Abstract
The activity of many transcriptional regulators is significantly altered by posttranslational modifications of specific sites. For example, the activity of the muscle-restricted transcription factor family myocyte enhancer factor 2 (MEF2) is tightly controlled by phosphorylation. This modification is responsible for either an increase or a decrease in transcriptional activity, depending on the specific amino acid residues that are phosphorylated by signal-dependent kinases.
Although mass spectrometry-based methods, such as precursor ion and neutral loss scans, are extremely useful for identifying unknown phosphopeptides from a complex mixture, they do not take advantage of any prior knowledge about the protein being investigated. Quite often a significant amount of information is available. This may include the primary sequence, type of phosphorylation (serine/threonine vs. tyrosine), or predicted phosphoacceptor sites (consensus peptide that is targeted by a kinase). This information can be used to predict precursor and fragment ion m/z values for a multiple reaction monitoring (MRM) experiment. By using these highly sensitive MRM experiments to trigger dependent product ion scans on a hybrid quadrupole linear ion-trap instrument, we were able to identify low levels of phosphorylation of MEF2A (a member of the MEF2 family), and α-casein. This method of monitoring protein phosphorylation at specific phosphoacceptor sites may prove useful in understanding the physiological regulation of protein function.
Keywords: phosphopeptides, LC-MS/MS, MEF2, linear ion trap, triple quadrupole
Mass spectrometry has become a powerful tool for investigating the posttranslational status of proteins. By taking advantage of the characteristic fragmentation pattern of phosphopeptides, it is possible to identify a phosphopeptide even when it is in a complex mixture of nonphosphorylated peptides.1 In many cases, it is possible to perform this analysis with no a priori knowledge as to the identity or characteristics of the protein under study. As impressive as this is, it does not take advantage of the wealth of biological knowledge that has been accumulated to date. By using this biological knowledge to design mass spectrometric experiments, it should be possible to achieve an even higher level of sensitivity. This targeted approach would be very suitable for investigating the posttranslational status of a specific protein.
For example, myocyte enhancer factor 2A (MEF2A) is a transcriptional regulator that is primarily involved in the development of cardiac and skeletal muscle cells. A significant amount of information is currently known about MEF2A, including such things as primary sequence, splice variants, cellular location, and evidence for phosphorylation at serine and threonine residues.2 Depending on the specific site, phosphorylation can affect the ability of MEF2A to activate transcription of muscle-specific genes,3 or it can target MEF2A for protein degradation.4 Clearly, tools are required that can help identify, and possibly quantify, the phosphorylation status of MEF2A during developmental and physiological processes.
MRM experiments, using a triple quadrupole instrument, are designed for obtaining the maximum sensitivity for detection of target compounds. This type of mass spectrometric experiment is widely used in detecting and quantifying drug and drug metabolites in the pharmaceutical industry.5 Knowing the mass and structure of the drug molecule, it is possible to predict the precursor m/z and a fragment m/z (MRM transition) for many common metabolites of this drug molecule. These MRM experiments can be used to screen for these metabolites, and trigger a dependent product ion scan to confirm the metabolite structure.6 The same principle can be applied to studying posttranslationally modified peptides. In this case, the mass and structure of the peptide is known. This information can be used to design an MRM experiment to specifically identify potential sites of posttranslational modifications in the protein of interest.
These highly sensitive MRM experiments were used to trigger dependent acquisition of product ion scans (MS/MS) using a hybrid quadrupole-linear ion trap instrument. This method was successful in identifying several phosphopeptides both from α-casein, and from the mammalian transcriptional regulator MEF2A.
MATERIALS AND METHODS
Unless otherwise noted, all chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Cell Culture and Transfections
COS7 cell cultures were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen/Gibco, Burlington, ON, Canada) containing penicillin, streptomycin (Gibco), and 10% fetal bovine serum (FBS, Atlanta Biologicals, Narcross, GA). Transfections were performed using the calcium phosphate precipitation method.
Tandem Affinity Purification
Cells were transfected with 20 μg of pCDNA4/TO/TAP, 2 μg of pMT3-p38, and 8 μg of pCDNA-MKK6 per 100-mm dish. Typically, 5 plates of cells were used for a single purification. The details of mammalian TAP-tagged protein purification are explained elsewhere.7,8 Briefly, cells were lysed by freeze/thawing. The lysate was passed over IgG resin and the beads were washed. Tagged proteins were eluted by cleaving with TEV protease (Invitrogen), then supplementing with Ca2+ and passed over calmodulin resin (Stratagene, LaJolla, CA) for a second round of purification. Proteins were finally eluted using SDS sample buffer and analyzed by SDS-PAGE. Proteins were visualized using Gelcode Blue (Pierce, Woburn, MA). The amount of protein in each band was estimated at 500 ng.
In-Gel Trypsin Digest
Protein bands were excised, cut into 1 mm3 pieces, and washed 3 times with 50% acetonitrile/25 mM ammonium bicarbonate for 15 min with shaking. Gel pieces were incubated with 50 mM ammonium bicarbonate + 10 mM dithiothreitol for 30 min at 50°C, washed with acetonitrile, then incubated with 50mM ammonium bicarbonate + 55 mM iodoacetamide (freshly made) for 20 minutes in the dark. The gel pieces were washed with acetonitrile, air dried, and rehydrated with 12.5 ng/μL trypsin (Promega, sequencing grade, Madison, WI) in 50 mM ammonium bicarbonate, then incubated at 37°C overnight. The liquid was transferred to a clean tube. Peptides were extracted once using 3% formic acid, 2 min at 65°C, followed by 20 min of shaking, 1 min of centrifugation, and combined with the liquid from the previous step.
For the in-solution digestion of casein, 5 mg of Sigma Aldrich C-6780 was weighed using a Mettler UMT2 scale (Fisher Scientific) and dissolved in 5 mL of 50 mM ammonium bicarbonate. The solution was brought up to 1 mM dithiothreitol and 50°C for 20 min. The solution was brought up to 5.5 mM iodoacetamide and left in the dark for 30 min. The solution was digested using trypsin (Promega, sequencing grade) at a substrate-to-enzyme ratio of 1:50 for 12 h at 37°C. The solution was diluted using LC solvent A (described below). The same set of dilutions was used for the experiments involving casein.
LC/MS
The digests were analyzed using an Agilent 1100 Nanoflow LC (G2226A) system connected to the nanoSpray source of a 4000 QTRAP. The 4000 QTRAP is a linear ion trap instrument, similar in principle to the QTRAP,9 only built on the API 4000 platform. In addition to being a linear ion trap instrument, it performs identically to an API 4000 for triple quadrupole-based scans. One microliter of the sample was directly loaded onto a 75 μm × 15 cm Vydac Everest column at 300 nL/min. Following 10 min of loading, the valve was switched so that the autosampler was bypassed. Solvent A was 2% acetonitrile 0.1% formic acid. Solvent B was 98% acetonitrile 0.1% formic acid. The gradient was as follows: 0 min, 2% B; 10 min, 2% B; 40 min, 45% B; 42 min, 80% B; 47 min, 80% B; 49 min, 2% B; 70 min, 2% B. The 4000 QTRAP was operated in MRM mode. These MRM transitions (50 msec dwell time) were used to trigger dependent linear ion trap scans: enhanced resolution and enhanced product ion (EPI) scans. An EPI scan is similar to a traditional product ion scan on a triple quadrupole instrument, in that the fragments are generated by accelerating ions into a collision cell. The difference, however, is that an EPI scan uses the linear ion trap to trap the resulting fragments and perform a mass scan.10 The total cycle time for this method was 3–5 sec. This data-dependent method is referred to as targeted MRM-IDA, for multiple reaction-monitoring information-dependent acquisition.
Software
The MRM transitions for α-casein and MEF2A were calculated using a prototype version of software from the Applied Research Group at MDS Sciex (Concord, ON, Canada). The software requires an amino acid sequence of the protein of interest, a starter method containing the LC conditions and an empty MRM-IDA experiment. The software will perform an in-silico digest of the protein and create a set of peptides each containing at least one possible site of modification. For each peptide, it will generate an MRM transition for the calculated m/z of the precursor ion and an appropriate fragment ion. The new method, specific for the protein of interest, is saved and submitted as a batch for data acquisition. The software is research grade. (Please contact the authors for information on obtaining and using this software.) The Agilent Nanoflow LC system and 4000 QTRAP were both controlled using Analyst 1.4. The combined information from each MRM–IDA experiment was used to perform Mascot searches against the SwissProt database.
RESULTS AND DISCUSSION
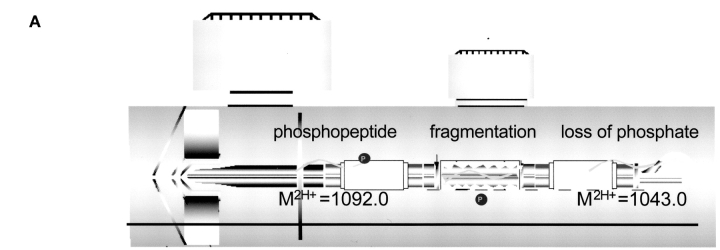
Triple quadrupole instruments are capable of performing MRM experiments.5 This involves using the first quadrupole in resolving mode (RF and DC voltages applied) so that a specific m/z is allowed to pass. These ions are accelerated into the collision cell (a quadrupole used in RF mode only) where they collide with gas molecules and fragment (collision-induced dissociation, CID). The third quadrupole is also operated in resolving mode so that it passes only one of the fragment ions from the target compound. (Fig. 1A). When combined with chromatography, this makes the mass spectrometer a highly specific mass detector for the target molecule. It is highly unlikely that isobaric compounds that may coelute with the target compound will also have an identical fragment mass. This makes MRM methods ideal for quantifying compounds in a complex biological matrix.
FIGURE 1.
A: Hybrid triple quadrupole and linear ion trap mass spectrometer. Quadrupole-specific scans, such as MRM, can be combined with linear ion trap MS/MS scans in a single experiment. B: Many MRM transitions can be monitored simultaneously, enabling researchers to look for potential phosphopeptides from a known protein sequence.
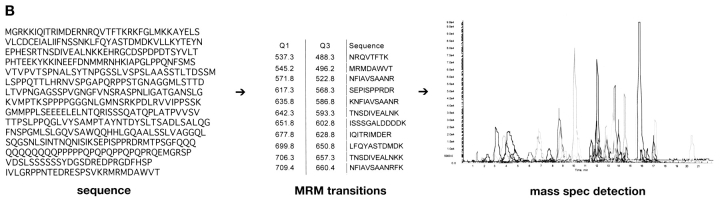
The same mass spectrometric methods can also be used for detecting peptides or posttranslationally modified peptides. For example, it is very likely that a phosphoserine-containing peptide, with a doubly charged m/z value of 1092.0, will lose this phosphate group during CID to form a doubly charged fragment at an m/z value of 1043.0 (Fig. 1A). A method using this specific MRM transition (Q1 = 1092.0 and Q3 = 1043.0) can be used to detect this phosphopeptide. If the collision cell incorporates a form of linear acceleration (Linac),11,12 which quickly transfers fragment ions out of the collision cell, it is possible to monitor a large number of these MRM transitions almost simultaneously. Therefore, it becomes possible to probe for several predicted phosphopeptides from a known protein sequence. This differs significantly from another mass spectrometric technique for identifying phosphopeptides, namely a neutral loss scan.13 In a neutral loss scan, Q1 scans a specified mass range while Q3 scans the same range, only it lags behind by a specified m/z (for example 49 m/z is useful for identifying doubly charged phosphopeptides). A neutral loss scan attempts to find any compound that will lose a functional group during CID, while an MRM scan looks for a target peptide to generate a specific fragment. A common problem with using a neutral loss scan to discover phosphopeptides is that many nonphosphopeptides will surreptitiously generate a similar neutral loss, causing the mass spectrometer to waste time identifying a nonmodified peptide. Or worse, a phosphopeptide will not generate the specified neutral loss, and therefore not be detected. A neutral loss scan is limited to investigating only one predicted fragmentation pathway (e.g., loss of 49 m/z), while a targeted MRM approach can investigate several predicted fragmentation pathways for the same peptide (loss of 49 m/z from 2+, loss of 32.7 m/z from 3+, formation of a b2 ion, etc.). Unlike most mass spectrometric methods, targeted MRM experiments benefit significantly from prior biological information or from knowledge of fragmentation pathways. Primary sequence, type of phosphorylation (serine/threonine or tyrosine), sequence motif (kinase consensus sites), predicted fragmentation pathways (b2, imminium ion, etc.) could all be used to calculate one, or several, MRM transitions that will identify the potential phosphopeptides (Fig. 1B). For example, phosphoserine or phosphothreonine peptides often display a neutral loss of 98 amu (49 m/z loss from a doubly charged precursor), while phosphotyrosine peptides often generate a 216 m/z imminium fragment ion.
A computer program was developed to simplify the process of developing an MRM method based on the primary amino acid sequence. The program performs a theoretical enzymatic digest of the protein, selects peptides containing a specified type of phosphorylation (S/T or Y), and calculates the Q1 and Q3 m/z values for various charge states and fragment ions. This program was used to create a method with 56 transitions, corresponding to potential phosphopeptides from α-casein. Likewise, for MEF2A, the program was used to create 58 transitions to search for potential phosphopeptides.
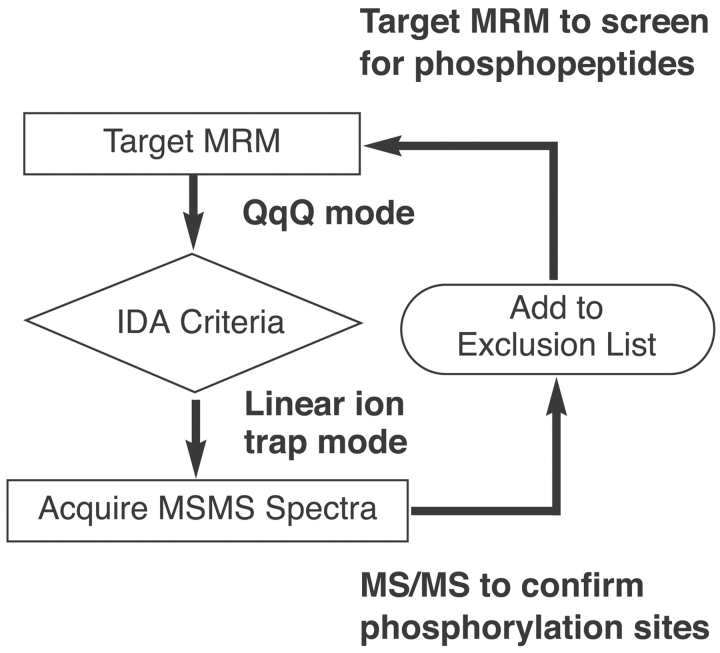
Although the signal from an MRM experiment is a good indication that the compound of interest is eluting from the column, it is not definitive proof that we have the appropriate peptide. To confirm the identity of the phosphopeptides being detected by MRM, an information-dependent acquisition (IDA) experiment was designed to obtain dependent MS and MS/MS spectra of the potential phosphopeptides (Fig. 2). In this way, when the signal corresponding to one of the MRM transitions reached a specified threshold, it triggered several linear ion trap scans.9,10 This included, an enhanced resolution scan (a high-resolution scan using the linear ion trap) to confirm the charge state and monoisotopic mass of the potential phosphopeptide, as well as an enhanced product ion scan (an MS/MS scan using the linear ion trap)13 to confirm the sequence of the peptide and the location of the phosphorylation.
FIGURE 2.
Targeted MRM triggers information-dependent acquisition (IDA). When the signal from an MRM transition reaches a specified threshold, it triggers a dependent acquisition of an MS/MS scan.
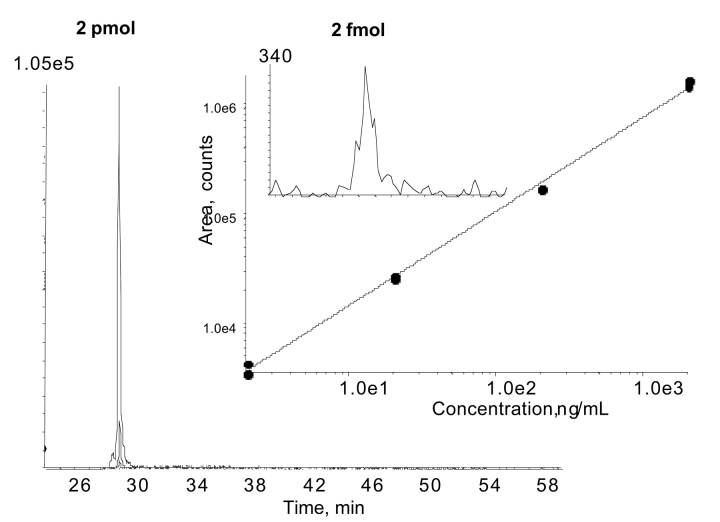
The specificity of an MRM scan means that there is very little chemical background. This permits very low levels of detection, as well as the ability to quantify over a wide dynamic range. As an example, a standard curve was performed using 1-μL injections of different concentrations of a tryptic digest of α-casein. A phosphopeptide from this digest (m/z = 76) demonstrated a linear response from 2 pmol down to 2 fmol on column (Fig. 3). The limit of detection from this experiment is estimated to be below 1 fmol. This example assumes that close to 100% of the peptide (YKVPQLEIVPN SAEER, 976 m/z) was phosphorylated. Other peptides displayed a similar result (data not shown).
FIGURE 3.
Quantitation using MRM is an established method. The MRM signal of a phosphopeptide from an α-casein digest was linear over several orders of magnitude (2–200 fmol) with an estimated limit of detection below 1 fmol. All data points are shown, two analyses for each concentration, 1 μL / injection.
The intensity, or area, of an MRM signal depends on several factors such as concentration but also includes ease of ionization and fragmentation efficiency.5 Therefore, it would be difficult to comment on the abundance of a particular phosphopeptide without some form of standardization. For example, once a particular phosphopeptide has been identified, it would be possible to synthesize a heavy isotope version of this phosphopeptide. Since this phosphopeptide has a structure identical to the native phosphopeptide but has a slightly higher mass, it would be possible to include this in the analysis as an internal standard. In this way, information about the absolute quantity14 of a phosphopeptide could be obtained. Alternatively, the intensity of an MRM for a particular phosphopeptide could be compared with that of a nonphosphorylated peptide from the same protein. This information could be used to determine the relative amount of phosphopeptide from one experiment to the next. This type of quantitative information concerning phosphorylation status is extremely valuable to biologists in determining the response of protein function to different physiological conditions.
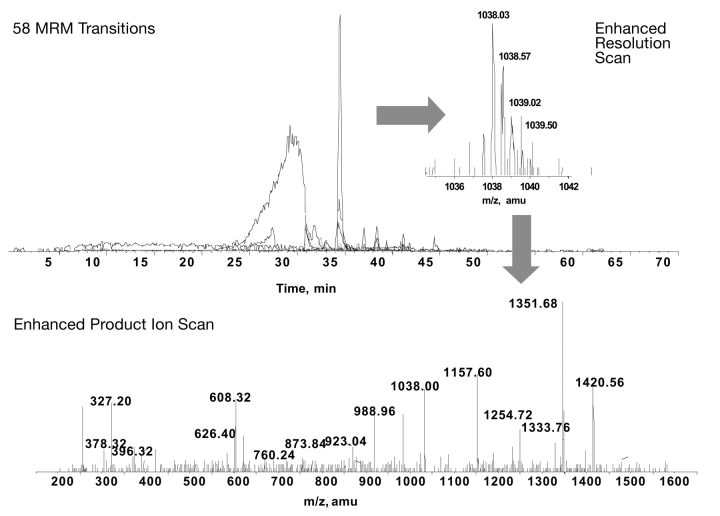
Our initial interest was to use the targeted MRM-IDA technique to identify phosphopeptides in MEF2A. Previous studies have identified several important phosphorylation sites using biochemical3,15 or mass spectrometric4 techniques. However, definitive MS/MS data to confirm the phosphorylated residue was missing for many of these potential phosphopeptides. In this study, we were able to identify six phosphopeptides (one is a missed cleavage) from mammalian-produced MEF2A that had been co-expressed with a known physiological regulator of MEF2A activity, p38 mitogen-activated protein (MAP) kinase. For each peptide, the MS/MS fragmentation pattern was used to identify the phosphorylated residue. Figure 4 shows an overlay of the extracted ion chromatograms for each of the 58 MRM transitions used to probe for potential phosphopeptides in MEF2A, and an example of the MS/MS fragmentation spectra that was triggered by one of the MRM transitions. This spectra identifies the sequence VMPTKphosSPPPPGGGNL GMNSR, a known phosphopeptide (Ser-255) involved in the targeted degradation of MEF2A.4
FIGURE 4.
Identification of phosphorylation sites in MEF2A. Fifty eight MRM transitions, corresponding to potential phosphorylation sites in MEF2A were monitored. The MS/MS spectra from one of the identified phosphopeptides is shown (VMPTKpSPPPPGGNLGMNSR). The peptide SEPIpSPPRDR did not resolve well chromatographically, but could be identified from time 25 to 30 min. Other phosphopeptides displayed good chromatographic peak shape.
The evidence for several other phosphopeptides from this same run are shown in Figure 5. The MS/MS spectra confirm the phosphorylation sites of the sequences SEPIphosSPPRDR (Ser-408) and GCDphosSPDPDTSYVLTPHTEEK (Ser-98). Both show evidence of phosphorylation at a MAP kinase consensus site (serine or threonine followed by a proline).
FIGURE 5.
The MS/MS spectra that identified several phosphopeptides from MEF2A. A–D: Sequence and phosphorylation site for each peptide.
Evidence for phosphorylation of the peptide GMMPPLphosSEEEELELNTQR (Ser-289) is shown in Figure 5C. This peptide is of particular interest because the phosphorylated residue (Ser-289) does not match the consensus for a MAP kinase site. Interestingly, it appears to be a strong candidate for a protein kinase CK2 site (serine or threonine followed by acidic residues). Further studies are needed to confirm if there is a link between p38 MAP kinase activation of MEF2A and CK2 phosphorylation of this site.
It has previously been shown that in vitro phosphorylation of Ser-453 (QEMGRphosSPVDSLSSSSSSY DGSDR) by p38 MAP kinase is possible.3 However, no endogenous function has been assigned to phosphorylation of this residue. This study conclusively demonstrates that Ser-453 is also phosphorylated in vivo, and likely has a role in the regulation of MEF2A activity (Fig. 5D).
It should be noted that previous studies have identified Thr-312 and Thr-319 as p38 MAP kinase-responsive sites in MEF2A. These sites are primarily responsible for an increased transcriptional activity of MEF2A. We were unable to identify this phosphopeptide by any mass spectrometric approach. Most likely this is as a result of both of these amino acid residues residing on a large tryptic peptide with a mass of 11,428.76 amu.
Compared with the traditional methods of phosphopeptide discovery (neutral loss and precursor ion scans), targeted MRM-IDA was successful in identifying more phosphopeptides from α-casein and from MEF2A. It is also significant to note that targeted MRM-IDA had fewer false positives, where dependent MS/MS scans were triggered for nonphosphorylated peptides (Table 1). This can be extremely important when attempting to identify a potential phosphopeptide from a complex mixture of nonphosphopeptides.
TABLE 1.
Phosphopeptides Identified by Targeted MRM-IDA
| α-Casein | |||
| Peptide | MH+ | Neg Prec 79 | Target MRM |
| DIGSESTEDQAMEDIK | 1846.71 | — | ✓ |
| DIGSESTEDQAMEDIK | 1926.00 | — | ✓ |
| YKVPQLEIVPNSAEER | 1950.95 | ✓ | ✓ |
| VPQLEIVPNSAEER | 1659.79 | ✓ | ✓ |
| TVDMESTEVFTK Oxidation (M) | 1481.60 | ✓ | ✓ |
| TVDMESTEVFTK | 1465.60 | ✓ | ✓ |
| KTVDMESTEVFTK | 1593.60 | ✓ | ✓ |
| KTVDMESTEVFTK Oxidation (M) | 1609.00 | ✓ | — |
| EQLSTSEENSK | 1330.50 | ✓ | ✓ |
| NMAINPSKENLCSTFCK Oxidation (M) | 2108.87 | — | ✓ |
| NMAINPSKENLCSTFCK | 2092.00 | ✓ | ✓ |
| 8 | 10 | ||
| MEF2A | |||
| Peptide | MH+ | Neutral Loss 49 | Target MRM |
| Nonphosphopeptides (false positives) | 16 | 3 | |
| SEPISPPR | 962.43 | — | ✓ |
| SEPSPPRDR | 1233.56 | ✓ | ✓ |
| VMPTKSPPPPGGGNLGMNSR | 2073.95 | ✓ | ✓ |
| GMMPPLSEEEELELNTQR | 2182.93 | ✓ | ✓ |
| GCDSPDPDTSYVLTPHTEEK | 2327.93 | ✓ | ✓ |
| QEMGRSPVDSLSSSSSSYDGSDR | 2514.00 | — | ✓ |
| Identified phosphopeptides | 4 | 6 | |
Targeted MRM-IDA method was able to identify more phosphopeptides than methods using neutral loss or precursor ion scans. This method was also more specific for phosphopeptide identification, in that it rarely triggered an MS/MS spectra for nonphosphorylated peptides. The phosphorylation site is indicated by an underscore.
CONCLUSIONS
By combining the triple quadrupole capability of an MRM scan with the MS/MS scan of a linear ion trap, it is possible to identify low femtomole levels of phosphopeptides. Biological knowledge, such as primary sequence or upstream kinases, can be used to design targeted MRM-IDA experiments for a specific protein of interest. In this case, we were able to identify several phosphopeptides from α-casein and from the mammalian transcription factor, MEF2A. There exists the potential for using this method to quantify target peptides, as MRM areas are linear over several orders of magnitude. This method of targeted MRM-IDA is a powerful tool for the identification and quantification of posttranslational modifications in proteins.
Acknowledgments
The authors would like to acknowledge Yves LeBlanc and Jane Zhao for insightful discussions and comments regarding linear ion trap technology. Research supported in part by grants from the Canadian Institutes of Health Research (CIHR), and the Natural Sciences and Engineering Research Council (NSERC) of Canada to J. C. McDermott.
REFERENCES
- 1.Mann M, Ong, S.E, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: Deciphering the phosphoproteome. Trends Biotechnol 2002;20:261–268. [DOI] [PubMed] [Google Scholar]
- 2.Naya FS, Olson E. MEF2: A transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol 1999;11: 683–688. [DOI] [PubMed] [Google Scholar]
- 3.Zhao M, New L, Kravchenko VV, et al. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol 1999;19:21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox DM, Du M, Marback M, et al. Phosphorylation motifs regulating the stability and function of myocyte enhanced factor 2A. J Biol Chem 2003;278: 15297–15303. [DOI] [PubMed] [Google Scholar]
- 5.Bruins AP, Covey TR, Henion JD. Ion spray interface for combined liquid chromatography/atmospheric pressure ionization mass spectrometry. Anal Chem 1987; 59:2642–2646. [Google Scholar]
- 6.Biesenthal T, Gamble T, Pace N, Impey G. Impurity and degradation product analysis: High sensitivity quantitative and qualitative analysis using a hybrid triple-quadrupole-linear ion trap mass spectrometer. 51st American Society for Mass Spectrometry Conference on Mass Spectrometry. Montreal, QC, Canada
- 7.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 1999;17:1030–1032. [DOI] [PubMed] [Google Scholar]
- 8.Cox DM, Du M, Guo X, Siu KW, McDermott JC. Tandem affinity purification of protein complexes from mammalian cells. Biotechniques 2002;33:267–270. [DOI] [PubMed] [Google Scholar]
- 9.Hager JW. A new linear ion trap mass spectrometer. Rapid Comm Mass Spec 2002;16:512–526. [Google Scholar]
- 10.Hager JW, LeBlanc JCY. Product ion scanning using a Q-q-Qlinear ion trap (Q TRAP™) mass spectrometer. Rapid Commun Mass Spectrom 2003;17:1056–1064. [DOI] [PubMed] [Google Scholar]
- 11.Mansori BA, Dyer EW, Lock CM, Bateman K, Thomson BA, Boyd RK. Analytical performance of a high pressure radio frequency-only quadrupole collision cell with an axial field applied by using conical rods. J Am Soc Mass Spectrom 1998;9:775–788. [Google Scholar]
- 12.Beaudry F, LeBlanc JCY, Coutu M, Brown NK. In vivo pharmacokinetic screening in cassette dosing experiments: The use of on-line prospekt® liquid chromatography/atmospheric pressure chemical ionization tandem mass spectrometry technology in drug discovery. Rapid Comm Mass Spec 1998;12:1216–1222. [DOI] [PubMed] [Google Scholar]
- 13.Shou W, Verma R, Annan RS, et al. Mapping phosphorylation sites in proteins by mass spectrometry. Methods Enzymol 2002;351:279–296. [DOI] [PubMed] [Google Scholar]
- 14.Lill J. Proteomic tools for quantitation by mass spectrometry. Mass Spectrom Rev 2003;22:182–194. [DOI] [PubMed] [Google Scholar]
- 15.Molkentin JD, Li L, Olson EN. Phosphorylation of the MADS-Box transcription factor MEF2C enhances its DNA binding activity. J Biol Chem 1996; 271: 17199–17204. [DOI] [PubMed] [Google Scholar]