Abstract
To detect diseases early in the general population, new diagnostic approaches are needed that have adequate sensitivity and specificity. Recent studies have used mass spectrometry to identify a serum proteomic pattern for breast and ovarian cancer. Serum contains 60–80 mg/mL protein, but 57–71% of this is serum albumin, and 8–26% are γ-globulins. These large proteins must be depleted before smaller, less-abundant proteins can be detected using mass spectrometry, but because serum albumin is known to act as a carrier for smaller proteins, removal of these molecules using columns or filtration may result in the loss of molecules of interest. The objective of this study was to develop a reproducible method to deplete serum samples of high-abundance proteins in order to analyze the less-abundant proteins present in serum. We used organic solvents to precipitate the large proteins out of solution. We also predicted that this would cause many smaller proteins to dissociate from their carrier molecules, allowing for detection of a larger number of peptides and small proteins. These treated samples were analyzed using capillary liquid chromatography coupled with electrospray ionization mass spectrometry. Analysis demonstrated reproducible results. Acetonitrile treatment clearly released many carrier-bound molecular species and was superior to ultrafiltration alone for serum proteomic analysis.
Keywords: biomarkers, serum, albumin, proteome, LC-MS
Many diseases, such as cancer, do not show obvious clinical symptoms until the disease is in advanced stages and difficult to treat successfully. Detection of such diseases in early stages can greatly reduce disease-related mortality.1,2 Traditionally, researchers have searched for disease biomarkers using a one-at-a-time approach. However, because of the complexity of many diseases, it is very likely that the use of multiple biomarkers (protein pattern diagnostics) in screening and diagnosis will be necessary to produce unequivocal results.1,3
Biomarkers are often low-molecular-weight proteins secreted into the bloodstream as a result of the disease process.3 Employing mass spectrometry (MS) in serum proteomic pattern diagnostics has a very high potential for discovering new, relevant disease-related biomarkers.4 Serum contains a total of 60–80 mg/mL of protein, with a concentration range spanning at least nine orders of magnitude. An estimated 10,000 different proteins typically exist in serum, the majority of which are low-molecular-weight species.5 The complexity of serum makes it a very informative source for developing a proteomic pattern; however, 65–97% of serum protein is serum albumin and immunoglobulins. The presence of these highly abundant proteins makes detection of the smaller, less-abundant serum proteins difficult.5,6
Some have used Cibacron dye and protein A/G columns to remove serum albumin and immunoglobulins from serum effectively, allowing detection of biomarkers present at very low concentrations.6,7 Molecular-weight cutoff filters have also been evaluated in separating the large, abundant proteins from the smaller, less-abundant ones.8 Albumin serves as a transport protein that binds, and thereby accumulates many low-abundance biomarkers that would otherwise be cleared from the blood by the kidney.3,5,9 Consequently, removal of serum albumin by filtration also inadvertently removes many small proteins and peptides of interest.
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), surface-enhanced laser desorption/ionization mass spectrometry (SELDI-MS), and liquid chromatography coupled to online electrospray mass spectrometry (cLC-MS) are methods that have been used for protein pattern development for several diseases. MALDI-MS employs no separation, and it is sensitive to organic and inorganic contaminants as evidenced by signal background problems.1 SELDI-MS is an affinity-based method that uses various stationary sorbants (hydrophobic, anionic, cationic, and metal binding) to selectively bind proteins of interest. Because many biomarkers are present at very low concentrations in biological fluids, they are not likely to be immobilized on the biochips used in SELDI-MS but may be washed away with all of the other protein species that do not bind to the active surface.10,11 Current SELDI instruments are also severely limited in their resolution. cLC-MS, while not as high throughput as MALDI-MS or SELDI-MS, is more effective for the detection of low-molecular weight, low-abundance proteins and peptides in a complex mixture.
In this report, we hypothesized that using acetonitrile (ACN) precipitation, which has been shown to effectively precipitate large, abundant proteins out of serum,12 is a superior method to remove serum albumin and immunoglobulins from serum because ACN not only denatures these large proteins allowing for their removal but should also cause the small, low-abundance proteins and peptides normally bound to these carrier proteins to dissociate, thus making them available for detection using cLC-MS. We report that this procedure was effective and allowed for the visualization of small proteins and peptides from serum. An average of approximately 4000 ionized species under 2.5 kDa, as well as some proteins between 2.5 and 10 kDa, were detected per serum sample analyzed.
MATERIALS AND METHODS
All reagents were purchased from Sigma (St. Louis, MO).
Sample Collection
After Institutional Review Board (IRB) approval was obtained at the University of Utah (IRB no. 10535), serum samples were obtained from women who presented to the Labor and Delivery suite with a pregnancy at or greater than 36 weeks gestation. Blood was obtained by antecubital venipuncture and after centrifugation at 3500 rpm for 15 min, serum was collected, aliquoted, and frozen at −80°C until further processing. Samples were also obtained at the Brigham Young University Student Health Center from healthy male volunteers.
Acetonitrile Precipitation
Briefly, the “standard” method12 involved adding 2 volumes of high-performance liquid chromatography (HPLC) grade ACN to 1 volume of serum followed by gentle mixing. After standing at room temperature for 30 min, samples were spun for 4 min at 12,000 rpm. An aliquot of supernatant was then lyophilized to 50 μL. The sample was reconstituted to its original volume by addition of water.
The “modified” method involved adding 2 volumes of HPLC grade ACN (400 μL) to 200 μL serum, vortexing vigorously for 5 sec, and allowing to stand at room temperature for 30 min. Samples were then spun for 10 min at 12,000 rpm in an IEC Micromax RF centrifuge (Thermo Electron, San Jose, CA) at room temperature. An aliquot of supernatant (550 μL) was then lyophilized to 200 μL in a vacuum centrifuge (CentriVap Concentrator, Labconco, Kansas City, MO). Supernatant protein concentration was determined using a Bio-Rad (Hercules, CA) microtiter plate protein assay according to manufacturer’s instructions. An aliquot containing 4 μg protein was transferred to a new microcentrifuge tube and lyophilized to near dryness. Twenty microliters of 88% formic acid, 2 μL of 5 pmol/μL mellitin, and 2 μL of 5 pmol/μL [Glu1]-fibrinopeptide was added to lyophilized samples. Samples were brought to 40 μL with HPLC water.
Centrifugal Ultrafiltration
One milliliter of serum was diluted with 5 mL of 25 mM ammonium bicarbonate and added to an appropriately conditioned 30-KDa nominal molecular weight limit cutoff centrifugal filter. After centrifugation, the filtrate was lyophilized to dryness and reconstituted to 40 μL with 20 μL 88% formic acid and 20 μL water. The retentate was reconstituted to its original 1-mL volume with HPLC grade water. Additionally, 200 μL of retentate were subjected to ACN precipitation and 1 μg of protein of each sample was loaded onto the column according to procedures outlined below.
cLC-MS Analysis
Capillary liquid chromatography, to fractionate or separate peptides and proteins, was performed using a 15 cm × 250 μm-i.d. capillary column, packed in-house using POROS R1 reversed-phase media (Applied Biosystems, Framingham, MA), employing a 2.2%/min gradient to an organic concentration of 60% ACN in 0.1% formic acid, followed by a 3.5%/min gradient up to a concentration of 95% organic phase. Chromatography used an LC Packings Ultimate Capillary HPLC pump system, with a fully automated micro autosampler (Dionex, Sunnyvale, CA), controlled by the mass spectrometer software (Analyst, Applied Biosystems). The LC is coupled directly to the MS. Effluent from the capillary column was directed into a QSTAR Pulsar i quadrupole orthogonal time-of-flight mass spectrometer through an ionspray source (Applied Biosystems). Data were collected for m/z 500–2500 over the entire range of the chromatogram (55 min). Data collection, processing, and preliminary formatting were accomplished using the Analyst QS software package with BioAnalyst add-ons (Applied Biosystems).
Normalization of Elution Times
Small differences in elution times were accounted for by normalizing them to the elution time of a molecular species with a unique m/z value, common to all samples.
RESULTS AND DISCUSSION
Our goal was to develop a protein precipitation method that had sufficient reproducibility to allow for accurate quantitative serum proteomic analysis in an effort to identify molecular species that differed between healthy and diseased states. The adequacy of this method was tested using cLC-MS. Additionally, we were interested in determining the residual molecular species and their molecular weights after a precipitation procedure.
Evaluation of Protein Precipitation Approaches
Organic solvents have long been used to precipitate proteins. Unlike other techniques, such as ultrafiltration or dialysis, this technique will typically result in many smaller proteins and peptides being released from large carrier proteins.
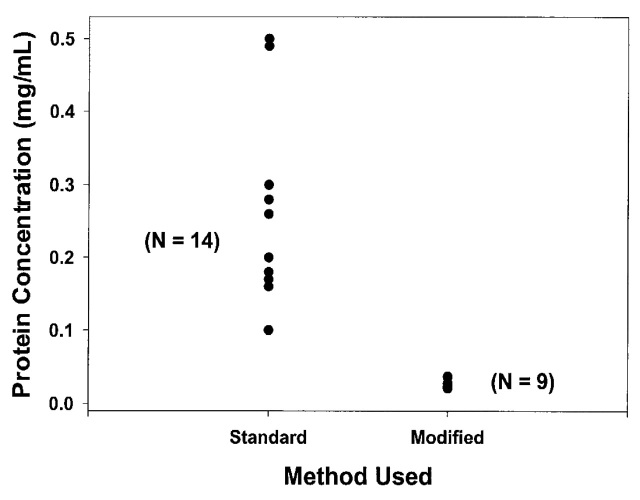
In our initial study two techniques were evaluated. The “standard” method, which was described in a previous publication,12 represented one of the first approaches to serum preparation for global serum proteomic analysis using MS. The “modified” method was a modification of that technique. The methods are described in detail in Materials and Methods and are summarized in Table 1. The results of the two approaches are shown in Figure 1. The standard method resulted in a mean protein level of 0.240 ± 0.124 mg/mL. The modified method produced a mean protein concentration of 0.028 ± 0.006 mg/mL which was significantly less than the standard method (Students unpaired t-test: p = 3.1 × 10−5). Moreover, protein concentrations remaining after the modified method were far less variable (method 1: coefficient of variation = 51.7%; method 2: coefficient of variation = 21.4%). It is beyond the aims and scope of these experiments to determine whether the two methods resulted in different proteins being precipitated or in more efficient removal of the same proteins. In addition, the original method left a cloudy solution after centrifugation, suggesting that some of the variability may result from an inadequately long or forceful centrifugation step. This unremoved precipitate is likely to be difficult to measure accurately in a protein assay and may contribute to the marked variability of residual protein concentration. Further reductions in the liquid portion of the supernatant, in particular taking the sample to dryness, resulted in a residue that was no longer appreciably soluble in the cLC mobile phase solvent.
TABLE 1.
Optimization of Acetonitrile Precipitation
| STANDARD METHOD | MODIFIED METHOD | |
| Mixing after addition of acetonitrile | Gentle Mixing | Vortexing |
| Cloudy supernatant | Clearer supernatant | |
| Higher protein concentration | Lower protein concentration | |
| Inconsistent protein concentration | Consistent protein concentration | |
| Centifugation | 4 min at 12,000 rpm | 10 min at 12,000 rpm |
| Cloudy supernatant | Clearer supernatant | |
| Concentrated volume of supernatant | 50 μL | 200 μL |
| Precipitate forms | No precipitate forms |
FIGURE 1.
Protein yield comparison of standard and modified acetonitrile precipitation. The left column shows that the standard method varied greatly in protein yield. The right column shows that the modified method resulted in a very reproducible protein yield.
cLC-MS Evaluation of Residual Proteins
Because of the improved reproducibility of the modified method, it was used for all subsequent experiments. The final volume of 200 μL was too large to inject onto our capillary column. Therefore, immediately prior to specimen introduction into the cLC-MS system, a volume of solution containing 4.0 μg of protein was aliquoted and concentrated to near dryness, avoiding complete dryness. Any precipitate that formed upon concentration was resolubilized by the addition of 20 μL of 88% formic acid. There was no appreciable protein loss in this step (data not shown).
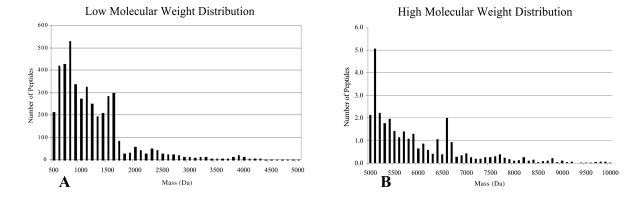
This precipitation method was further evaluated to determine the molecular weight range and number of molecular species present after treatment. Given the large number of molecular species, the whole elution interval was divided into 5-min time frames and the mass spectra were averaged for that period. The LC-MS reconstruct tool in BioAnalyst was then applied to the averaged spectrum. Output from the several 5-min intervals was compiled to capture all species over the entire sample run. Figure 2 represents the average of 49 different specimens. While the most abundant species were below a molecular weight of 2000 Da (Fig. 2A), many species with molecular weight greater than 2000 Da (Fig. 2B) were still present.
FIGURE 2.
Molecular weight distribution of species observed in acetonitrile supernatant. cLC-MS was used to analyze 49 serum specimens after acetonitrile precipitation. A: This histogram shows the distribution of all molecular species ranging from 500 to 5000 Da. B: This histogram shows the distribution of all molecular species ranging from 5 to 10 kDa. The majority of the species present were below 2000 Da.
It is important to note that close inspection of the actual unmanipulated mass spectra to the software LC-MS reconstructed data demonstrated that the LC-MS reconstruct tool occasionally omitted species, and did so variably, making it unreliable for use in clinical studies where quantification is critical.
Value of Acetonitrile Precipitation
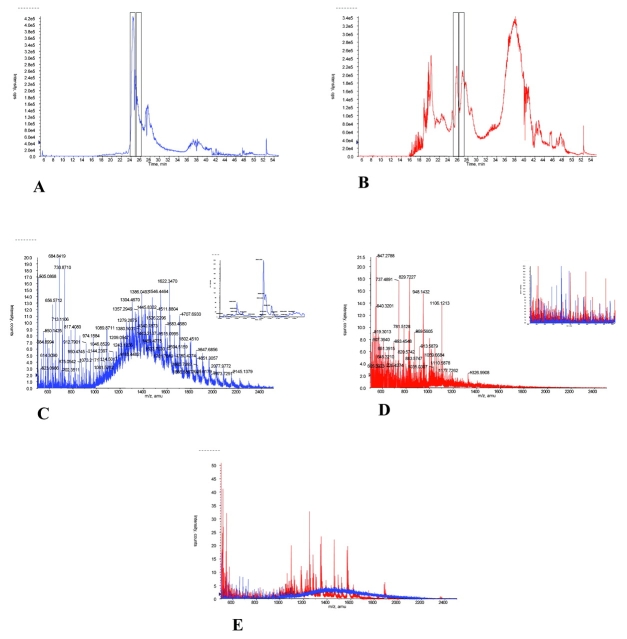
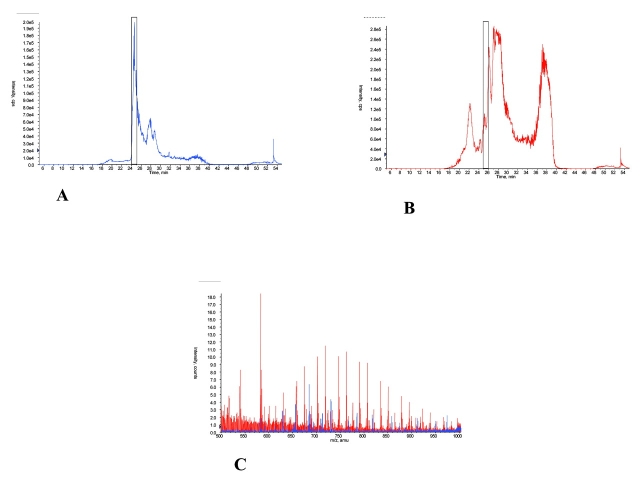
The effects of ACN precipitation of serum proteins were evaluated by direct comparison of pre- and post-precipitation cLC-MS chromatograms. Comparisons of untreated and treated serum demonstrated that many more molecular species were observable by MS after treatment. This is evident in Figures 3A and B, which show an expanded number of molecular species in the total ion chromatograms (TIC) after treatment (Fig. 3B). Moreover, the mass spectrum of the untreated specimen was dominated by the extensive envelope (see Fig. 3C), which peaked between an m/z of 1300 to 1400, representing a species with a molecular weight of 60 to 70 kDa (most likely albumin or related compounds) and which masked many molecular species. Even the region of lower m/z (see Fig. 3D, inset) had more species revealed by ACN treatment. These findings appear to confirm the hypothesis that organic solvent precipitation of proteins would also release smaller, protein-bound species. While it is true that larger proteins present in the serum specimen are sacrificed by ACN treatment, it is also true (as clearly evident in Fig. 3E) that higher molecular-weight species are obscured by the highly abundant albumin and that lower molecular-weight species are both more abundant and more observable after treatment (see Figs. 3C, D, as well as E).
FIGURE 3.
Comparison of serum proteins (1 μg) before and after acetonitrile (ACN) precipitation. Total ion chromatograms for serum proteins before (A) and after (B) acetonitrile are presented. The averaged mass spectra for the cLC elution (taken from 24 to 25 min) of a serum specimen prior to ACN precipitation (C) and after ACN precipitation (D) are shown. Inset in C shows the protein mass graph of the Baysein protein reconstruct performed on the mass spectrum from 900 to 2500 m/z. Inset in D shows the expanded (500–1000 m/z) mass spectra overlay of the pre- and post-treatment samples. E: The overlay of the mass spectra for species present between 25 and 26 min of elution for the two samples is shown. Before ACN precipitation is shown in blue, after ACN precipitation is shown in red.
In summary, the ACN pretreatment actually provides broader mass spectral access to the serum proteome and makes more likely the development of an informative and diagnostic pattern for defining differing states in the human. It should also be noted that most growth, stimulatory, regulatory, and pathology-responsive factors are small proteins and peptides. Consequently, the additional ability of serum proteomics to actually define the mediators of disease may be more successful after treatments that more fully expose this lower molecular-weight subproteome.
Ultrafiltration Versus Precipitation
Centrifugal ultrafiltration is a common method used to remove serum albumin, immunoglobulins, and other abundant, high-molecular-weight proteins from a serum specimen.13 Such treatment is often necessary before chromatography or capillary electrophoretic techniques can be employed. Although this method is selective for low-molecular-weight peptides and proteins in a sample, the resulting filtrate may not allow for a thorough examination of the entire low-molecular-weight serum proteome. Small proteins and peptides are commonly bound by large carrier proteins to both moderate their free and biologically active concentration and to prevent them being rapidly cleared from the blood by the kidney. The experiments described in the previous section suggested strongly that there was an increase in low-molecular-weight species after the addition of ACN to denature serum proteins. A second line of evidence for this is provided by comparison of the ACN precipitation with ultrafiltration of the same serum.
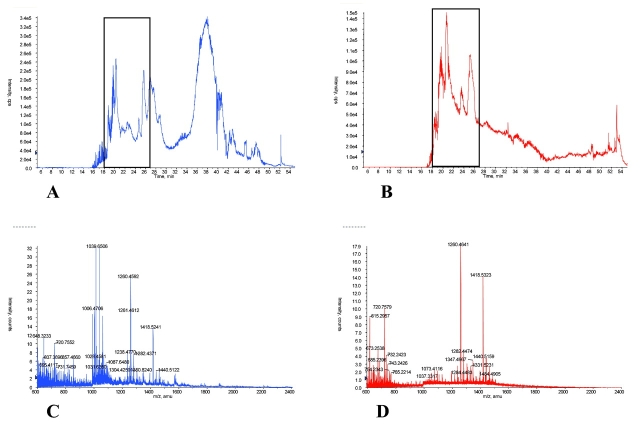
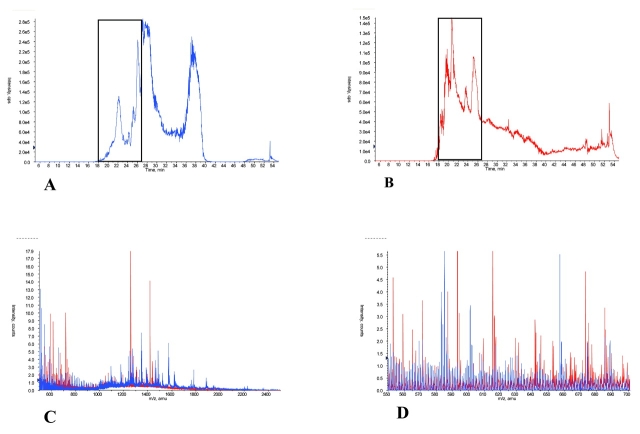
Figure 4 provides cLC-MS results for equal quantity protein injections (1 μg) of post-ACN-precipitated serum (Fig. 4A) with the filtrate obtained after a 30,000-molecular-weight cutoff ultrafiltration (Fig. 4B) of the same specimen. The general TIC patterns for both samples were somewhat similar, except for the broad peak at 38 min in the ACN-treated specimen. Figures 4C and D show the averaged mass spectra of species in 9 min of eluate (18–27 min) as indicated in Figs. 4A and B, respectively. Comparison of Figures 4C and D shows that most of the peaks present in the ultrafiltrate were also present in the ACN-precipitated serum, but the ACN-treated specimen contained many additional molecular species, some quite abundant, that did not appear in the ultrafiltrate mass spectrum. This strongly supports the hypothesis that the ACN precipitation method liberated protein-bound species, making them available for cLC-MS analysis.
FIGURE 4.
LC-MS of acetonitrile (ACN) supernatant compared with 30K nominal molecular weight filtrate. Total ion chromatograms of ACN-precipitated serum (A) and reconstituted filtrate (B) are shown. C and D show the averaged mass spectra overlay from 18 to 27 min cLC elution for the serum and the filtrate respectively. ACN-precipitated serum is shown in blue, the filtrate is shown in red. Comparison of C and D shows that the ACN precipitation method was superior to the centrifugal ultrafiltration technique in both number of low-molecular-weight species and intensity of peaks.
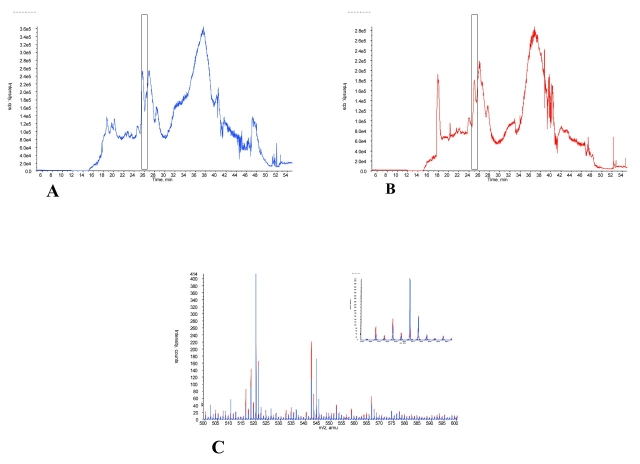
In order to confirm that ACN precipitation does indeed release and allow for analysis of more small proteins and peptides attached to serum albumin and other carrier proteins, additional experiments were carried out. The first of these involved the treatment of the large, retained proteins, present after ultrafiltration, with ACN to release potentially bound species. Figure 5 provides typical results from one set of experiments. In this approach the retentate obtained after a 30,000-molecular-weight cutoff ultrafiltration of serum was explored by cLC-MS. The TIC of retentate prior to ACN precipitation (Fig. 5A) and after ACN precipitation (Fig. 5B) are shown. Figure 5C shows the averaged mass spectra overlay for MS data collected from 25 to 26 min elution (as indicated in Figs. 5A and B). The retentate prior to ACN precipitation is shown in blue and the ACN-precipitated retentate is shown in red. As predicted, the retentate prior to ACN precipitation has a TIC spectrum very similar to that of unfiltered serum prior to ACN precipitation, and the mass spectrum is dominated by large-molecular-weight proteins. Prior ultrafiltration would have effectively removed unbound small proteins and peptides from the retentate. The appearance, then, of additional low-molecular-weight species after ACN precipitation would be evidence of the dissociation of bound, small proteins and peptides from carrier proteins. This is strikingly evident in Figure 5C, which shows dramatically increased numbers of small-molecular-weight proteins and peptides after ACN treatment of the retentate, and confirms that this approach makes available many more small, low-abundance proteins and peptides for proteomic analysis. When these results were extended to a longer time region of the cLC chromatogram, similar results were obtained (data not shown).
FIGURE 5.
Examination of 30K nominal molecular weight retentate before and after acetonitrile precipitation. Total ion chromatograms of 1 μg of ultrafiltration retentate prior to acetonitrile precipitation (A) and retentate afteracetonitrile precipitation (B) are shown. C: The averaged mass spectra overlay for species present in eluate from 25 to 26 min of the cLC separation step are shown. Retentate prior to acetonitrile precipitation is shown in blue, acetonitrile-precipitated retentate is shown in red. The presence of the high-abundance proteins masked the presence of information-rich small peptides.
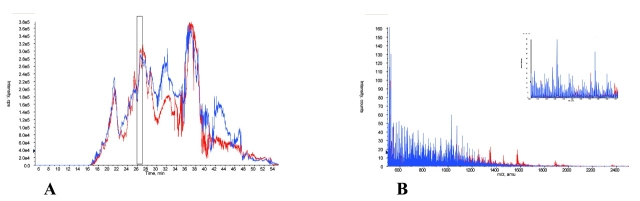
An additional approach is summarized in Figure 6 which compares the mass spectra of ACN-precipitated retentate with the untreated filtrate obtained after ultrafiltration of a single serum. Figure 6A and B show the TIC of the ACN-precipitated retentate and the filtrate, respectively. Figure 6C displays the averaged mass spectra overlay of 9 min of eluate (18–27 min) of the ACN-precipitated retentate (blue) and the filtrate (red). This overlay shows that ACN treatment produced many additional molecular species. Interestingly, while both procedures yield small proteins and peptides, the MS spectrum from the ACN precipitation of the retentate differed significantly from that of the filtrate, indicating that two subsets of small proteins and peptides exist and that those in the ACN-precipitated retentate were not representative of those that routinely passed through the filter.
FIGURE 6.
Comparison of acetonitrile (ACN)-precipitated retentate with filtrate. Total ion chromatograms of 1 μg of ACN-precipitated retentate (A) and 1 μg of filtrate (B) from 30 K nominal molecular weight ultrafiltration are shown. C: The averaged mass spectral overlay for 18 to 27 min of the cLC chromatogram is presented. ACN-precipitated retentate is shown in blue; filtrate is shown in red. D: An expanded region (550–700 m/z) of C is shown.
Reproducibility
Not only is reproducibility in the precipitation step important, but reproducibility through the entire cLC-MS analysis is necessary for useful diagnostic applications of serum proteomics. This was tested. The comprehensive approach of ACN treatment with cLC-MS analysis was carried out on serum specimens from a healthy patient, prepared and run on the same day. In addition, comprehensive analysis of specimens from healthy patients prepared and run on different days was assessed.
Mass spectra obtained from different serum specimens from the same patient prepared and run on the same day were virtually indistinguishable (data not shown). Mass spectra obtained from different specimens from different healthy subjects prepared and run on different days were also remarkably similar after normalization of elution times (see Fig. 7).
FIGURE 7.
Reproducibility of LC-MS day to day. Total ion chromatograms of acetonitrile-precipitated serum of two different healthy subjects are shown. A: Total ion chromatogram of subject 1 serum (blue), prepared and run on day 1. B: Total ion chromatogram of subject 2 serum (red), prepared and run on day 2. C: An expanded mass spectral overlay (500–600 m/z) with inlay expanded to 550–560 m/z of both patients from a 1 min cLC window that had been calibrated for variations in elution time is shown (blue: 25.4–26.4 min; red: 24.8–25.8 min).
The sensitivity of the results of our approach to variation in specimen handling was also explored (see Fig. 8). Figure 8A displays an overlay of the TIC of ACN-precipitated serum thawed just prior to use (blue) compared with the same specimen left at room temperature overnight (red). Figure 8B shows the averaged mass spectra overlay of molecular species in 1 min of cLC eluate (26–27 min) of the same freshly processed serum (blue) and serum processed after 24 h at room temperature (red). Despite the delay, the mass spectra matched closely with modest changes.
FIGURE 8.
Reproducibility of LC-MS with variable sample handling. A: An overlay of the total ion chromatogram of acetonitirile (ACN)-precipitated serum thawed just prior to analysis (blue) with the total ion chromatogram of serum left at room temperature overnight (red) is shown. B: An overlay of the averaged mass spectrum for cLC elution time of 26–27 min of the ACN-precipitated fresh serum (blue) with the averaged mass spectrum (same retention time) of the room temperature incubated serum (red) is provided. The inset in B shows an expanded mass spectrum (1000–1100 m/z). Even with delayed handling of the serum, the mass spectra of the samples matched closely with only slight variation.
Application of Method
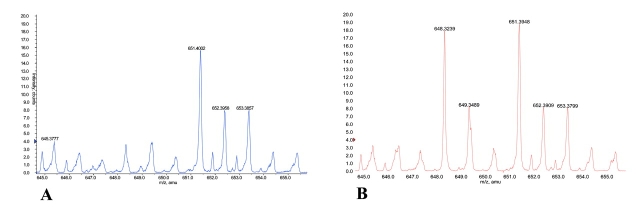
As proof of principle of this approach, we applied this method to individuals representing two differing clinical states. Figure 9 shows a narrow region of the mass spectra (645–655 m/z) obtained after ACN treatment and cLC separation. A restricted region of the cLC chromatogram for each specimen was selected and an averaged mass spectrum developed for each. Figure 9A represents analysis of a specimen from a pregnant woman with an uncomplicated pregnancy. Figure 9B displays the results of a specimen from a pregnant woman of comparable gestational age who had pre-eclampsia, a severe complication of pregnancy. There was an additional molecular species at m/z = 648.3 in the pre-eclamptic specimen. This is one of several differences found. It is premature to know if the difference is consistently seen in other pre-eclamptic patients. Nevertheless, it points out the very real potential of identifying differences between patient populations that might allow for the accurate diagnosis or prediction of disease, or even the eventual identification of mechanistic candidates.
FIGURE 9.
Variability of LC-MS from one individual to another. The same retention time and m/z ranges were selected from cLC runs of a serum specimen of patient A (normal) and patient B (case). As shown, the variations in the make up of the serum from individual to individual can be detected. These differences represent a potential source of serum biomarkers.
SUMMARY
In order to use the information-rich proteomic analysis of serum in a diagnostic manner, it is essential that the method used to prepare the sample provide reproducible results. The new method of ACN precipitation described here adequately and reproducibly precipitates the noninformative, abundant, high-molecular-weight proteins. Moreover, this approach increases the number of low-molecular-weight species that can be analyzed by cLC-MS. We demonstrate, importantly, that the new method of ACN precipitation disrupts binding of small proteins and peptides from the large carrier proteins, providing a larger set of low-molecular-weight species for analysis. Most regulatory molecules have been shown to be small proteins and peptides and to be protein-bound in serum; hence, the possibility of developing a diagnostic pattern and indentifying potential mediators of disease is increased using this approach. The combination of protein precipitation coupled with cLC-MS appears capable of meaningful evaluation of a substantial portion of the serum proteome.
REFERENCES
- 1.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem 2002;48:1296–1304. [PubMed] [Google Scholar]
- 2.Petricoin EF, Liotta LA. Clinical applications of proteomics. J Nutr 2003;133, 2476S–2484S. [DOI] [PubMed] [Google Scholar]
- 3.Petricoin EF, Paweletz CP, Liotta LA. Clinical applications of proteomics: Proteomic pattern diagnostics. J Mammary Gland Biol Neoplasia 2002;7:433–440. [DOI] [PubMed] [Google Scholar]
- 4.Petricoin EF, Liotta LA. Counterpoint—The vision for a new diagnostic paradigm. Clin Chem 2003;49:1276–1278. [DOI] [PubMed] [Google Scholar]
- 5.Adkins JN, Varnum SM, Auberry KJ, et al. Toward a human blood serum proteome: Analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics 2002;1:947–955. [DOI] [PubMed] [Google Scholar]
- 6.Govorukhina, N, Keizer-Gunnink A, van der Zee AG, de Jong S, de Bruijn HW, Bischoff R. Sample preparation of human serum for the analysis of tumor markers. Comparison of different approaches for albumin and gamma-globulin depletion. J Chromatogr A 2003;1009:171–178. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed N, Barker G, Oliva K, et al. An approach to remove albumin for the proteomic analysis of low abundance biomarkers in human serum. Proteomics 2003;3:1980–1987. [DOI] [PubMed] [Google Scholar]
- 8.Georgiou HM, Rice GE, Baker MS. Proteomic analysis of human plasma: Failure of centrifugal ultrafiltration to remove albumin and other high molecular weight proteins. Proteomics 2001;1:1503–1506. [DOI] [PubMed] [Google Scholar]
- 9.Liotta LA, Ferrari M, Petricoin E. Clinical proteomics: Written in blood. Nature 2003;425:905. [DOI] [PubMed] [Google Scholar]
- 10.Diamandis E. Point-proteomic patterns in biological fluids: Do they represent the future of cancer diagnostics? Clin Chem 2003;49:1272–1275. [DOI] [PubMed] [Google Scholar]
- 11.Banez LL, Prasanna P, Sun L, et al. Diagnostic potential of serum proteomic patterns in prostate cancer. J Urol 2003;170:442–6. [DOI] [PubMed] [Google Scholar]
- 12.Alpert A, Shukla A. Precipitation of large, high-abundance proteins from serum with organic solvents. ABRF 2003: Translating biology using proteomics and functional genomics. Poster no. P111-W, 2003.
- 13.Tirumalai R, Chan C, Prieto D, Issaq H, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics 2003;2:1096–1103. [DOI] [PubMed] [Google Scholar]