Abstract
Four commercially available immobilized metal ion affinity chromatography (IMAC) methods for phosphopeptide enrichment were compared using small volumes and concentrations of phosphopeptide mixtures with or without extra-added bovine serum albumin (BSA) nonphosphorylated peptides. Addition of abundant tryptic BSA peptides to the phosphopeptide mixture increases the demand for selective IMAC capture. While SwellGel gallium Discs, IPAC Metal Chelating Resin, and ZipTipMC Pipette Tips allow for the possibility of enriching phosphopeptides, the Gyrolab MALDI IMAC1 also presents the possibility of verifying existing phosphopeptides after a dephosphorylation step. Phosphate-containing peptides are identified through a mass shift between phosphorylated and dephosphorylated spectra of 80 Da (or multiples of 80 Da). This verification is useful if the degree of phosphorylation is low in the sample or if the ionization is unfavorable, which often is the case for phosphopeptides. A peptide mixture in which phosphorylated serine, threonine, and tyrosine were represented was diluted in steps and thereafter enriched using the four different IMAC methods prior to analyses with matrix assisted laser desorption/ionization mass spectrometry. The enrichment of phosphopeptides using SwellGel Gallium Discs or Gyrolab MALDI IMAC1 was not significantly affected by the addition of abundant BSA peptides added to the sample mixture, and the achieved detection limits using these techniques were also the lowest. All four of the included phosphopeptides were detected by MALDI-MS only after enrichment using the Gyrolab MALDI IMAC1 compact disc (CD) and detection down to low femtomole levels was possible. Furthermore, selectivity, reproducibility, and detection for a number of other phosphopeptides using the IMAC CD are reported herein. For example, two phosphopeptides sent out in a worldwide survey performed by the Proteomics Research Group (PRG03) of the Association of Biomolecular Resource Facilities (ABRF) were detected and verified by means of the 80 Da mass shift achieved by on-column dephosphorylation.
Keywords: proteomics, phosphorylation, phosphopeptide, compact disc, microfluidics, microlaboratory, immobilized metal affinity chromatography, IMAC, alkaline phosphatase, MALDI mass spectrometry, and dephosphorylation
One of the most frequent posttranslational modifications on proteins is phosphorylation. It has been estimated that between 20 and 50% of all proteins are subjected to this modification.1–4 Since the modification often is transiently expressed, investigation of each protein under as many conditions as possible is of vital importance in order to obtain a clear picture of how and when the protein becomes modified. This, however, is not an easy task since no real high throughput methods for screening of phosphorylations are available, so far. A novel approach based on compact disc (CD) technology was recently reported.5–7 In the CD, liquid is driven through capillary microstructures by centrifugal force. In the previously reported application, portions of the microstructures were packed with reversed-phase resin for cleanup and preparation of peptide samples before crystallization on the CD and analysis by matrix assisted laser desorption/ionization mass spectrometry (MALDI-MS). As compared with off-line methods, the CD technology offers better recovery since it integrates sample preparation and MS analysis in a single platform format. The improved sample handling is combined with higher throughput via parallel processing of up to 96 samples per CD, which is an important aspect in proteomics. In this paper we report the comparison of several commercial phosphopeptide purification kits (ZipTipMC Pipette Tips, SwellGel Gallium Discs, and IPAC resin) with Gyrolab MALDI IMAC1 technology.8 The latter method makes use of a readymade CD containing 96 immobilized metal ion affinity chromatography (IMAC) columns of 16 nL volume each. The samples are loaded automatically from a 96-well microplate into the CD where the phosphopeptides are trapped on a Ga(III)-charged column. Enriched phosphopeptides are eluted onto a restricted desorption area (200 × 400 μm) located towards the perimeter of the CD. After processing the CD, it is introduced into the ion source of a MALDI mass spectrometer and mass spectra are acquired in automatic mode. In order to increase the specificity of the analysis, up to 48 samples are analyzed in duplicates with one of the replicates being enzymatically dephosphorylated. The criterion for phosphorylation is a shift of 80 Da (or multiples of 80 Da) between two peaks in the two sets of samples. Additionally, with a MALDI time of flight (TOF)/TOF instrument, fragmentation patterns of both the phosphorylated and dephosphorylated peptide can be obtained to further consolidate the detection of a phosphopeptide as well as establishing its amino acid sequence.
EXPERIMENTAL
Samples
Bovine serum albumin (BSA) and bovine β-casein, (Sigma, St. Louis, MO) were digested according to the following procedure: The protein absorbance at λ = 280 nm was measured and concentration was calculated through the use of the protein-specific extinction coefficient. The proteins were then diluted in 50 mM NH4HCO3 to 0.78 mg/mL. Several aliquots of 1 mg (1282 μL) were incubated for 15 min at 50°C after addition of 25 μL of 45 mM dithiothreitol to reduce disulfide bonds. Alkylation was performed by incubation of the aliquots for 15 min at room temperature after addition of 25 μL of 100 mM iodoacetamide. Proteins were digested with 200 μL of sequencing grade modified trypsin (Promega) by incubation at 37°C and terminated before 24 h by addition of 80 μL of 10% aqueous trifluoracetic acid (TFA) and 242 μL of 15% acetonitrile.
Four phosphopeptides were mixed and used as stock solution. Tryptic digest of β-casein includes one singly and one quadruply phosphorylated peptide (m/z 2059.8, FQpSEEQQQTEDELQDK and m/z 3121.3, RELEELNVPGEIVEpSLpSpSpSEESITR; p denotes a phosphorylated amino acid). Synthetic phosphorylated platelet-derived growth factor receptor (PDGF-r) peptide (m/z 1405.6, pYQQVDEEFLR) was a gift by Ulf Hellman, Ludwig Institute for Cancer Research, Uppsala, Sweden. CaM kinase II substrate 281–291 (m/z 1437.6, MHRQEpTVDCLK), also a synthetic phosphorylated peptide, was purchased from AnaSpec, San Jose, CA (masses are denoted as monoisotopic and without protonation).
The stock solution (dilution 1) containing 1 pmol/μL of β-casein digest, 5 pmol/μL of CaM kinase II substrate, and 2.5 pmol/μL of PDGF-r peptide was prepared both without or mixed with 4 pmol/μL BSA digest. Aliquots were stored at −20°C until used and a fresh aliquot was used for each experiment. Dilution 1 was further diluted 10 times, called dilution 2; and 10 times again, called dilution 3. These three dilutions, both with and without digested BSA, generated a total of six mixtures which were used to evaluate the different IMAC enrichment protocols.
SwellGel Gallium Discs
The method was essentially carried out according to the manufacturers Phosphopeptide Isolation Kit instructions (www.piercenet.com). Briefly, the sample was diluted in 0.1 M acetic acid. Before applying the sample, it was assured that the SwellGel was at the bottom of the spin column. The sample, either in 1 μL or 20 μL, was added to the SwellGel Gallium Disc and the sample-resin mixture was incubated at room temperature for 5 min. When analyzing 1 μL of sample, a preloading buffer of 20 μL was placed on top of the column and the sample was added into this liquid. Thereby the total loading volume was kept at recommended volume and the total amount loaded was the same for all compared techniques, but additional losses to surfaces were limited. All solutions following sample application were passed through the columns by centrifugation at 3000 rpm (720 × g) for 1 min. The columns were washed twice to remove nonbound peptides: first with 50 μL of 0.1% acetic acid, and then with 50 μL of 0.1% acetic acid containing 10% acetonitrile. To equilibrate the SwellGel before elution, 75 μL Milli-Q (Millipore, Billerica, MA) water was added. The column was transferred into a new collection tube and incubated for 5 min with 20 μL of elution buffer, 100 mM NH4HCO3, pH 9. After centrifugation, the eluted sample was rendered acidic with 5 μL of 10% TFA. Samples were desalted using ZipTipC18 (Millipore) before crystallization and analysis with MALDI-MS. The ZipTipC18 purified peptides were eluted onto a steel target using 2 μL 50% acetonitrile, 0.1% TFA with 20 mg/mL dihydroxybenzoic acid (DHB).
Eprogen IPAC Metal Chelating Resin
The method was essentially carried out according to the modified protocol of Kaffashan and Chenhui.9 Briefly, the compact reaction column (CRC) was fitted with a CRC lower filter (both purchased from USB (Cleveland, OH). Solutions were passed through the columns by centrifugation at 3000 rpm (720 × g) for 1 min. Ten microliters of IPAC resin slurry were resuspended in the spin columns and washed by adding 50 μL of Milli-Q water. Metal ions were immobilized on the IPAC resin support by passing 30 μL of 50 mM metal containing solution through the column, such as 50 mM ferric chloride (FeCl3), or 50 mM gallium nitrate [Ga(NO3)3] in Milli-Q grade water. In order to remove unbound metal ions, the column was washed with 50 μL of Milli-Q water. Columns were equilibrated through rinsing with 50 μL of 0.1 M acetic acid solution, and thereafter, 20 μL of sample diluted in 1.6 M acetic acid were loaded and allowed to interact with the beads for 15 min. As with the SwellGel, when analyzing 1 μL of sample, a volume of 20 μL sample dilution buffer was placed on top of the column and the sample was mixed into the preloaded buffer in order to keep the total loading volume and amount the same for all comparisons and to avoid losses to additional surfaces. After sample application, the column was washed twice with 50 μL washing solution containing 30% acetonitrile, 0.1% acetic acid, and finally eluted with 20 μL of 200 mM K2HPO4, pH 8.5. The eluted sample was acidified with 5 μL of 10% TFA, and desalted according to the ZipTipC18 protocol before crystallization and analysis with MALDI-MS. The ZipTipC18 purified peptides were eluted directly onto a steel target using 2 μL 50% acetonitrile, 0.1% TFA with 20 mg/mL DHB.
Millipore ZipTipMC
The method was carried out according to the Millipore User Guide for ZipTipMC Pipettes (www.millipore.com). Briefly, tip equilibration included aspiration and dispensing 10 μL of freshly prepared 0.1% acetic acid with 50% acetonitrile (three times to waste). All dilutions were performed using Milli-Q water. Charging the tips was accomplished by performing 10 cycles of aspiration and dispensing of 10 μL metal ion solution such as ferric chloride (FeCl3), or gallium nitrate [Ga(NO3)3], nickel chloride (NiCl2), or cupric sulphate (CuSO4), all dissolved in 10 mM HCl. Two washing steps were included by aspirating and dispensing three times to waste: first 10 μL of Milli-Q water, followed by 10 μL of 1.0% acetic acid containing 10% acetonitrile. Before sample application, 10 μL of binding solution (0.1% acetic acid) was rinsed/aspirated 5 times over the column bed. The samples (1 μL) were applied by pipetting the sample onto the top of the column. After proper placement, the sample was aspirated and dispensed with 10 μL of binding buffer in 10 cycles for maximum binding. The column was washed once with 0.1% acetic acid, followed by 0.1% acetic acid in 10% acetonitrile, and finally with Milli-Q water. All washing solutions were aspirated and dispensed in three cycles before going to waste. The phosphopeptides were eluted from the column through aspirating and dispensing the elution solution (20 μL of freshly prepared 0.3 N ammonium hydroxide solution) in 10 cycles. The eluted sample was acidified with 5 μL of 10% TFA, and further sample cleanup was achieved by ZipTipC18 pipette tips. The ZipTipC18 purified peptides were eluted directly onto a steel target using 2 μL 50% acetonitrile, 0.1% TFA with 20 mg/mL DHB.
Gyrolab MALDI IMAC1
A CD microlaboratory (Gyrolab MALDI IMAC1, Gyros AB, Uppsala, Sweden) containing prepacked IMAC columns (16 nL volume, Poros MC 20 chelated with Ga3+) was used for sample preparation (www.gyros.com). Within the CD, liquid flow through different compartments and channels is controlled by a combination of centrifugal force, hydrophobic barriers, channel geometry, and packed columns. Properly adjusted rotational speed allows for directed flow within the channels.
To start with, the columns were conditioned with a solution containing 50% acetonitrile and 0.6% acetic acid in water. The sample was diluted with the same solution and 1 μL of sample solution was applied to the microstructures on the CD. Each sample was processed in duplicate, one for phosphopeptide analysis and one for analysis after dephosphorylation. Rinses using about 50 column volumes of 50% isopropanol, 5% acetic acid, followed by equal amounts of 30% acetonitrile, 5% acetic acid, were used to wash off nonphosphorylated peptides unspecifically bound to the IMAC particles or CD walls. Dephosphorylation was accomplished through addition of 200 nL of 0.1 U/μL alkaline phosphatase (Aldrich, Milwaukee, WI) bovine intestinal mucosa, P6774) in 50% acetonitrile/25 mM ammonium bicarbonate, pH 9.5, during a period of approximately 20 min. Peptides were eluted from the column using 200 nL of 50% acetonitrile/1% H3PO4 containing 10 mg/mL DHB as MALDI matrix. In order to allow the solvent to evaporate slowly and form a homogenous layer of crystals, a controlled spin was applied to direct the eluate into a dedicated open MALDI target area on the CD.
Mass Spectrometry
For mass spectrometry, Voyager DE-Pro (Applied Biosystems, Foster City, CA) and Ultraflex (Bruker Daltonics, Billerica, MA) instruments were employed, operated in reflectron mode. Before on-CD MALDI analysis, the CD was cut into two halves when using the Ultraflex or six pieces when using the Voyager DE-Pro by aid of a precision cutter. The respective CD pieces were then mounted into the special made CD-target holders for each instrument.
RESULTS
SwellGel Gallium Discs
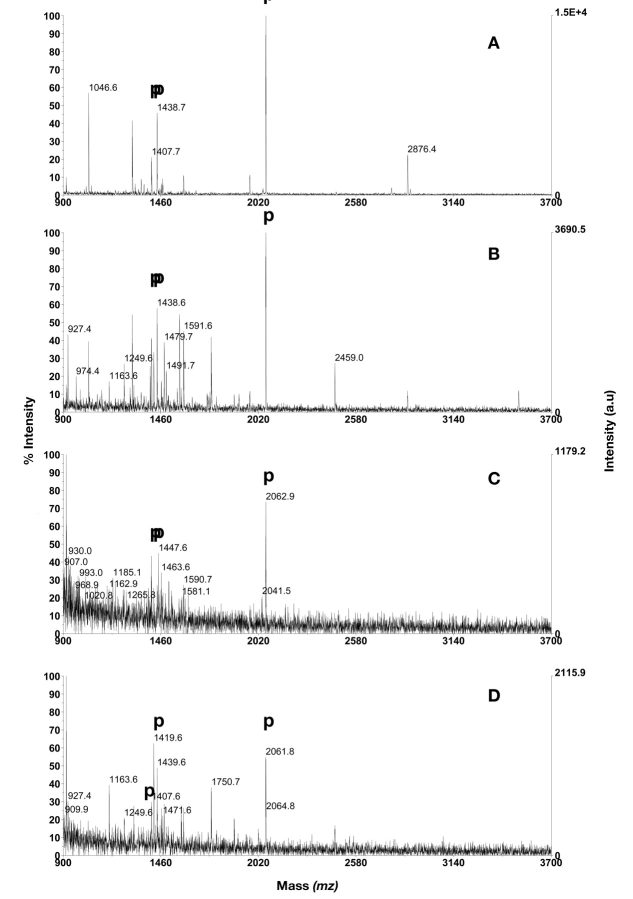
When 20 μL sample was loaded, three out of four phosphopeptides were detected in all three dilutions, and addition of BSA digest did not interfere with the phosphopeptides isolation efficiency, even though a number of BSA peptides were observed. This implies that detection of 200 fmol β-casein digest, 1 pmol CaM kinase II substrate, and 500 fmol of the PDGF-r peptide is possible using the SwellGel protocol. The quadruply phosphorylated β-casein peptide (m/z 3121.3) was not detected at any concentration, independent of added BSA digest. When the same mixtures were analyzed using 1 μL loading volume, which was mixed into 20 μL loading buffer placed on top of the column, the same three peptides were detected in dilutions 1 and 2 (Fig. 1) but none of the phosphopeptides was detected in dilution 3. In dilution 2, with added BSA digest, a large number of BSA peptides were detected, but this did not seem to affect the detection of phosphopeptides (not shown). These results are summarized in Table 1.
FIGURE 1.
Phosphopeptide purification using SwellGel Gallium Disc. A: 1 μL of dilution 1; B: 1 μL of dilution 1 in mixture with BSA digest in excess; C: 1 μL of dilution 2; D: 1 μL of dilution 2 in mixture with BSA digest in excess. Phosphopeptides are annotated with p. PDGF-r at m/z 1406.6, CaM kinase II substrate at m/z 1438.6, β-casein peptides at m/z 2060.8 and 3122.3.
TABLE 1.
Isolation Efficiency for Phosphopeptides by Different IMAC Methods
| Number of Detected Phosphopeptides | ||||||
| +/− BSA | Dilution 1 | Dilution 2 | Dilution 3 | Replicas | ||
| SwellGel Gallium Discs | 1 μL | − | 3 | 3 | − | 1 |
| + | 3 | 3 | − | 1 | ||
| 20 μL | − | 3 | 3 | 3 | 2 | |
| + | 3 | 3 | 3 | 2 | ||
| IPAC Metal Chelating Resin | 1 μL | − | 2 | − | − | 1 |
| + | − | − | − | 1 | ||
| 20 μL | − | 3(3) | 3(2) | 1.5(2) | * | |
| + | 1 | 0.5 | 1 | 2 | ||
| ZipTipMC Pipette Tips | 1 μL | − | 2 | 1 | − | 2 |
| + | 2 | 0.5 | − | 2 | ||
| Gyrolab MALDI IMAC1 | 1 μL | − | 4 | 4 | 2.7 | 3 |
| + | 4 | 4 | 3.3 | 3 | ||
*Replicas within parentheses.
IPAC Metal Chelating Resin
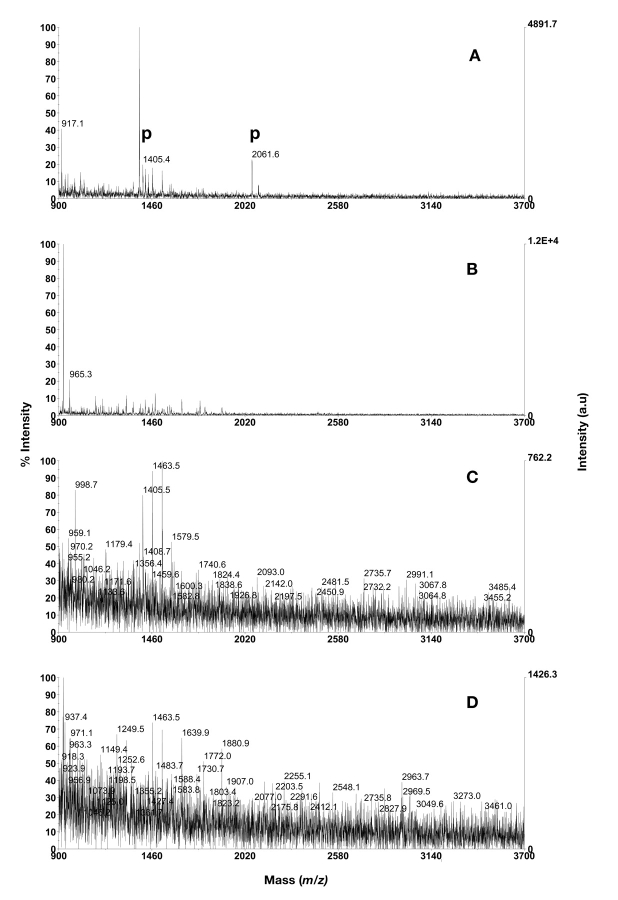
In initial experiments the IPAC resin was packed into spin columns and activated with Ga3+ or Fe3+. Since Ga3+ activation resulted in higher intensities and cleaner spectra of the phosphopeptides it was used in the IPAC following evaluation. When 20 μL of dilution 1 was loaded onto the column, an average of three phosphopeptides could be detected. In the triplicate analysis, all four phosphopeptides were detected in the first experiment, whilst three were detected in the second, and two in the third. In dilution 2, three out of four phosphopeptides could be enriched and detected; the quadruply phosphorylated β-casein peptide (m/z 3121.3) was not detected. In dilution 3 (also 20 μL loading volume), one or two phosphopeptides were detected in the duplicate analysis. When BSA digest was added to the mixtures, an average of 1, 0.5, and 1 phosphopeptides, respectively, could be detected in the three dilutions (analyzed in duplicates, data not shown). When 1 μL was loaded to the column, two phosphopeptides were detected in dilution 1 if BSA digest was not present, but none if BSA was present (Fig. 2). In dilutions 2 and 3, no phosphopeptides were found (Table 1, data not shown).
FIGURE 2.
Phosphopeptide purification using Ga3+ activated IPAC metal chelating resin. A: 1 μL of dilution 1; B: 1 μL of dilution 1 in mixture with BSA digest in excess; C: 1 μL of dilution 2; D: 1 μL of dilution 2 in mixture with BSA digest in excess. Phosphopeptides are annotated with p. PDGF-r at m/z 1406.6, CaM kinase II substrate at m/z 1438.6, β-casein peptides at m/z 2060.8 and 3122.3.
ZipTipMC Pipette Tips
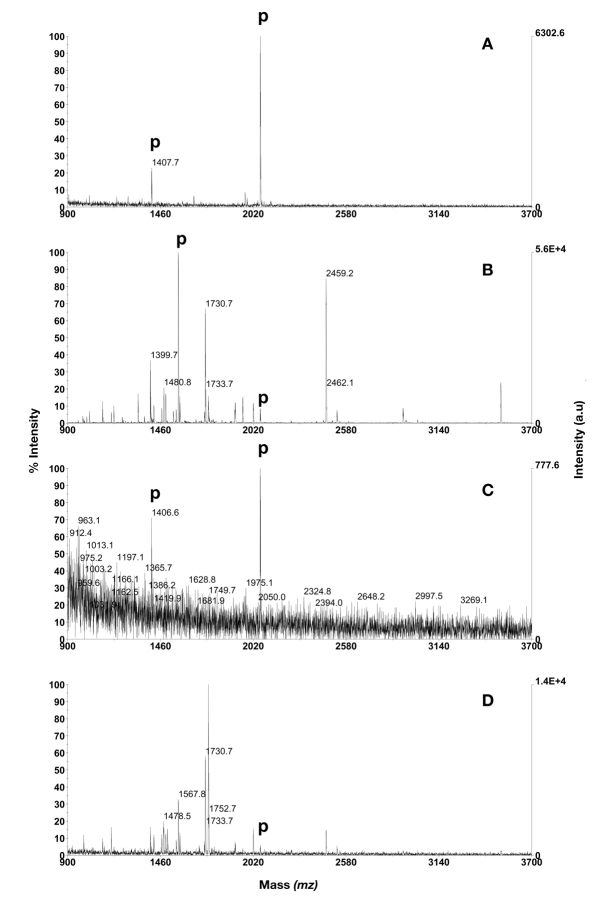
Two phosphopeptides were detected at the highest concentration (1 μL of dilution 1) for all of the immobilized ions initially evaluated, which included Ga3+, Fe3+, Ni2+, and Cu2+, but the signal intensity and purity was best when gallium was chelated (Fig. 3). Therefore, activation with gallium was found to be the best choice among the metal ions tested for further evaluation using ZipTipMC. Analysis of 1 μL of dilution 2, without BSA peptides, resulted in detection of two or none peptides (duplicate), but in dilution 3 none of the phosphopeptides were detected. Furthermore, when BSA peptides were added to the mixture, two phosphopeptides could be found in dilution 1, whereas an average of 0.5 (duplicate) were detected in dilution 2 and none in dilution 3. The quadruply phosphorylated β-casein peptide (m/z 3121.3) and CaM kinase II substrate (m/z 1437.6) were not detected in any dilution (Table 1).
FIGURE 3.
Phosphopeptide purification using Ga3+ activated ZipTipMC Pipette Tips. A: 1 μL of dilution 1. B: 1 μL of dilution 1 in mixture with BSA digest in excess. C: 1 μL of dilution 2. D: 1 μL of dilution 2 in mixture with BSA digest in excess. Phosphopeptides are annotated with p. PDGF-r at m/z 1406.6, CaM kinase II substrate at m/z 1438.6, β-casein peptides at m/z 2060.8 and 3122.3.
Gyrolab MALDI IMAC1
One microliter volumes of the three dilutions were analyzed on Gyrolab MALDI IMAC1. All four peptides were detected in the two highest concentrations with or without BSA digest added to the mixtures. At dilution 3, without BSA digest (representing 10 fmol β-casein), both β-casein peptides and the PDGF-r peptide could be detected, whereas the peptide CaM kinase II substrate was missing (Fig. 4E, Table 1). The average number of detected phosphopeptides (triplicates) was 2.7 in dilution 3 without BSA peptides added, and 3.3 when BSA peptides were added. All four peptides could however be detected in one spectrum when BSA digest was added, yet with low intensity (Fig. 4F). The Gyrolab MALDI IMAC1 method was the only purification technique that made it possible to detect the selected phosphopeptides at this low concentration (dilution 3) when 1 μL sample was loaded. Approximately the same number of detected phosphopeptides were obtained using SwellGel Gallium Discs if 20 μL were loaded, indicating a sensitivity difference of approximately 20 times between the two techniques (Table 1).
FIGURE 4.
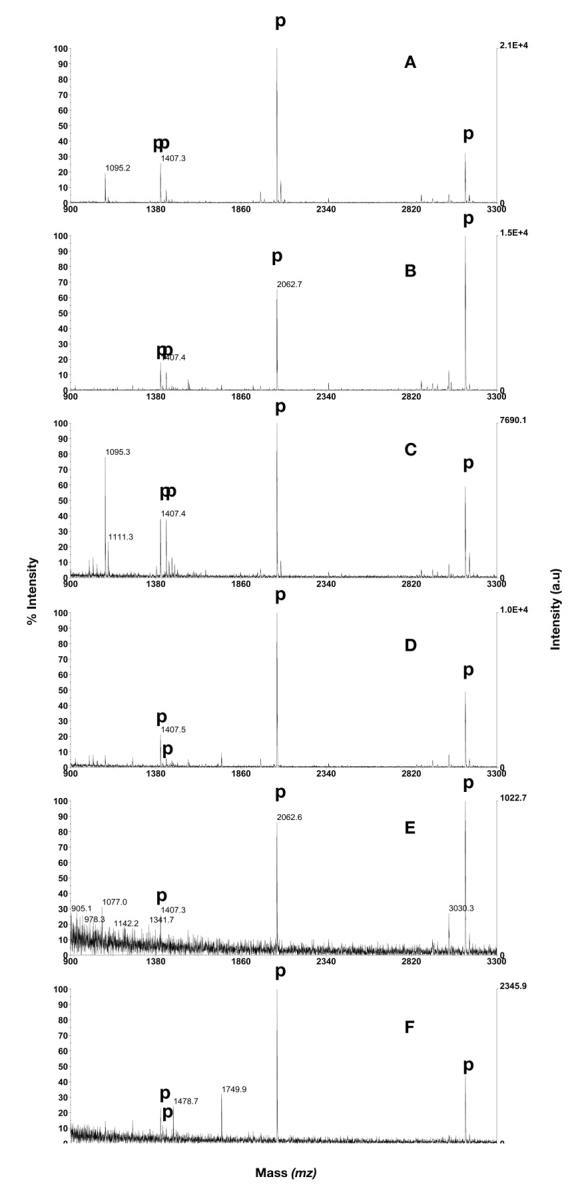
Phosphopeptide purification using Gyrolab MALDI IMAC1 containing Ga3+ activated Poros MC 20. A: 1 μL of dilution 1. B: 1 μL of dilution 1 in mixture with BSA digest in excess. C: 1 μL of dilution 2. D: 1 μL of dilution 2 in mixture with BSA digest in excess. E: 1 μL of dilution 3. F: 1 μL of dilution 3 in mixture with BSA digest in excess. Phosphopeptides are annotated with p. PDGF-r at m/z 1406.6, CaM kinase II substrate at m/z 1438.6, β-casein peptides at m/z 2060.8 and 3122.3.
Gyrolab MALDI IMAC1 Reproducibility
In order to verify the robustness of the method, eight CDs were processed loading the same phosphopeptide mixture (totally 200 fmol BSA digest, 100 fmol CaM kinase II substrate peptide, 50 fmol β-casein, and 25 fmol PDGF-r peptide) in all 96 structures. The outcome is summarized with respect to isolation success, which takes into consideration both the outcome from the phosphorylation and dephosphorylation experiment (Table 2). A sample was considered successfully analyzed when all four of the respective forms of the peptides were detected as phosphorylated and dephosphorylated. Different instrument and climate parameters, such as number of samples, MALDI instrument used, and relative humidity during CD processing, were applied to stress the robustness of the system. In seven of eight evaluated CDs, complete elution and co-crystallization of the phosphopeptides within the dedicated MALDI desorption area were observed. In the eighth CD the crystallization success was 98.9%. The average success rate of detection in the MS monitoring the phosphopeptides was slightly higher than the one observed when monitoring the dephosphorylated peptides, 98.6% versus 95.8%. The total success rate ranged between 86.7 and 100% for the eight different CDs.
TABLE 2.
Isolation Efficiency of Phosphopeptide and Subsequent Dephosphorylation Verification Performed on Gyrolab MALDI IMAC1 Followed by Analysis Using Two Different MALDI Mass Spectrometers
| Summary | CDs Analyzed in Mass Spec 1 | CDs Analyzed in Mass Spec 2 | ||||||
| Temperature (°C) | 23 | 24 | 24 | 23 | 23 | 24 | 24 | 23 |
| Humidity (%) | 23 | 52 | 52 | 26 | 22 | 21 | 42 | 21 |
| Sample crystals (%) | 100 | 100 | 100 | 100 | 98.9 | 100 | 100 | 100 |
| Calibrant crystalls (%) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Isolation efficency Phos-dePhos (%) | 93.3 | 95.5 | 100 | 90.9 | 86.7 | 93.3 | 96.2 | 100 |
The peptides tested were singly phosphorylated (pTyr) synthetic PDGF-r peptide (1405.6 Da), singly phosphorylated (pThr) CaM kinase II substrate peptide (1437.6 Da), singly phosphorylated (pSer) β-casein peptide (2059.8 Da), and a quadruply phosphorylated (4pSer) β-casein peptide (3121.3 Da). All samples—50 fmol β-casein, 100 fmol CaM kinase II substrate peptide, and 25 fmol PDGF-r peptide—were mixed with 200 fmol BSA tryptic peptides. A sample is considered successfully analyzed when all four phosphopeptides are detected as phosphorylated and as dephosphorylated in the duplicate analysis.
Gyrolab MALDI IMAC1 Selectivity
A tryptic digest of BSA without miscleavages theoretically contains 35 different peptides in the mass range m/z 900–3500. These peptides contain 29 aspartic acid and 46 glutamic acid residues unevenly distributed in the peptides. It is clear that many of these peptides will be negatively charged at neutral pH and in fact the average pI of them is 5.3 (median 4.5) as calculated using GPMAW (ChemSW, Fairfield, CA, http://www.chemsw.com/). Since acidic peptides often are contaminants in spectra recorded after IMAC enrichment of phosphopeptides, a mixture containing phosphopeptides and tryptic digest of BSA was used for specificity measurements. Nonspecifically retained peptides can be used as a measure of the column efficiency to selectively enrich phosphopeptides. The possibility of detecting phosphopeptides using MALDI-MS after IMAC purification increases with the total phosphopeptide amount loaded but decreases with the content of nonphosphorylated peptides competing for IMAC binding sites. At very low concentrations this could possibly be balanced by the carrier effect the additional peptides would constitute.
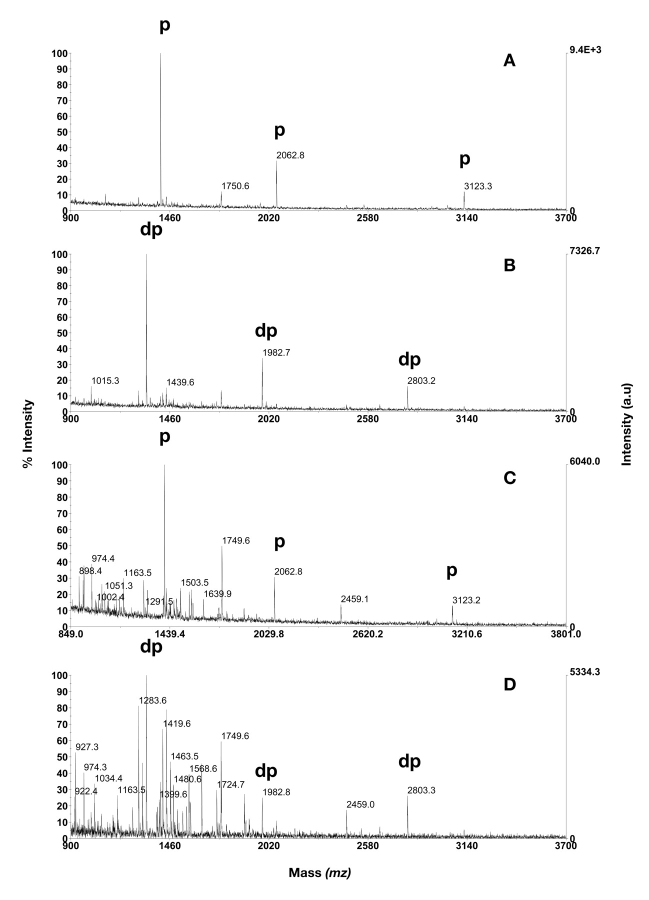
A mixture containing the PDGF-r peptide and digest of β-casein was mixed with digested BSA in a molar ratio of 1:1:9 or 1:1:99, such that the phosphopeptide concentration was kept constant. Thus, the three phosphopeptides were accounting for 10% or 1% of the molar content, corresponding to a total of 30 fmol/μL in both cases. Of these mixtures, 1 μL volumes were analyzed on Gyrolab MALDI IMAC1, which then corresponded to 30 fmol of each phosphopeptide and 270 or 2970 fmol of BSA peptides. All three phosphopeptides could be enriched and dephosphorylated on-column at both BSA peptide concentrations (Fig. 5). When the ]phosphopeptides constituted 10% of the total amount loaded, their corresponding signal intensities were high and therefore easy to find in the spectra (Fig. 5A and B). However, when the BSA peptide content was increased to 99%, a high degree of contaminating peptides was detected (Fig. 5C and D).
FIGURE 5.
Phosphopeptide isolation and enzymatic verification by usage of Gyrolab MALDI IMAC1 before MALDI analysis. A: 30 fmol β-casein digest, 270 fmol BSA digest. B: dephosphorylation of sample depicted in A. C: 30 fmol β-casein digest, 2970 fmol BSA digest. D: dephosphorylation of sample depicted in C. Phosphopeptides are annotated with p, dephosphorylated phosphopeptides with dp.
In a separate test where β-casein and BSA were digested, and mixed to a final concentration of 50 fmol/μL β-casein and 5000 fmol/μL BSA, the included phosphopeptides could be detected and verified through enzymatic dephosphorylation in 75% of the occasions (n = 16) employing automatic MALDI acquisition and automatic peak comparison, using a script for finding peak-shifts of 80 (or multiples of 80 Da) between phosphorylated and dephosphorylated spectra (data not shown).
DISCUSSION
A mixture containing four phosphopeptides representing pSer, pThr, and pTyr as well as singly and quadruply phosphorylated peptides was prepared and diluted 10 and 100 times in order to compare the efficiency of four different IMAC-based methods for enrichment of phosphopeptides. The mixtures were analyzed in an experimental setup involving three different concentrations, in two different sample volumes (for two of the methods) as well as with and without the addition of BSA digest. As the manufacturers of IPAC resin and SwellGel disc are recommending approximately 20 μL as the lowest loading volume, some adjustments had to be made. Both these methods were evaluated using a loading volume of 20 μL or with 1 μL sample added into 20 μL loading buffer placed on top of the columns. In this way, the volume, concentration, and amount could be kept the same for all methods evaluated and nonspecific adsorption of peptides to extra tube walls, pipette tips, etc., could be minimized.
Another difference between the four methods is the solvent used for elution of the phosphopeptides; while SwellGels, IPAC resin and ZipTipMC are eluted using a high pH eluent according to the manufacturers protocol, Gyrolab IMAC columns are eluted using acidic solvent containing DHB matrix. We have combined a phosphopeptide elution mixture using DHB dissolved in 50% acetonitrile and 1% phosphoric acid. DHB in itself has previously been reported to exhibit an elution effect on monophosphorylated peptides.10 This mixture has been powerful for elution of all investigated phosphopeptides. The difference in elution buffer between the methods introduces a difference in posttreatment of the phosphopeptide-containing eluate. In case of the CD, the peptides were directly eluted into a dedicated desorption area in the outer rim of the CD whereas the other methods required a buffer change to lower the pH before crystallization. To achieve this, ZipTipC18 was used and additional peptide losses to the hydrophobic resin cannot be excluded.
Generally, all methods were able to enrich phosphopeptides, but to a varying extent. At higher concentrations, two to four phosphopeptides could be detected using the different techniques, with ZipTipMC being the least successful with respect to the number of retained and eluted phosphopeptides. Interestingly, all of the methods, except Gyrolab MALDI IMAC1, had a problem enriching and/or releasing the heavy β-casein peptide at m/z 3121.3. It has also been reported that multiply phosphorylated peptides are difficult to detect after IMAC enrichment, which has been attributed to strong binding of these peptides to the IMAC resin.10,11 The only exception was for the IPAC resin, in which all four peptides were detected in one of the triplicate samples when using a loading volume of 20 μL (i.e., 20 times more peptides loaded) and the highest concentration. Two, three, and four phosphopeptides, respectively, were detected in the sample analyzed in triplicate. Whether this difference in number of detected peptides is due to improper sample handling, instable IPAC material, unregular metal activation, loss of peptides after ZipTipC18, or inhomogeneous peptide distribution in the MALDI spot on the target plate remains unclear. The absence or addition of BSA digest did not negatively affect the enrichment of phosphopeptides at the highest concentration investigated. However, at lower concentrations, none of the methods but Gyrolab IMAC were able to enrich all the phosphopeptides. In the evaluation of dilution 3, the lowest concentration investigated, an average of about three phosphopeptides were detected by both Gyrolab IMAC and SwellGel Gallium Discs with similar signal to noise ratios but with different loading volumes, 1 μL or 20 μL, respectively, indicating a 20 times difference in sensitivity after enrichment between the techniques. The increased sensitivity might be explained by the smaller desorption area for the CD (200 × 400 μm) as compared with the steel target φ1500 μm), the different elution protocol, the lack of postpurification for the CD or the significantly smaller column (16 nL), or a combination thereof.
Since the CD approach combines monitoring of both phosphopeptides and their individual enzymatically dephosphorylated species, the calculated mass-shift can be used for valuable phosphopeptide verification. β-casein phosphopeptides are generally considered to be uncomplicated to detect since they ionize well during MALDI. More important is the possibility to enable enzyme verification when the phosphopeptides ionize poorly and therefore are detected just above the background noise.
The robustness of the Gyrolab method was tested using eight CDs, or a total of 768 individual samples (i.e., 8 × 48 × 2 samples), which were analyzed using the same sample loaded in all individual inlets. An analysis was counted successful if all four phosphopeptides could be detected both as phosphorylated in the first sample and as dephosphorylated in the duplicate sample. The outcome, called isolation success, in percent is summarized in Table 2. Two MS instruments from different manufacturers were used for the analysis. The detection of enriched phosphopeptides was more successful than the detection of dephosphorylated peptides that were IMAC-enriched and enzymatically dephosphorylated. Considering the fact that the dephosphorylated phosphopeptides have been treated with additional steps in the CD (i.e., pH equilibration before addition of enzyme followed by dephosphorylation), it is likely that the lower detection success is linked to additional losses during these steps. Also, we observed a slower dephosphorylation of the pThr-containing peptide, CaM kinase II substrate peptide, which in some cases caused the dephosphorylated peptide to be missing in the dephosphorylated spectrum, which directly affected the isolation success.
IMAC enrichment methods are commonly evaluated including trypsin digests of α-, β-casein, and ovalbumin,11,12 which generate acidic phosphopeptides. IMAC is therefore sometimes considered to preferentially enrich acidic phosphopeptides, but also to enrich nonphosphorylated Glu- and Asp-containing peptides. Ficarro et al.13 previously suggested a solution to the problem of unspecifically binding acidic peptides. By methylation of the acidic residues the metal-acidic residue interaction was limited, which largely decreased nonspecifically bound peptides and thereby increased the possibility for selective phosphopeptide isolation. Nonspecifically bound peptides could also be washed off the column using a large volume of acidified organic solvents, which does not jeopardize the retention of phosphopeptides. In the CD approach, washes with 5% acetic acid in 50% 2-propanol and 5% acetic acid in 30% acetonitrile effectively removed large quantities of nonphosphorylated but acidic peptides from the IMAC column. Yet, in complex mixtures such as cell lysates, serum, or other body fluids, the affinity of IMAC for phosphopeptides might not be high enough for effective enrichment of only phosphate-containing peptides. In such cases, prepurification is required in order to facilitate the analysis of multiprotein-containing samples. Table 3 presents some peptides detected by MALDI-MS after processing in Gyrolab MALDI IMAC1. The average pI for these peptides is 5.2, i.e., most of them are acidic. However, three of the investigated phosphopeptides were neutral or basic, and were isolated in mixtures containing an abundance of other nonphosphorylated peptides. These results indicate that IMAC effectively can be used for enrichment of phosphopeptides independent of peptide pI.
TABLE 3.
pI, Molecular Weight, and Sequence of Detected Phosphopeptides Using Gyrolab MALDI IMAC1
| Name of Peptide | m/z [m + H]+ | Sequence | pI |
| PDGF-r peptide | 1405.6 | pYQQVDEEFLR | 3.9 |
| CaM kinase II substrate | 1438.6 | MHRQEpTVDCLK | 7.1 |
| β-casein peptide | 2060.8 | FQpSEEQQQTEDELQDK | 3.5 |
| β-casein peptide | 3122.3 | RELEELNVPGEIVEpSLpSpSpSEESITR | 3.9 |
| MBP peptide | 1651.8 | NIVpTPRpTPPPSQGK | 11.5 |
| ABRF PRG03 heavy peptide | 2027.0 | THILLFLPKpSVSDYEGK | 7.5 |
| ABRF PRG03 light peptide | 964.4 | SVpSDYEGK | 4.1 |
| Fetuin peptide | 1408.5 | CDSSPDpSAEDVR | 3.5 |
| α-casein peptide | 1660.8 | VPQLEIVPNpSAEER | 4.0 |
| α-casein peptide | 1951.9 | YKVPQLEIVPNpSAEER | 4.6 |
| Ovalbumin peptide | 2088.9 | EVVGpSAEAGVDAASVSEEFR | 3.6 |
| Ovalbumin peptide | 2891.3 | FDKLPGFGDpSIEAQCGTSVNVHSSLR | 5.3 |
The Gyrolab IMAC1 method was further tested to enrich two synthetic, but naturally occurring neutral phosphopeptides, which were supplied by the Proteomics Research Group (PRG03) of the Association of Biomolecular Resource Facilities (ABRF). This peptide mixture was sent out to a number of researchers participating in a global survey concerning phosphopeptide analysis. The outcome of the survey is available at www.abrf.org/ResearchGroups/Proteomics/EPosters/ABRF_PRG03.pdf. We analyzed the PRG03 sample after the results of the study were made public, thus the sequence and phosphorylation sites were known to us when we carried out our analysis. The sample was dissolved in 10 μL 10% ethanol, 0.1% TFA, and identified using Gyrolab MALDI SP1 for purification followed by MALDI-MS.5 The sample was thereafter diluted with 10 μL acetonitrile, giving a final concentration of 250 fmol digest/μL containing 50 fmol/μL of the phosphopeptides. One microliter of the sample was transferred in duplicate by the workstation to the IMAC-CD. Figure 6 shows that the two phosphopeptides were both enriched and dephosphorylated by aid of the Gyrolab IMAC1 method. Interestingly, one of the two peptides is slightly basic (pI of the peptide’s backbone is 7.5, calculated using GPMAW), again indicating that retention and elution of other than acidic peptides is possible using the CD method. This indication was further confirmed when the basic miscleaved tryptic peptide (pI 11.5) of myelin basic protein was enriched from a mixture containing a four-time excess of BSA tryptic peptides. The doubly phosphorylated peptide was robustly detected down to 100 fmol loaded onto the IMAC CD.
FIGURE 6.
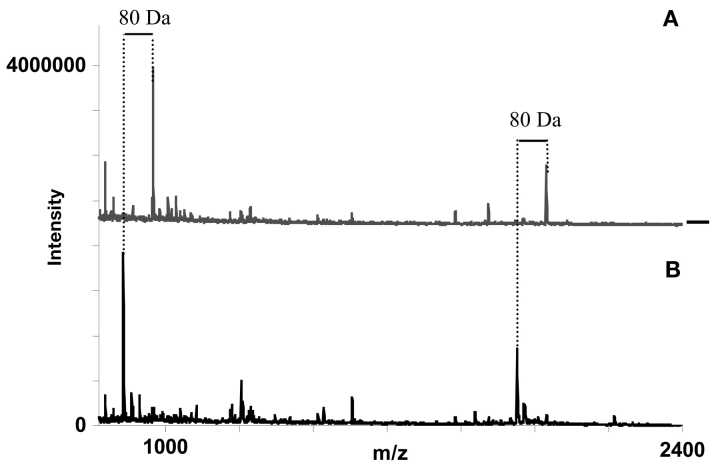
Enrichment and enzymatic verification of phosphopeptides in the ABRF Proteomics Research Group 2003 (PRG03) sample used for their global survey. We analyzed the PRG03 sample after the result of the study was made public. The sample is genuine but the sequence and phosphorylation sites were known to us when we carried out our analysis. Two phosphate-containing peptides were identified by mass shifts of 80 Da between phosphorylated and dephosphorylated sample. Peptide sequences were determined as SVSDYEGK and THILLFLPKSVSDYEGK by a search of their molecular weights against the amino acid sequence of the identified protein. A: Phosphorylated sample. B: Dephosphorylated sample.
Acknowledgments
We are grateful to the ABRF Proteomics Research Group (Chair, Kaye D. Speicher) for supplying the PRG03 phosphopeptide sample; to Ulf Hellman, Ludwig Institute for Cancer Research, Uppsala, for supplying the phosphorylated synthetic PDGF-r peptide; and to Allan Stensballe and Ole Jensen, University of Southern Denmark, Odense, for help and suggestions.
REFERENCES
- 1.Zolnierowicz S, Bollen M. Protein phosphorylation and protein phosphatases. De Panne, Belgium, September 19–24, 1999. EMBO J 2000;19:483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinna LA, Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta 1996;1314: 191–225. [DOI] [PubMed] [Google Scholar]
- 3.Williams DM, Cole PA. Kinase chips hit the proteomics era. Trends Biochem Sci 2001;26:271–273. [DOI] [PubMed] [Google Scholar]
- 4.Mann M, Ong SE, Gronborg M, et al. Analysis of protein phosphorylation using mass spectrometry: Deciphering the phosphoproteome. Trends Biotechnol 2002;20:261–268. [DOI] [PubMed] [Google Scholar]
- 5.Gustafsson M, Hirschberg D, Palmberg C, Jornvall H Bergman T. Integrated sample preparation and MALDI mass spectrometry on a microfluidic compact disk. Anal Chem 2004;76:345–350. [DOI] [PubMed] [Google Scholar]
- 6.Hirschberg D, Tryggvason S, Gustafsson M, et al. Identification of endothelial proteins by MALDI-MS using a compact disc microfluidic system. Protein J 2004;23: 263–271. [DOI] [PubMed] [Google Scholar]
- 7.Roblick UJ, Hirschberg D, Habermann JK, et al. Sequential proteome alterations during genesis and progression of colon cancer. Cell Mol Life Sci 2004; 61:1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kånge R, Sennerfors T, Werner E, et al. Detection and identification of phosphopeptides captured by IMAC and prepared for MALDI MS analysis within a CD microlaboratory. Proceedings of the 51th ASMS Conference on Mass Spectrometry and Allied Topics, 2003.
- 9.Kaffashan A, Chenhui Z. Evaluation of commercially available IMAC kits: Millipore ZipTipMC, Eprogen IPAC beads and Pierce Swellgel Gallium Chelated Disks. Proceedings of the 51th ASMS Conference on Mass Spectrometry and Allied Topics, 2003.
- 10.Hart SR, Waterfield MD, Burlingame AL, Cramer R. Factors governing the solubilization of phosphopeptides retained on ferric NTA IMAC beads and their analysis by MALDI TOFMS. J Am Soc Mass Spectrom 2002;13: 1042–1051. [DOI] [PubMed] [Google Scholar]
- 11.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem 1999;71:2883–2892. [DOI] [PubMed] [Google Scholar]
- 12.Stensballe A, Andersen S, Jensen ON. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics 2001;1:207–222. [DOI] [PubMed] [Google Scholar]
- 13.Ficarro SB, McCleland ML, Stukenberg PT, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol 2002;20:301–305. [DOI] [PubMed] [Google Scholar]