in this medium was the exceptionally high level of Leu. The expected concentration was 59 μg/mL, and the measured concentration was 990 μg/mL. The other amino acids were measured close to their expected levels. The same results were obtained when using our other eluent conditions—where selectivity may be modified for some compounds—and when using greater sample dilutions. We surveyed five media manufacturers and found that only one manufacturer performs quality control measurements of amino acid content in their final formulation.
FIGURE 6.
Separation of amino acids in Dulbecco’s Modified Eagle’s F-12 Ham Mixture (10-fold dilution, 25 μL) using the 15/8 gradient method (Table 1, Method 3). (A) Full scale, and (B) expanded view of the baseline.
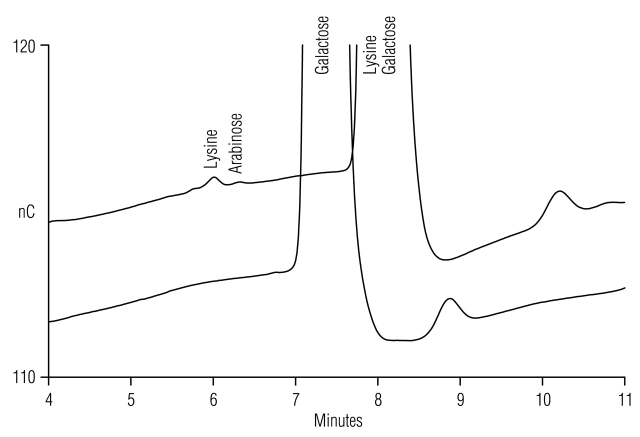
All the expected amino acids in L-15 (Leibovitz) medium are identified in Table 2. In Figure 7, Lys is shown to coelute with galactose at 15 mM NaOH (Method 3, condition 15/8) and resolve at 20 mM NaOH (Method 3, condition 20/8). L-15 contains galactose instead of the commonly used glucose (dextrose) as a carbon source during cell culture. Gln was omitted from this media, Asp and Glu are not normal ingredients of this formulation, and as expected, none were detected.
FIGURE 7.
Separation of Lys from galactose in L-15 (Leibovitz) medium (100-fold dilution, 25 μL) by adjusting the gradient method from 15/8 (upper trace) to 20/8 (lower trace) (Table 1, Method 3).
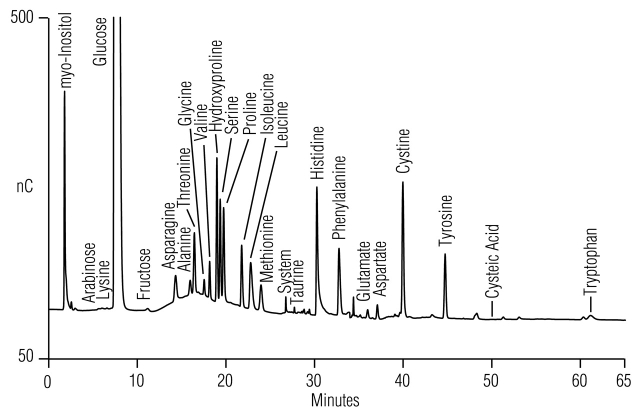
Figure 8 shows that the 15/8 method (Method 3) separates all expected amino acids—except Cys—in McCoy’s 5A medium, including hydroxyproline (Table 2).
FIGURE 8.
Separation of amino acids and carbohydrates in McCoy’s 5A medium (10-fold dilution, 25 μL) using the 15/8 gradient method (Table 1, Method 3).
In all the media studied using the improved AAA-Direct gradient methods, there were numerous unidentified peaks. We speculate that with additional studies, some of the unknown peaks will be identified as important constituents of media (e.g., specific vitamins). We believe the improved resolution of these peaks using the gradient methods described in this paper enables future discoveries.
SUMMARY
The revised gradient methods improved resolution of His, Phe, Asp, Glu, and cystine from minor system peaks, and maltose from the major system peak (compare Figs. 2 and 3). The addition of NaOH to the eluent channels sensitive to bioburden improved system robustness by maintaining sterility, and thereby reduced minor system peaks. The addition of an acetic-acid column wash significantly reduced carryover of His, Asp, and Glu. These changes were compatible with and improved determinations of amino acids in cell culture and fermentation broth media (Figs. 4–8).
Acknowledgments
AAA-Direct is a trademark of the Dionex Corporation. BioLC, AminoPac, and Chromeleon are registered trademarks of the Dionex Corporation.
REFERENCES
- 1.Dionex Corporation. Determination of amino acids in cell cultures and fermentation broths. Application Note 150, LPN 1538. July2003.
- 2.Hanko VP, Rohrer JS. Determination of amino acids in cell culture and fermentation broth media using anion-exchange chromatography with integrated pulsed amperometric detection. Anal Biochem 2004;324:29–38. [DOI] [PubMed] [Google Scholar]
- 3.Dionex Corporation. Installation instructions and troubleshooting guide for the AAA-Direct Amino Acid Analysis System. Document No. 031481, Version 8; 11August2003.
- 4.Paul J. Cell and Tissue Culture, 5th ed. New York: Churchill Livingstone; 1975.