Abstract
In this work we present a hybrid linear trap/Fourier transform ion cyclotron resonance (ICR) mass spectrometer to perform protein sequencing using the bottom-up approach. We demonstrate that incorporation of the linear trap greatly enhances the overall performance of the hybrid system for the study of complex peptide mixtures separated by fast high-performance liquid chromatography gradients. The ability to detect in the linear trap enables employment of automatic gain control to greatly reduce space charging in the ICR cell irregardless of ion flux. Resulting accurate mass measurements of 2 ppm or better using external calibration are achieved for the base peak as well as ions at 2% relative abundance. The linear trap is used to perform ion accumulation and activation prior to detection in the ICR cell which increases the scan rate. The increased duty cycle allows for data-dependent mass analysis of coeluting peptides to be acquired increasing protein sequence coverage without increasing the gradient length. In addition, the linear trap could be used as an ion detection device to perform simultaneous detection of tandem mass spectra with full scan mass spectral detection in the ICR cell resulting in the fastest scan cycles for performing bottom-up sequencing of protein digests. Comparisons of protein sequence coverage are presented for product ion detection in the linear trap and ICR cell.
Keywords: Linear quadrupole ion trap, Fourier transform mass spectrometry, bottom-up protein sequencing, HPLC
Combining high-performance separations with Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) provides a powerful tool for proteomics analysis using the bottom-up approach.1–4 The bottom-up approach utilizes two prominent workflow methods: one performs an initial protein separation using 2-dimensional gel electrophoresis5 followed by excision of the protein(s) spot and then enzymatic digestion; the second method performs enzymatic digestion on the whole cell lysate followed by 2D high-performance liquid chromatography (HPLC).3 Both methods utilize mass spectrometry to generate full scan and/or tandem mass spectra that are then matched against computer-predicted spectra from theoretical digestion of protein databases resulting in identification and sequencing. The high complexity of the resulting peptide mixture requires either fast scan cycles to acquire mass spectral data for coeluting peptides or long chromatographic gradients to improve chromatographic resolution.
The FT-ICR mass spectrometer is unmatched in mass resolution as well as accurate mass measurement capability. High mass accuracy and resolution provide a means to separate analytical signal from background at or near the detection limit. Previous reports have shown subfemtomole sensitivity by coupling electrospray ionization to an FT-ICR mass spectrometer.6 In the past, the primary drawback to performing bottom-up proteomics on FT-ICR mass spectrometers was poor sequence coverage attributed to low duty cycle, defined as the percentage of the total experimental time used to sample the continuous ion beam.7 A pulse of collisional cooling gas was needed to trap peptides formed by electrospray ionization in the ICR cell. Consequently, long pump-down times were required following the trapping stages of the experiment prior to performing high resolution mass measurements.8 These long cycle times prohibited employment of the FT-ICR for on-line analysis of enzymatic protein digests.
Recent advances in FT-ICR instrumentation have taken advantage of external ion accumulation optics to enhance the measured ion signal, duty cycle, and dynamic range.6,7,9,10 Utilization of external ion accumulation optics was shown to increase instrument duty cycle to nearly 100% for detection of 500 amol of the peptide hHRHL.6 Increases in the duty cycle can be attributed to the ability to detect a packet of ions in the ICR cell while accumulation is done in the external multipole. Senko and co-workers showed that mass resolving power also increased using external ion collection compared with performing all mass spectral events in the Penning trap for bovine ubiquitin.7 Increased resolving power was attributed to ion-neutral collisions in the octopole, reducing radial and axial ion velocity and resulting in a more homogenous ion packet prior to transfer and detection in the ICR cell.
In addition to ion accumulation, use of external multipoles enables activation of the ion ensemble prior to mass analysis. The two primary methods of ion activation incorporate infrared multiphoton dissociation (IRMPD)11,12 and ion-neutral collisions.9,13 IRMPD has proven to be an excellent method for generating b- and y-type fragment ions used for database searching, resulting in further structural confirmation of posttranslational modifications and/or structural irregularities without dramatically increasing the time consumption of the mass spectral scan. To maintain fast scanning rates, the radius of the ion population trapped in the ICR cell is increased to allow an infrared laser beam to transverse the ICR cell and excite ions in the external octopole. This results in detection of the ions in the ICR cell while preparing the next packet of ions for detection following dissociation. The primary drawback with utilizing IRMPD for ion activation is that it still requires isolation of the precursor ion prior to ion activation to perform true tandem mass spectrometry, which must be done in the ICR cell. Without precursor ion isolation, sequencing the resulting product ion spectrum may be complex due to coeluting peptides.
Collision-induced dissociation via the use of external octopole rf potentials was used to sequence complex mixtures of peptides. Belov et al. utilized different combinations of rf and dc fields on the octopole rods to manipulate the radial potential well, resulting in collision-induced dissociation on selected mass-to-charge ratios, molecular weights, and/or charge states of precursor ions.9 A mixture of gramicidin S, bradykinin, and angiotensin I was used as a test solution. By increasing the axial well depth from 5 to 6.5 V, only the singly charged precursor ions were observed to dissociate. A further increase to 8 V resulted in dissociation of all precursor ions following 500 msec of accumulation time.
The primary drawbacks of the above methods are the inability of the external rf multipoles to manipulate ions in simple ways such as isolation and MSn, and the lack of ion detection capability. Although Belov et al. showed that specific ions could be selected and excited, isolation of the precursor ion was not achieved9; thus true tandem mass spectral capability was not exhibited. Without isolation of the precursor ion prior to activation, the resulting spectrum contains a mixture of precursor and product ions. In addition, the flux of ions generated from an LC separation of protein digests varies unpredictably due to single or multiple peptides eluting. In most cases, ion production may vary between two and four orders of magnitude, resulting in varying number of ions in the ICR cell prior to detection. Fluctuations in charge density are detrimental to high performance of the FT-ICR in regards to high mass accuracy.14
Solving some of these problems is a new hybrid linear trap/FT-ICR mass spectrometer.15,16 The LTQ-FT mass spectrometer (Thermo Electron, Bremen, Germany) combines a 2D linear ion trap mass spectrometer coupled to a 7-Tesla FT-ICR mass spectrometer via a series of transfer octopoles, providing many advantages over previous FT-ICR systems. Similar to the external ion optics presented above, the 2D linear ion trap is positioned in the low vacuum region of the manifold which facilitates accumulation of greater charge density, without sacrificing ion trapping ability. The linear trap pressure is maintained with helium, which is used for trapping and for collisional activation of the trapped ions. In addition, the linear trap is equipped with dual orthogonal detection systems that allows it to operate as a separate mass spectrometer capable of performing numerous mass spectral experiments (e.g., secondary ion MS, MSn, selective reaction monitoring) that operate on extremely fast time scales. For example, an MS spectra generally requires 0.2 sec while MS/MS spectra can be recorded in 0.4 sec. Employment of automatic gain control,17 which prescans the ion current briefly prior to performing the analytical scan, allows for accumulation of a constant ion count for each scan. The prescan measures the ion flux for a fixed period of time and uses the ion current to calculate the optimum accumulation time for reaching a target value preset by the user. For ion detection in the ICR cell, a constant ion count results in accurate mass measurements without the need for internal calibrants while maintaining mass accuracies of approximately 2 ppm.
Here, we report an introductory study utilizing multiple scan functions of the LTQ-FT mass spectrometer for the analysis of protein digests separated by HPLC. Initial experiments are done to compare scan speeds for peptide detection of both full scan MS and MS/MS in the ICR cell as well as MS/MS detection in the linear trap, which utilizes parallel detection. Operating the LTQ-FT in parallel detection mode enables simultaneous detection of both the ICR cell (MS) and linear trap (MS/MS) for the fastest scan speeds to perform data-dependent mass analysis. Peptide identification using both database searching for peptide mass fingerprinting as well as sequencing are shown for an enzymatic digest of horse myoglobin.18 The benefits of accurate mass analysis for full scan and full scan MS/MS spectra are presented and show that the greater sequencing capability results in increased confidence in peptide matches that would be rejected using previous search criteria.
EXPERIMENTAL
Sample Preparation
Equine myoglobin was purchased from Sigma (St. Louis, MO) and digested using Promega trypsin (Madison, WI) according to standard procedures. The initial protein digest solution (1 mg/mL) was diluted in 0.1% trifluoroacetic acid to a final concentration of 1 pmol/μL prior to loading onto a Hypersil-Keystone BetaBasic 100 × 2.1 mm HPLC column (Thermo Electron, Bellefonte, PA). A total of 20 μL was injected into a flow rate of 120 μL/min using a Surveyor LC system (Thermo Electron, San Jose, CA). A binary solvent system consisting of (A) 0.1% formic acid and (B) 99.9% acetonitrile (0.1% formic acid) was used for a linear 25-min gradient increasing the composition of (B) from 5 to 60%.
Electrospray Ionization FT-MS MS/MS
All experiments were performed using an LTQ-FT hybrid linear trap/7-T FT-ICR mass spectrometer (Thermo Electron, Bremen, Germany) equipped with an open-ended cylindrical cell controlled by LTQ Tune 1.0 (Thermo Electron, Bremen, Germany). The Xcalibur software package enables detection in both the linear trap as well as the ICR cell simultaneously, increasing the duty cycle of the instrument. Two sets of experimental data were acquired for the enzymatic myoglobin digest analysis. The first acquired both MS and MS/MS ion detection in the ICR cell with a resolution setting of 50,000 (at m/z 400) for MS data and 25,000 (at m/z 400) for product ion detection. The second experiment utilized parallel data acquisition of MS and MS/MS by employing both the ICR cell and linear trap. Simultaneous detection is accomplished by filling the ICR cell for the full scan MS scan event; then a preview of the transient is taken and the precursor masses are queried by the linear trap so that MS/MS experiments can take place in the time required for transient detection to be completed. Once the simultaneous detection is recorded, the next full scan MS event in the ICR cell can be initiated. Collisional activation for both experiments was performed in the linear trap using helium as the target gas, qexcite = 0.25, 25% normalized collision energy and an activation time of 30 msec.
Selection of all precursor ions was done using data-dependent scanning. Data-dependent scanning uses computation interrogation of the proceeding MS scan to direct subsequent tandem mass spectral scan events. Both experiments described above utilized three data-dependent MS/MS scan events covering the three most intense ions from the MS spectrum prior to initiating another MS scan. Following the acquisition of an MS/MS scan event, the precursor mass-to-charge value was placed on an exclusion list for the next 30 sec of experimental time to increase the number of precursor ions analyzed by tandem mass spectral analysis.
Database Searching
All full scan MS/MS spectral data were searched using the Bioworks version 3.1 SR1 suite of programs (Thermo Electron, San Jose, CA). The nonredundant database was downloaded from the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.ih.gov/blast/db/fasta). The nonredundant database was used for initial protein identification for tandem mass spectral data acquired in the ICR cell as well as the linear trap. Evaluation of total protein coverage was done by creating a protein subset database consisting of equine proteins only. Database searching parameters assumed proteolysis was performed using trypsin with the possibility of one internal cleavage residue. A precursor mass tolerance of 1 Da was used to search the resulting DTA files against computer-predicted fragments from the equine database. The current version of Bioworks used for this analysis does not factor in high resolution/mass accuracy data for either the precursor mass or product ions, hence the large mass tolerance used.
All Bioworks 3.1 output files were further filtered according to cross-correlation (Xcorr) scores as a function of charge states to increase confidence limits. Specifically, the Xcorr cutoff values used to determine acceptable peptide matches were 1.5 for digest fragments originating from precursor ions with a charge state of one, 2.0 for digest fragments originating from doubly charged precursor ions, and 2.5 for triply charged ions.1,18,19 Evaluation of coverage was reported as percent coverage of amino acids identified.
Additionally, full scan accurate mass spectral data was searched to determine sequence coverage. The program Protein Prospector (http://prospector.ucsf.edu) was used to search full scan mass spectra for protein identification and sequence coverage. The search parameters incorporated mass accuracy tolerance of 5 ppm and a mass range of 500–3500 Da and was searched against a mammalian database. The molecular weight search (MOWSE) score was used for evaluation of sequence coverage.
RESULTS AND DISCUSSION
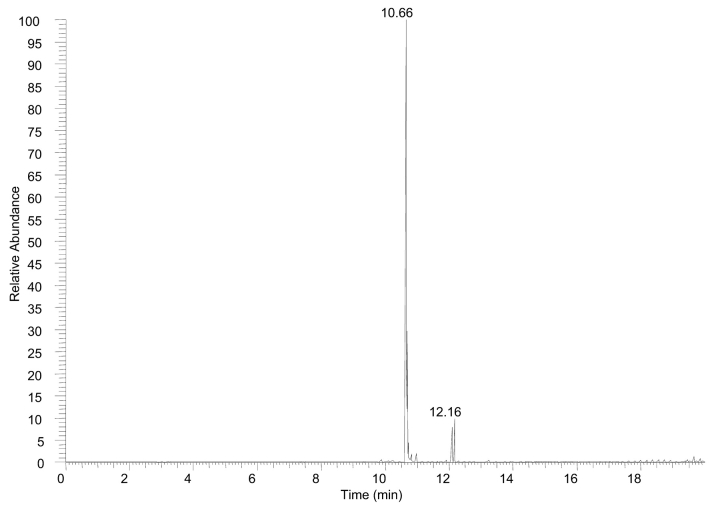
The present study demonstrates the capabilities of the Finnigan LTQ-FT mass spectrometer to perform full scan and full scan tandem MS experiments in both the linear trap as well as the ICR cell to sequence known peptides and enzymatic digests on an LC time scale. Initial experiments performed on a single peptide were done to demonstrate the time frame of full scan MS and MS/MS events detected in the ICR cell prior to pursuing enzymatic digest analysis. Figure 1 shows a base peak plot for 20 pmol of angiotensin I on column. The total chromatographic peak width is approximately 30 sec (at base) resulting in seven full scan mass spectra and 18 full scan tandem mass spectra triggered via data-dependent acquisition mode. The average time for completion of the four scan event cycle is 7.1 sec with all ion detection performed in the ICR cell. At the leading and tailing edge of the elution peak where the ion flux is low, the time for completion of the cycle averaged 9.6 sec whereas the apex of the elution profile averaged only 5.4 sec. The fastest scan cycle accounted for only 1.35 sec of experimental time, which consisted of 0.45 sec for the full scan and 0.29, 0.30, and 0.31 sec for each of the subsequent data-dependent MS/MS scan events.
FIGURE 1.
Base peak plot of angiotensin I with 20 pmol loaded on column. All ion detection was performed in the ICR cell.
The dramatic difference in cycle time is attributed to automatic gain control, which monitors ion flux for each scan and adjusts the time required to fill the ion trap to a user-defined ion population target prior to completion of the individual scan event. At points of high ion flux (apex of the elution peak) the time required to fill the linear trap prior to proceeding with either ion detection or collisional activation is greatly reduced compared with the leading and tailing edge of the elution peak without reducing spectral quality.
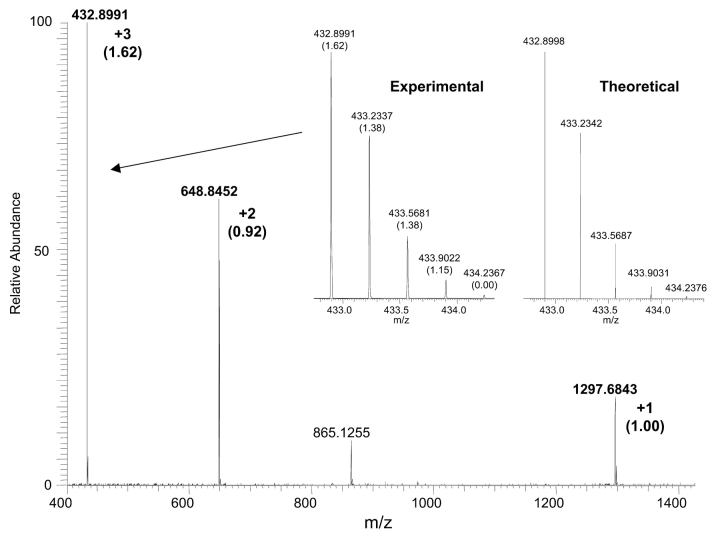
The full scan mass spectrum for the angiotensin I peptide resulting from one scan is shown in Figure 2. Each of the three charge states is labeled along with the corresponding mass accuracy (in ppm) as determined from the Xcalibur software package. The mass accuracy for each of the charge states is better than 2 ppm using external calibration regardless of the measured ion intensity. For example, the +1 charge state is approximately 20% of the +3 charge state yet the measured mass error is only 1 ppm. Of primary interest is the wide mass range that can be used for data acquisition without sacrificing mass accuracies. The inset shows a narrow mass range for the +3 charge state of angiotensin I compared with the theoretical distribution for the +3 charge state. The accurate mass measurements and corresponding mass errors (in ppm) for each of the isotopes are labeled. Accurate mass determination of isotopes is not straightforward. The isotopes contain 13C, 15N, 2H, 18O, etc., and the mixture of isotopes has to be considered. At low resolution the isotopic mixtures are not resolved and the “accurate mass” is a weighted average of isotopic contributions. At ultrahigh resolution the isotopes can be resolved and precise, accurate mass measurements can be achieved. The experimental isotopic distribution shows excellent mass accuracy for all isotopes greater than 2% ion intensity of the base peak and excellent agreement with the predicted isotopic distribution.
FIGURE 2.
Full scan mass spectrum of angiotensin I showing the +1, +2, and +3 charge states labeled with their mass error in ppm (in parentheses) for the 12C isotope. The inset shows an expanded mass range of the experimental and theoretical +3 charge state distribution with the corresponding mass errors for each isotope.
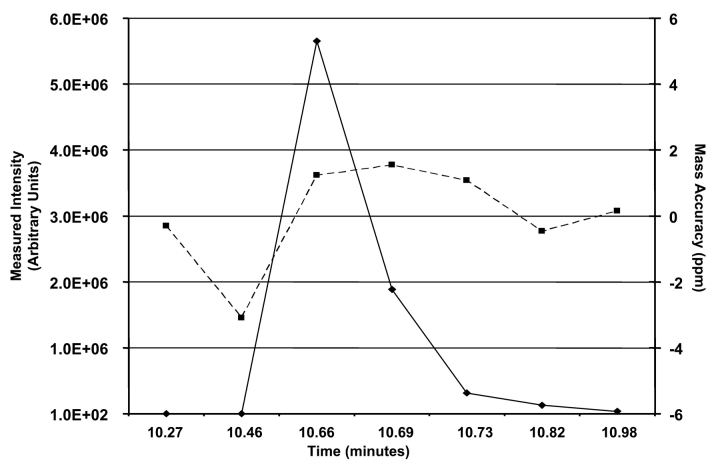
The capability of the LTQ-FT mass spectrometer to maintain high mass accuracies at various ion intensities during the elution of a peak is demonstrated for the +2 charge state of angiotensin I. Figure 3 shows a plot of the m/z 648 ion intensity for each full scan mass spectrum and the corresponding mass error. The seven full scan mass spectra used in Figure 3 represent the full scan mass spectral profile of angiotensin I eluting off of the HPLC column acquired between three data-dependent full scan MS/MS events. Despite the measured ion intensity of the m/z 648 ion ranging from 516 counts at a retention time of 10.46 min to an intensity of approximately 6 million at a retention time of 10.66 min, the mass accuracy was measured at −3 ppm for the weakest signal and 1.23 ppm for the most intense signal. The large mass error was attributed to the low ion flux for the +2 charge state of angiotensin during at the leading edge of the elution peak. Excluding the data point with the greatest mass error, the six remaining measurements vary no greater than approximately 1.8 ppm. Similar measurements for the more intense +3 charge state showed a variance of only 2.1 ppm despite a measured ion intensity difference of 8 million counts.
FIGURE 3.
Plot of seven successive ion intensity measurements from full scan mass spectra data as a function of elution time for the +2 charge state (m/z 648) of angiotensin I. The mass accuracy of the measured 12C isotope for the +2 charge state is plotted for each full scan mass spectrum.
Accurate mass analysis can also be beneficial for assigning product ions. Using data-dependent scanning, the +2 and +3 charge states of angiotensin I were analyzed by full scan MS/MS events without employing a mass list. That is, MS/MS scan events for each ion were performed without directing the mass spectrometer to select the corresponding mass-to-charge values, provided the measured ion intensity was greater than a user-defined threshold. Despite the different ion flux for each precursor ion, comparative full scan MS/MS spectra were acquired for sequence verification. Table 1 lists the fragment ions and mass assignments resulting from collision-induced dissociation of each charge state. In addition to the measured mass, the theoretical mass calculated using SEQUEST, the mass difference, and the relative intensities for each fragment are listed. The relative intensities were calculated from the base peak in each separate MS/MS spectra and not combined. All of the measured mass values for the fragment ions displayed mass accuracies better than 2 ppm for both charge states except for the y8 ion from the +2 charge state and the b8 ion from the +3 charge state which had mass errors of −3.71 and 3.31 ppm, respectively. Note the small change in mass assignments despite initiating the tandem mass spectral event from different precursor ion intensities. For example, the m/z 784 ion, which is identified as the b6+ product ion, has a measured intensity of 7.5E4 resulting from dissociation of the +2 charge state compared with 2.6E4 from the +3 charge state, yet the mass difference between the two charge states is 0.6 mmu.
TABLE 1.
b- And y-Type Ions Assigned for the +2 and +3 Full Scan Angiotensin I MS/MS Spectra from On-Line Analysis
| Ion Assignment | Theoretical m/z | Experimental m/z | Mass Difference (ppm) | Relative Intensity (%) |
| +2 Charge State | ||||
| b4+ | 534.2671 | 534.2670 | 0.19 | 4.39 |
| b9+2 | 583.2987 | 583.2988 | −0.17 | 38.78 |
| b5+ | 647.3511 | 647.3522 | −1.70 | 2.02 |
| b6+ | 784.4100 | 784.4067 | 0.38 | 100 |
| b7+ | 881.4628 | 881.4639 | −1.25 | 2.56 |
| b8+ | 1028.5312 | 1028.5331 | −1.85 | 32.54 |
| y2+ | 269.1608 | 269.1609 | −0.37 | 8.85 |
| y4+ | 513.2820 | 513.2823 | −0.58 | 43.34 |
| y6+ | 763.4244 | 763.4250 | −0.79 | 2.03 |
| y7+ | 926.4883 | 926.4891 | −0.86 | 1.24 |
| y8+ | 1025.5567 | 1025.5605 | −3.71 | 2.23 |
| +3 Charge State | ||||
| b6+2 | 392.7087 | 392.7085 | 1.37 | 32.74 |
| b4+ | 534.2671 | 534.2667 | 0.75 | 12.06 |
| b9+2 | 583.2987 | 583.2979 | 1.37 | 91.87 |
| b5+ | 647.3511 | 647.3501 | 1.54 | 88.65 |
| b6+ | 784.4100 | 784.4091 | 1.15 | 26.56 |
| b8+ | 1028.5312 | 1028.5278 | 3.31 | 2.59 |
| y2+ | 269.1608 | 269.1609 | −0.37 | 5.88 |
| y5+2 | 325.6741 | 325.6735 | 1.84 | 4.05 |
| y3+ | 416.2292 | 416.2289 | 0.72 | 3.31 |
| y8+ | 513.2820 | 513.2816 | 0.78 | 100 |
The relative intensity reported is for each separate MS/MS spectrum.
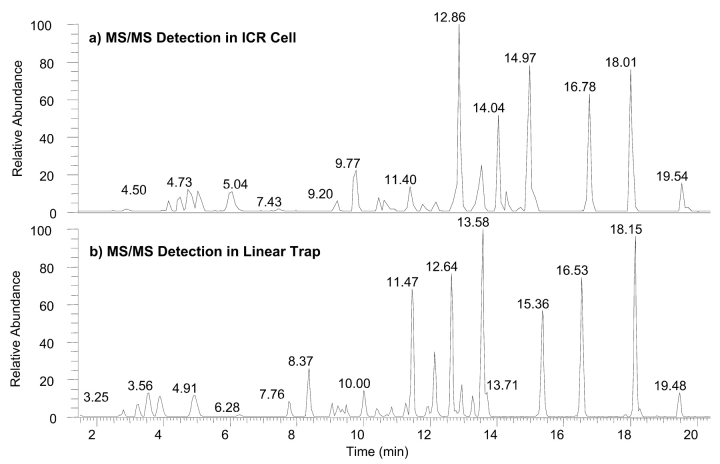
High resolution/mass accuracy analysis of a complex mixture of peptides was done to evaluate the effectiveness for improving sequence coverage using full scan MS and MS/MS spectra. A standard myoglobin enzymatic digest was analyzed under identical conditions as the angiotensin I sample, i.e., the experimental sequence consisted of a full scan mass spectrum triggering three successive full scan MS/MS events using data-dependent/dynamic exclusion experimental settings. Both experiments used full scan mass spectral detection in the ICR cell with a resolution setting of 50,000 and 25,000 for detection of full scan MS/MS spectra. In addition, the same protein digest sample was analyzed employing the linear trap for all tandem mass spectral detection while maintaining full scan mass spectral detection in the ICR cell using a 50,000 resolution setting. Figure 4 shows comparative base peak plots of the myoglobin enzymatic digest with full scan MS/MS detection in the ICR cell (Fig. 4a) and linear trap (Fig. 4b). The traces show comparative elution profiles for both experiments. Note that both profiles show similar peak widths and fine structure of coeluting peptides. The reproducibility of chromatographic response for ion detection in the ICR cell is indicative of comparable experimental time frames for full and tandem mass spectral experiments.
FIGURE 4.
Base peak plots of an HPLC chromatogram using a Big 3 data-dependent/dynamic exclusion experiment for the analysis of 20 pmol of myoglobin enzymatic digest with full scan MS/MS spectral acquisition in (a) the ICR cell and (b) the linear ion trap. All MS2 events for both experiments were triggered using full scan mass spectral data acquired in the ICR cell.
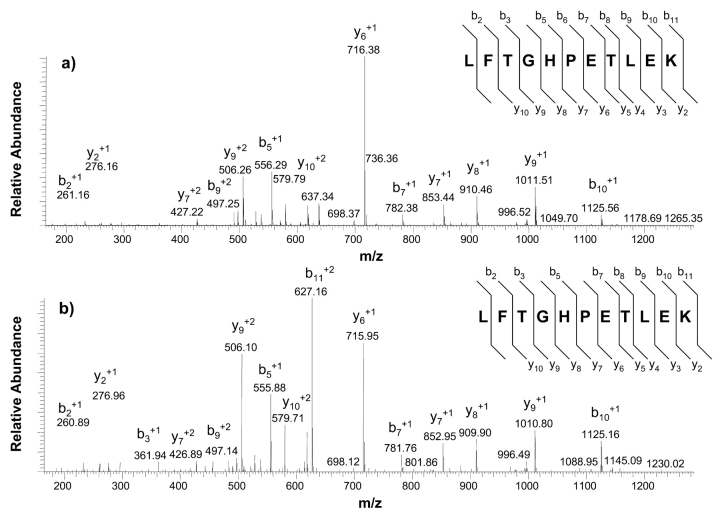
Figure 5 shows a full scan MS/MS spectrum for the myoglobin proteolytic peptide LFTGHPETLEK (T3) obtained in the ICR cell (Fig. 5a) and the linear trap (Fig. 5b). As with the full scan mass spectral data, the full scan MS/MS spectra show similar product ions due to all ion activation occurring in the linear trap. The differences in the detected mass-to-charge values are attributed to the high mass accuracy capabilities afforded by the ICR cell. The sequence coverage reported for product ion detection in the ICR cell shows that mass accuracies for all product ions were better than 2 ppm except for the b6 and b8 ions, which were measured to have mass accuracies of 2.3 and −3.5 ppm. Note the y-series that was identified for both modes of product ion detection which covers the y2–y10 which would be more than sufficient to perform de novo sequencing if necessary.2,18,19
FIGURE 5.
Comparative full scan MS/MS spectra of the +2 charge state of the peptide LFTGHPETLEK with detection in the a) ICR cell and b) linear trap. Identification of b- and y-type fragments was done using SEQUEST.
By performing all full scan MS/MS scan events in the linear trap, the time consumed for the data-dependent events is greatly reduced, increasing the number of data-dependent MS/MS events that can be performed. The time scale of the experimental cycle for full scan mass spectral detection in the ICR cell was 0.92 sec. Each of the three successive full scan data-dependent MS/MS scan events took 0.20 sec. Thus, the time needed to complete a full scan mass spectral event using a resolution setting of 50,000 allows for almost five MS/MS scan events to be collected in parallel; that is, simultaneously performing full scan MS analysis in the ICR cell and MS/MS detection in the linear trap. Parallel detection can alleviate poor chromatographic resolution and may increase protein sequence coverage by triggering more data-dependent MS/MS scan events from different precursor ions.
Benefits of high resolution/mass accuracy were evaluated by employing database searching for both modes of ion detection as well as high mass accuracy analysis of intact peptides for total protein coverage. Search results performed using Bioworks 3.1 SR1 resulted in identification of a mutant strand of horse myoglobin as the top hit. The top-hit horse myoglobin sequence contained an asparagine residue instead of leucine at position 104, resulting in a mass difference of only 0.96 Da for the +1 charge state of the peptide YNEFISDAIIHVLHSK (T17, MW 1885 Da). The current version of Bioworks does not enable high mass accuracy (< 50 ppm) as a search parameter for either full scan or full scan MS/MS spectra; thus the failure to differentiate the mass defect. Assuming the mutation is correct according to Bioworks search result, coverage for product ion detection in the linear trap was 86% and 73% for detection in the ICR cell. Reported coverage for both modes of detection omitted potential hits that did not meet the cross-correlation criteria outlined above.
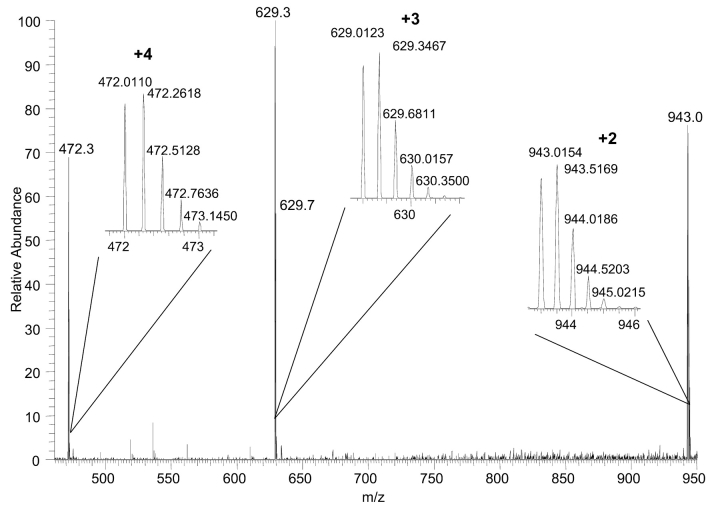
A closer inspection of the T17 peptide (residues 103–118) was done using the high resolution/high mass accuracy measurements from the LTQ-FT. Figure 6 shows high-resolution full scan mass spectra of the +2, +3, and the +4 charge states for T17 peptide with a retention time at 20.33 min. In addition, magnified mass ranges for the three ions of interest are shown. Note the isotope containing one 13C is more intense than the monoisotopic peak for all three charge states, which is attributed to identification of the mutant sequence for the T17 peptide. Spectral acquisition in low-resolution mode or in protein search engines that do not incorporate high-resolution data fails to take isotopic distribution into account and yields molecular ion identification to that of the mutant version of the myoglobin sequence. Table 2 lists the experimentally measured masses and mass errors for each of the three charge states using the monoisotopic peak as detected in the ICR cell. The native and mutant myoglobin sequences were used to calculate the theoretical masses for comparison. Clearly, the native peptide sequence showed significantly better mass accuracies for all charge states compared with that of the mutated sequence, with each charge state showing a sub ppm mass error compared with mass errors in excess of 20 ppm for the mutant sequence.
FIGURE 6.
Full scan mass spectrum for T17 peptide with a retention time of 20.33 min. An expanded mass range for each of the three most intense ions demonstrates charge state assignment.
TABLE 2.
Experimental and Theoretical 12C Mass-to-Charge Values for the +2, +3, and +4 Charge States of the Native and Mutant T17 Peptide Sequences
| Theoretical m/za | |||
| Charge State | Experimental m/z | YLEFISDAIIHVLHSK | YNEFISDAIIHVLHSK |
| +2 | 943.0154 | 943.0146 (0.85) | 943.4940 (24.27) |
| +3 | 629.0123 | 629.0121 (0.32) | 629.3317 (28.83) |
| +4 | 472.0110 | 472.0109 (0.21) | 472.2506 (23.72) |
aMass errors in ppm shown in parentheses.
Further analysis of the full scan MS/MS spectrum for all three of the charge states was performed using high mass accuracy detection in the ICR cell. Table 3 lists the measured mass for the identified b-type product ions and its corresponding theoretical mass and mass accuracies for the native as well as the mutant T17 peptide. The mass accuracies for the native sequence are better than 2 ppm except for the b10 and b11 product ions. Accurate mass analysis for the mutant sequence lists errors in excess of 500 ppm. In addition, the b2 product ion which is the site of modification shows a mass accuracy of 1.80 ppm for the native sequence and 3449 ppm for the mutant.
TABLE 3.
b-Type Ions for the Proposed Sequence of the Native and Mutant Position 104 for Horse Myoglobin
| YLEFISDAIIHVLHSK | YNEFISDAIIHVLHSK | |||||
| b-Ion Sequence | Product Ion | Experimental m/z | Theoretical m/z | Mass Accuracy (ppm) | Theoretical m/z | Mass Accuracy (ppm) |
| L/N | 2 | 277.1542 | 277.1547 | 1.80 | 278.1135 | 3449.31 |
| F | 4 | 553.2653 | 553.2657 | 0.72 | 554.2245 | 1730.71 |
| I | 5 | 666.3483 | 666.3497 | 2.10 | 667.3086 | 1439.06 |
| D | 9 | 868.4081 | 868.4087 | 0.69 | 869.3676 | 1103.68 |
| I | 10 | 1052.5265 | 1052.5299 | 3.23 | 1053.4887 | 913.35 |
| I | 11 | 1165.6099 | 1165.6139 | 3.43 | 1166.5728 | 825.41 |
| H | 12 | 1302.6725 | 1302.6729 | 0.31 | 1303.6317 | 735.79 |
| V | 13 | 1401.7400 | 1401.7413 | 0.93 | 1402.7001 | 684.47 |
| L | 13 | 1514.8230 | 1514.8253 | 1.52 | 1515.7842 | 634.13 |
| H+2 | 14 | 826.4457 | 826.4458 | 0.12 | 826.9252 | 579.86 |
| S+2 | 15 | 869.9600 | 869.9618 | 2.07 | 870.4412 | 552.82 |
| K+2 | 16 | 934.0076 | 934.0093 | 1.82 | 934.4887 | 514.83 |
Accurate mass assignments were obtained using SEQUEST browser.
Database searching of MS/MS spectra is the primary source of protein identification/sequencing. Full scan MS/MS spectra acquired in both the linear trap and ICR cell were used for determining protein identification and coverage through database searching. Bioworks search results from tandem mass spectral data acquired in the linear trap showed 13% better amino acid residue coverage compared with product ion detection in the ICR cell following output filtering. Discrepancies in sequence coverage were attributed to additional fragment ions identified using ion trap detection compared with that for the data collected in the ICR cell. For example, tandem mass spectral data for the +1 charge state of the T7 peptide showed identification of 8 out of 10 possible b- and y-type ions and a cross-correlation score of 1.5. The Bioworks cross-correlation score resulting from ICR data acquisition was 1.1 despite identifying 7 out of 10 possible fragment ions. Closer inspection of the full scan MS/MS data acquired in the ICR cell showed that mass accuracies for the seven identified fragments were better than 2.5 ppm. In addition to the identified fragments, two additional product ions labeled as the b2 (m/z 231) and y5 ion (m/z 607) were identified despite having measured ion intensities of 1.5% and 3.3% of the base peak. The measured mass errors for each ion were 1.3 and 2.6 ppm, respectively. Inspection of the remaining peptides in the +1 charge state showed similar mass measurement accuracies for product ions, resulting in an increased confidence in the search results using lower cross-correlation scores. Thus, if the criterion used for filtering cross-correlation scores was reduced for only the +1 charge states by 0.5, protein coverage increased to 90% using data acquired in the ICR cell.
In addition to using full scan MS/MS spectra for database searching, full scan mass spectra were used to determine sequence coverage. All full scan mass spectra were searched using the MOWSE algorithm with 5 ppm constraints. The top five protein matches from the data were all myoglobin with horse being the best match. The MOWSE score was 5.6E7 compared with the next possible match with a score of 1.3E4 which was rabbit myoglobin. All peptides matched to rabbit myoglobin protein contained the same peptides as those attributed to the horse myoglobin protein, indicating a conserved sequence for 6 of the 14 molecular weight entries. Based on the high confidence of the full scan mass spectral search, mass accuracy measurements were made for all charge states of identified and predicted peptide masses for the enzymatic digest of horse myoglobin. Table 4 lists theoretical and experimental masses for peptide identification along with the corresponding mass errors for all charge states detected. All results show mass accuracies better than 1.0 ppm except for the +1 charge states for the T1 and T3 peptides, which had mass errors of −1.4 and 1.6 ppm. Incorporating all identified peptides revealed 93% sequence coverage. The high mass accuracies measured for both the precursor ions as well as the product ions yields high confidence in the identified sequences despite the cross-correlation scores.
TABLE 4.
Comparative Myoglobin Tryptic Peptide Mass Accuracies for All Detected Charge States from Full Scan Mass Spectral Data Acquired in the ICR Cell at a Resolution Setting of 50,000
| Peptide Identification | Sequence | Charge State | Theoretical m/z | Experimental m/z | Mass Error (ppm) |
| T1 | GLSDGEWQQVLNVWGK | +2 | 908.4549 | 908.4563 | −1.4 |
| +3 | 605.9723 | 605.9732 | −0.9 | ||
| T2 | VEADIAGHGQEVLIR | +2 | 803.9310 | 803.9306 | 0.4 |
| +3 | 536.2898 | 536.2895 | 0.3 | ||
| T3 | LFTGHPETLEK | +1 | 1271.6630 | 1271.6614 | 1.6 |
| +2 | 636.3352 | 636.3346 | 0.6 | ||
| +3 | 424.5592 | 424.5589 | 0.3 | ||
| T7 | TEAEMK | +1 | 708.3233 | 708.3229 | 0.4 |
| T8 | ASEDLK | +1 | 662.3356 | 662.3352 | 0.4 |
| T10–T11 | HGTVVLTALGGILKK | +2 | 753.9720 | 753.9724 | −0.4 |
| +3 | 502.9837 | 502.9837 | 0.0 | ||
| T13–T14 | GHHEAELKPLAQSHATK | +2 | 927.4845 | 927.4842 | 0.3 |
| +3 | 618.6587 | 618.6585 | 0.2 | ||
| +4 | 464.2459 | 464.2456 | 0.3 | ||
| T14 | PLAQSHATK | +1 | 952.5211 | 952.5204 | 0.7 |
| +2 | 476.7642 | 476.7639 | 0.3 | ||
| T15–T16 | HKIPIK | +1 | 735.4876 | 735.4869 | 0.7 |
| T17 | YLEFISDAIIHVLHSK | +2 | 943.0146 | 943.0154 | 0.9 |
| +3 | 629.0121 | 629.0123 | 0.3 | ||
| +4 | 472.0109 | 472.0110 | 0.2 | ||
| T18 | HPGDFGADAQGAMTK | +2 | 751.8383 | 751.8380 | 0.3 |
| +3 | 501.5613 | 501.5611 | 0.2 | ||
| T19 | ALELFR | +1 | 748.4352 | 748.4344 | 0.8 |
| T20 | NDIAAK | +1 | 631.3410 | 631.3408 | 0.2 |
| T21–T22 | YKELGFQG | +1 | 941.4727 | 941.4719 | 0.8 |
A comparison of protein coverage using each ion detection method was done to evaluate the benefits of accurate mass measurements. Figure 7 shows the equine myoglobin sequence with corresponding coverage resulting from MS/MS detection in the linear trap, ICR cell, and ICR detection of full scan mass spectra. The greatest coverage was reported using full scan mass spectral data that showed 93.4% coverage, whereas MS/MS detection in the ICR showed 90.1% coverage compared with MS/MS detection in the linear trap of 83.2%. The coverage reported for MS/MS detection in the linear trap assumed that T17 peptide had the mutant sequence. It is of primary interest that fewer MS/MS scans were performed in the ICR cell than were acquired using tandem mass spectral detection in the linear trap (1088 to 1794), yet better coverage was obtained with greater confidence in the peptide identification.
FIGURE 7.
Coverage report generated using MS/MS data from the linear trap (solid lines), ICR cell (dashed lines), and full scan accurate mass data from the ICR cell (dash-dotted line). The coverage reported for the linear trap data satisfied the Xcorr vs. charge state criteria outlined in the Experimental section while the coverage reported for the ICR data relaxed the Xcorr criteria by 0.5 for each charge state.
CONCLUSIONS
In this work we explored three methods of bottom-up peptide detection and sequencing for total protein coverage using a hybrid linear trap/FT-ICR mass spectrometer on an LC time scale. Incorporation of a linear trap mass spectrometer on the front end provides several advantages that greatly increase scan time and instrument performance compared with that of previous FT-ICR mass spectrometers. Using the two orthogonal electron multipliers on the linear trap enables prescan ion counting to regulate the analytical ion population (incorporation of automatic gain control), regardless of ion flux during an HPLC gradient separation. In addition to controlling the ion population, the linear trap is instrumental in isolating and collisionally activating the precursor ions prior to detection in either the linear trap or the ICR cell. Due to a constant ion population in the ICR cell, external calibration was used for all mass measurements resulting in mass accuracies better than 2 ppm for full scan MS and MS2 spectra. Simultaneous detection of full scan MS data in the ICR cell and full scan MS2 data in the linear trap yielded the fastest scan times while maintaining high confidence limits for peptide identification. This approach offers a simple and robust method for increasing protein coverage by incorporating accurate mass measurements of precursor ions and the option of performing simultaneous tandem mass spectral sequencing in the linear trap or acquiring accurate mass spectral information on resulting product ions. Both options were demonstrated on chromatographic time scale, demonstrating the utility for complex proteomics applications.
REFERENCES
- 1.Link AJ, Eng J, Schieltz DM, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 1999;17:676–682. [DOI] [PubMed] [Google Scholar]
- 2.Wenner BR, Lynn BC. Factors that affect ion trap data-dependent MS/MS in proteomics. J Am Soc Mass Spectrom 2004;15:150–157. [DOI] [PubMed] [Google Scholar]
- 3.Peng J, Gygi, SP. Proteomics: The move to mixtures. J Mass Spectrom 2001;36:1083–1091. [DOI] [PubMed] [Google Scholar]
- 4.Washburn MP, Wolters D, Yates JR, III. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 2001;19: 242–247. [DOI] [PubMed] [Google Scholar]
- 5.AebersoldR, Goodlett DR. Mass spectrometry in proteomics. Chem Rev 2001;101:269–296. [DOI] [PubMed] [Google Scholar]
- 6.Quenzer TL, Emmett MR, Hendrickson CL, Kelly PH, Marshall AG. High sensitivity Fourier transform ion cyclotron resonance mass spectrometry for biological analysis with nano-LC and microelectrospray ionization. Anal Chem 2001;73:1721–1725. [DOI] [PubMed] [Google Scholar]
- 7.Senko MW, HendricksonCL, Emmett MR, Stone D-H S, Marshall AG. External accumulation of ions for enhanced electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom 1997;8:970–976. [Google Scholar]
- 8.Beu SC, Senko MW, Quinn JP, McLafferty FW. Improved Fourier-transform ion-cyclotron-resonance mass spectrometry of large biomolecules. J Am Soc Mass Spectrom 1993;4:190–192. [DOI] [PubMed] [Google Scholar]
- 9.Belov ME, Nikolaev EN, Anderson GA, et al. Design and performance of an ESI interface for selective external ion accumulation coupled to a Fourier transform ion cyclotron mass spectrometer. Anal Chem 2001;73: 253–261. [DOI] [PubMed] [Google Scholar]
- 10.Sannes-Lowery K, Griffey RH, Kruppa GH, Speir JP, Hofstadler SA. Multipole storage assisted dissociation, a novel in-source dissociation technique for electrospray ionization generated ions. Rapid Commun Mass Spectrom 1998;12:1957–1961. [DOI] [PubMed] [Google Scholar]
- 11.Hofstadler SA, Sannes-Lowery K, Griffey RH. Infrared multiphoton dissociation in an external ion reservoir. Anal Chem 1999;71:2067–2070. [DOI] [PubMed] [Google Scholar]
- 12.Hofstadler SA, Drader JJ, Gaus H, Hannis JC, Sannes-Lowery K. Alternative approaches to infrared multiphoton dissociation in an external ion reservoir. J Am Soc Mass Spectrom 2003;14:1413–1423. [DOI] [PubMed] [Google Scholar]
- 13.Masselon C, Anderson GA, Harkewicz R, Bruce JE, Pasa-Tolic L, Smith RD. Accurate mass multiplexed tandem mass spectrometry for high-throughput polypeptide identification from mixtures. Anal Chem 2000;72: 1918–1924. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Marshall AG. An off-center cubic ion trap for Fourier transform ion cyclotron resonance mass spectrometry. Int J Mass Spectrom Ion Processes 1994;133: 29–38. [Google Scholar]
- 15.Peterman SM, Dufresne CP, Horning S, Pfaff H, Ueckert T. On-line identification of posttranslational modifications and sequence irregularities using a hybrid linear trap/FT-ICR mass spectrometer. ABRF 2004, Portland, OR, P50-M.
- 16.Syka JEP, Marto JA, Bai DL, et al. Novel linear quadrupole ion trap/FT mass spectrometer: Performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J Proteome Res 2004;3:621–626. [DOI] [PubMed] [Google Scholar]
- 17.Stafford GC, Taylor DM, Bradshaw SC. Method of increasing the dynamic range and sensitivity of a quadrupole ion trap mass spectrometer. US Patent #5,107,109, 1992.
- 18.Kapp EA, Schutz F, Reid GE, et al. Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal Chem 2003;75:6251–6264. [DOI] [PubMed] [Google Scholar]
- 19.Moore RE, Young MK, Lee TD. Qscore: An algorithm for evaluating SEQUEST database search results. J Am Soc Mass Spectrom 2002;13:378–386. [DOI] [PubMed] [Google Scholar]