Abstract
Glycosylation is one of the most important posttranslational modifications affecting the functions of proteins and cell activities. Mass spectrometry (MS) has proven to be an effective tool for structural glycobiology and has helped gain an understanding of glycoprotein-mediated diseases. Although electro-spray ionization-tandem MS remains widely recognized as an effective means for oligosaccharide characterization, the hydrophilic nature of glycans has often caused the poor ionization efficiency requiring either derivatization or nanoelectrospray to improve detection sensitivity. In this report we describe the use of a chip-based infusion nanoelectrospray platform coupled with the hybrid triple quadrupole/linear ion trap for identification and characterization of glycosylation in complex mixtures. The high-mannose-type N-glycosylation in ribonuclease B was used to map the glycosylation site and obtain glycan structures. Using the chip-based nanoelectro-spray with precursor ion scanning linear ion trap MS, we were able to map the glycosylation site and obtain the glycan structures in ribonuclease B at 100 fmol/μL in a single analysis. In addition, a new, low-abundant glycoform with an additional hexose (Hex10GlcNAc2) attached to ribonuclease B was discovered. The results reported here demonstrate that the chip-based infusion nanoelectrospray ionization coupled to a quadrupole/linear ion trap platform is a valuable system, as it provides high sensitivity and stability for nanoelectrospray analysis, and allows extended acquisition time for completing precursor ion scanning and subsequent MS2 and MS3 information in a single analysis.
Keywords: Chip-based nanoelectrospray, ribonuclease B, glycosylation, precursor ion scanning, quadrupole linear ion trap, mass spectrometry
Glycosylation is one of the most common cotranslational and posttranslational modifications. It is estimated that 50% of all proteins are glycosylated.1 Glycosylation of proteins plays a vital role in biological processes such as molecular recognition and intracell and intercell signaling. Glycans can influence protein function by affecting its folding, solubility, stability, and recognition of binding partners. Alterations in protein glycosylation have often been associated with various diseases.2–4 Consequently, understanding the detailed structure of glycoproteins would provide insight to aid in biomedical research and drug discovery. Typically, oligosaccharides can be linked to proteins via oxygen atoms of serine or threonine side chains (O-glycosylation) or to the nitrogen atom of asparagine side chain (N-glycosylation).
N-Linked glycosylation is well documented in the literature and the structures of N-linked glycans have been studied extensively.5,6 Glycosylation of the asparagine residue appears in the consensus sequence Asn-Xxx-Ser/Thr and occurs in the endoplasmic reticulum and Golgi apparatus of the cell and involves a complex series of reactions. The large core oligosaccharides are added to proteins in the endoplasmic reticulum and further trimmed by glycosidases and mannosidases. The spectrum of glycoforms remains rather uniform until the glycoproteins reach the medial stacks of the Golgi apparatus, where structural diversification is introduced by a series of nonuniform modifications. Generally, all N-linked oligosaccharides can be divided into three classes; the high-mannose type, the complex type, and the less common hybrid type. The type of sugar attached to a protein will depend on the cell in which the protein is expressed and on the physiological status of the cell.7,8 O-linked glycosylation is initiated in the Golgi by the addition of a single sugar, typically N-acetylgalactosamine in mammals, to serine or threonine. O-oligosaccharides vary in size from a single N-acetylgalactosamine residue to oligosaccharides comparable in size to N-linked glycans. O-linked glycans are divided into domains, namely the core and the antenna. Since the antenna of O- and N-linked glycans is synthesized in a similar manner, they often carry the same terminal structures. There are at least seven O-oligosaccharide core structures, four of which (core types 1, 2, 3, and 4) are particularly widespread in mammals.9
Structural elucidation of glycans is historically difficult as they are typically highly heterogeneous, and chemically complex with branching, different configuration, and isoforms. Unlike other biopolymers, glycans are structurally branched in a nonlinear nature and with a variety of types of intersaccharide linkages. Generally, a comprehensive analysis of a glycoprotein consists of identification of glycosylated peptides; identification of the location of the glycosylation sites; and elucidation of the glycan structure where the constituent of monosaccharides and their sequence and branching pattern have to be determined.10
Mass spectrometry (MS) has been an important tool for the characterization of glycosylation including fast atom bombardment, matrix-assisted laser desorption ionization, and electrospray ionization using a wide variety of mass analyzers.9 Generally, the intact glycoprotein is isolated and the glycan structure characterized either after its release from the protein or while still linked to the protein. Many strategies have been developed to analyze the structure of the glycans including derivatization techniques of the reducing ends and protection of the functional groups.11,12 The development of separation techniques for glycan and glycopeptides has also been an important part of structural glycobiology. These include the use of porous graphitic carbon columns for the separation of oligosaccharides,13 capillary electrophoresis, and reversed-phase high-performance liquid chromatography.14–16
Electrospray ionization-tandem mass spectrometry remains widely recognized as an effective means for oligosaccharide characterization, particularly with the introduction of nanoelectrospray technology for improving ionization efficiency to overcome the challenge from the hydrophilic nature of sugar compounds.17,18 Nanoelectrospray ionization also offers low sample consumption due to its low flow rate which provides extended analysis time for completing multistage fragmentation analyses on oligosaccharides.18 The ion trap mass analyzer, with its capabilities of performing multiple stages of fragmentation, is one of the most useful systems in carbohydrate structure elucidation.19–21 Recently, precursor ion scanning for selectively detecting glycan-derived fragment ions has been widely used for glycan analysis.22–24 As further MS/MS studies of the identified glycopeptides are performed, the precursor ion scanning approach will be particularly useful in the analysis of samples containing unseparated peptides and glycopeptides, minimizing sample handling and loss.
In this article, we describe the use of a chip-based infusion nanoelectrospray ionization platform in combination with precursor ion scanning quadrupole linear ion trap MS for identification and characterization of glycosylation in complex mixtures without prior separation or modification of the samples. We employed a fully automated nanoelectrospray system (NanoMate 100, Advion BioSciences, Inc.) developed and recently characterized.25,26 This robotic infusion system serves as an automated liquid handling device and nanoelectrospray ionization ion source. The system removes the manual, tedious alignment processes of conventional nanoelectrospray ionization while offering a one-time spray optimization as well as enhanced spray stability and reproducibility.27,28 We report the benefits realized by using precursor ion scanning with MS3 capability of the hybrid triple quadrapole linear ion trap with extended analysis times to enable the detailed examination and mapping of glycosylation sites and glycan structures, and the discovery of an additional hexose site for N-linked high-mannose-type structure of ribonuclease B (RNase B).
MATERIALS AND METHODS
Bovine RNase B was purchased from Sigma (St. Louis, MO). Modified trypsin was from Promega (Madison, WI). All other chemical reagents, unless otherwise noted, were obtained from Aldrich (Milwaukee, WI).
Reduction, Alkylation, and Tryptic Digestion
One milligram of RNase B was reconstituted in 100 μL of solution containing 20 mM Tris-HCl pH 7.8, 6 M guanidine-HCl, and 10 mM dithiothreitol. The mixture was incubated for 30 min at 50°C. Fifty microliters of 0.2 M iodoacetamide and 50 μL of 0.2 M ammonium bicarbonate, pH 7.8, were then added to this solution. The mixture was incubated at room temperature in the dark for 2 h.
The alkylated solution was dialyzed against 20 mM ammonium bicarbonate, pH 7.8, overnight at 4°C using a Slide-A-Lyzer Mini Dialysis Unit from Pierce (Rockford, IL).
The dialyzed protein was digested by adding trypsin (0.52 mg/mL, Promega) at a 1:60 ratio (w/w) in 50 mM ammonium bicarbonate, pH 7.8, and incubated at 37°C overnight.
Infusion Nanoelectrospray and Mass Spectrometric Analysis
RNase B digest samples were made at a concentration of 1 pmol/ μL and 100 fmol/μL in 50% methanol with 0.1% formic acid prior to MS analysis. The NanoMate 100 (Advion BioSciences)26,27 was coupled to the front of the ABI/MDS Sciex 4000 Q Trap via a mounting bracket and 5 μL samples were analyzed via fully automated chip-based nanoelectrospray using spray voltages and sample delivery pressures of 1.55–1.75 kV and 0.2–0.4 psi, respectively, for positive ion mode. The estimated flow rate is approximately 200 nL/min.
The sample was analyzed in the tune mode of Analyst 1.4 software using an ABI/MDS Sciex 4000 Q Trap mass spectrometer. Nitrogen was used as the curtain (value of 10) and collision gas (set to high). Heated nanoelectrospray ionization source temperature was set to 40°C. The declustering potential was set at 20–40 V to minimize in-source fragmentation. The 4000 Q Trap was operated in positive LIT-mode at enhanced MS for survey scan (m/z 400–2000), for precursor ion scan (m/z 400–1700), for enhanced product ion scan, and MS3 scan modes (m/z 100–2000) for each sample. The scan speed was set to 1000 Da/sec for each scan mode and at least 2–5 min of data (over 100 scans) were summed for each spectrum. The collision energy was 28 eV in product ion scan and 50 eV for MS3 scan with 100 msec excitation time. The trap fill-time was 5 msec in the enhanced MS mode, 20 msec in both product ion and MS3 scan modes or with Dynamic Fill Time on. Precursor ion scans at m/z 163.06, 204.08, and 678.37 were acquired at a step size of 0.2 Da across a mass range of m/z 400–1700.
RESULTS AND DISCUSSION
Identification of Glycopeptides in RNase B Digest by Precursor Ion Scanning
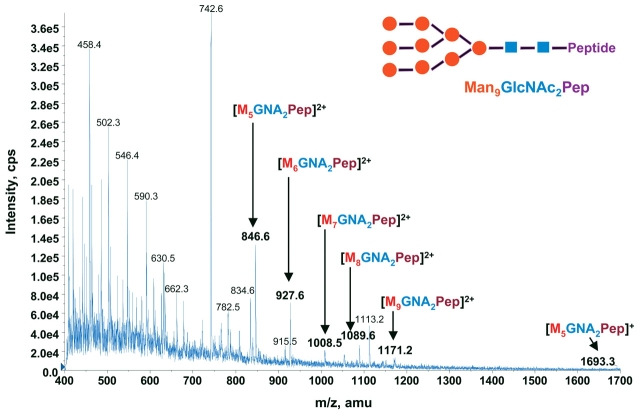
The tryptic digests of bovine pancreatic RNase B at 100 fmol/μL were analyzed by chip-based infusion nanoelectrospray mass spectrometry as described in Materials and Methods. The survey enhanced MS scan mass spectrum (Figure 1) shows the complex peptide mass fingerprint of RNase B. The total ion current (TIC) of 100 fmol/μL RNase B digest acquired for 3 min by chip-based nanoESI/4000 Q Trap indicates a sustained, constant, and efficiency ionization as demonstrated by the high intensity of ion current (data not shown). By summing across the entire recorded TIC, a spectrum of high signal/noise ratio was obtained (Fig. 1). The spectrum includes a pattern (81 m/z difference for the doubly charged species) of potential glyco-isoform ions that previously were not identified by data-dependent MS/MS analysis, although the fragmentation pattern of those ions appeared to be of high quality.21 To verify that those unidentified ions are glycopeptides, we used the hybrid triple quadrupole/LIT feature of the 4000 Q Trap for performing precursor ion scan to selectively identify glycan-derived fragment ions in the RNase B digest mixture. The characteristic fragment ions used for this experiment are the marker oxonium ions of hexose at m/z 163 and of N-acetylhexoamine (HexNAc) at m/z 204, and the larger oxonium ion (Y1+ at m/z 678.37) as reported recently.22 The larger oxonium ion used as a reporter fragment ion for precursor ion scanning in glycosylation studies is based on several observations:
FIGURE 1.
Survey scan (enhanced MS) mass spectrum (m/z 400–1700) of bovine RNase B tryptic digest at a concentration of 100 fmol/μL as described in Materials and Methods.
glycans are extremely labile and the collision energy required to induce the cleavage of the glycosidic bonds is usually far below the energy used for fragmentation of the peptide backbone;
under optimum collision energy conditions, glycopeptides are fragmented predominately within the carbohydrate moiety generating Y and B fragment ions;29 and
fairly high collision energy is required for cleavage of the linkage between the reducing terminal Glc-NAc and peptide (Y1).
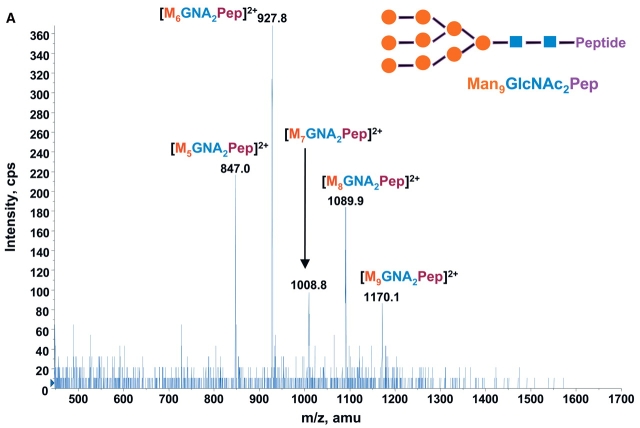
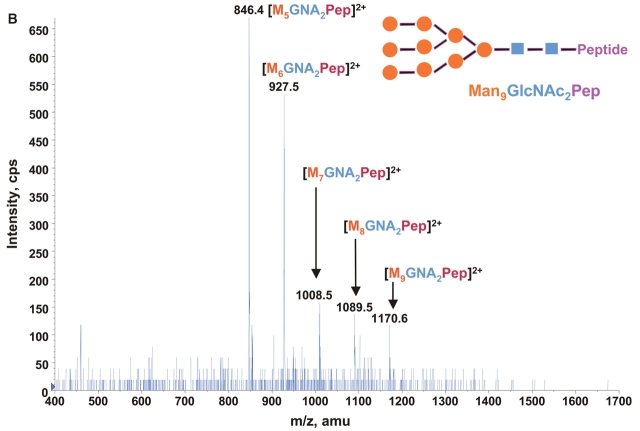
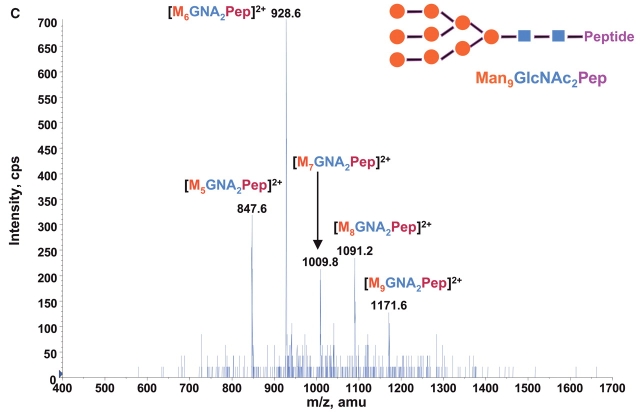
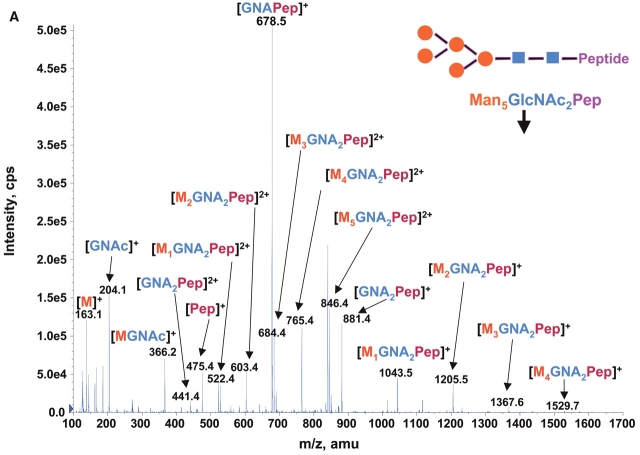
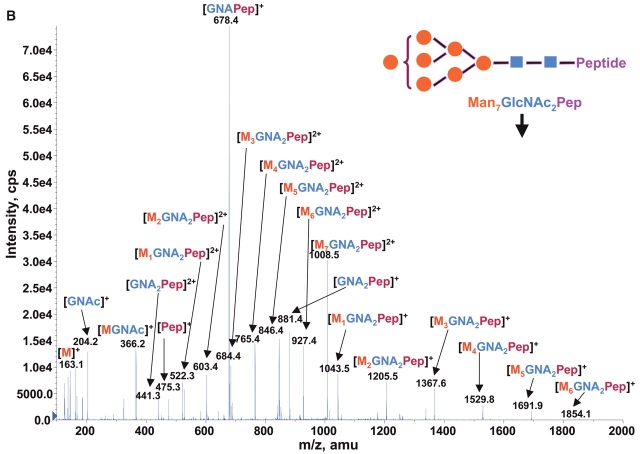
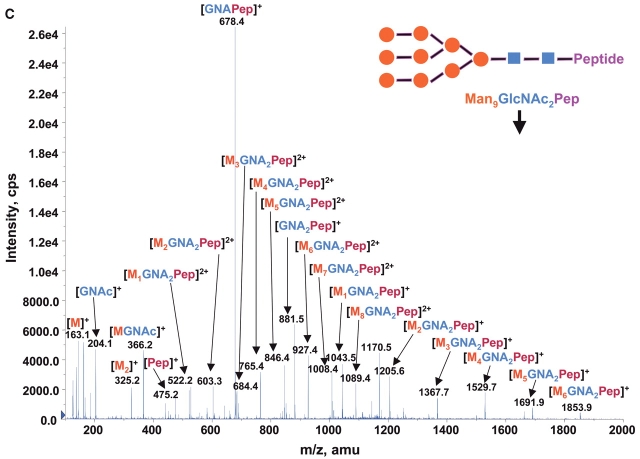
Consequently at the normal condition for glycopeptide fragmentation, Y1 ion is often becoming one of the most abundant Y type product ions.22 As glycans are heterogeneous and Y1 ions share the same mass and core peptide for all related glycopetide isoforms, the precursor ion scan for glycopeptide ions that fragment to yield Y1 ion can be used not only for potentially increasing the detection sensitivity but more importantly for improving the detection selectivity for glycopeptides from a common glycosylation site. Combining the precursor ion scan on the sugar oxonium ions (at m/z 163 and m/z 204), which is used to identify all the glycopeptides and free glycans in the sample, one can find multiple glycosylation sites in the complex samples. Two precursor ion spectra of m/z 163 and m/z 204 for 100 fmol/μL RNase B digest are shown in Figures 2A and B. As expected, the two precursor ion spectra show the same glycosylation pattern with five high-intensity ions having mass difference of 81 Da. This pattern found in precursor ion scan is consistent with the finding in the enhanced MS spectrum of the same sample (Fig. 1), indicating that the RNase B digest contains a typical high-mannose-type glycopeptides. A precursor ion spectrum of Y1 ion was also performed for the same digest at m/z 678.37 which was determined from the mass of the only possible N-linked glycosylation motif, a tetra-peptide in RNase tryptic digest (474.3 Da) plus the mass of the protonated reducing terminal GlcNAc (204 Da). The spectrum (Fig. 2C) also shows the same heterogeneous glycosylation pattern ions. Compared with the two previous precursor ion spectra (Figs. 2A and B), the Y1 ion spectrum yields relatively higher intensity ion signals for the most detected glycosylation ions. Given that the precursor ion spectrum of the Y1 ion compared with those from the two sugar oxonium reporter ions were identical, this confirms that there is only a single glycosylation site in the RNase B protein.
FIGURE 2.
Precursor ion scan mass spectra (m/z 400–1700) of bovine RNase B tryptic digest at a concentration of 100 fmol/μL (A) for oxonium ions of hexose at m/z 163, (B) for oxonium ions of N-acetylhexoamine (HexNAc) at m/z 204, and (C) for the larger oxonium ion (Y1+, reducing terminal HexNAc plus core peptide at m/z 678.4).
Characterizing the Glycopeptide Isoforms, Sequence, and Glycosylation Site
All ions of significant abundance in the precursor ion experiment were sequentially selected for product ion experiments to further gain the structure information of the glycopeptide isoforms. Collision-induced fragmentation of the five doubly charged ions, differing by 81 m/z, confirms the presence of a high-mannose-type glycosylation. Three selected examples of product ion spectra for the doubly charged precursors at m/z 846.4, 1008.4, and 1170.4 are shown in Figures 3A–C, respectively. All spectra show predominately high m/z Y-type fragment ions as well as few low m/z B ions indicative of fragmentation within the carbohydrate. Those visible low m/z fragment ions, such as m/z 163, 204, 325, and 366, not only provide the evidence indicating two different types of sugar moiety (hexose and HexNAc) in the selected glycopeptide, but also demonstrate that the 4000 Q Trap differs from conventional 3D and linear ion traps by completely eliminating the low mass cut-off. Although there is a sugar-free peptide ion at m/z 475 being detected in all MS/MS spectra, the intensity of the ion is relatively low and does not appear to be fragmented at all (Fig. 3). In contrast, the Y1+ ion, at m/z 678.4 is always the most abundant Y-type ion in all product ion spectra, consistent with the finding that precursor ion scans of Y1 ion are the most sensitive precursor ion scan and confirming the identity in the core peptide of the glycopeptides. As shown in Figure 3, the singly and doubly charged Y-type fragment ions also suggest that the sequence of the N-linked core pentasaccharide exists in all glycopeptide isoforms.
FIGURE 3.
Enhanced product ion mass spectra (m/z 100–2000) derived by collision-induced dissociation of the (M + 2H)2+glycopeptide precursor ions of the RNase B tryptic digest at a concentration of 1 pmol/μL, (A) m/z = 846.4, (B) m/z = 1008.4, and (C) m/z = 1170.4.
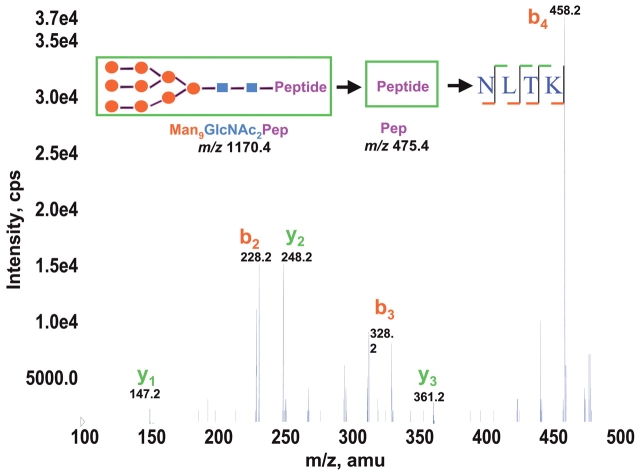
As described above, product ion scans of detected glycopeptides show high-quality spectra and result in useful information for the characterization of the glycosylation. However, since glycopeptides tend to fragment only at their oligosaccharide structure in tandem mass spectra at relatively low collision energy (28 eV in this work), it is often difficult to obtain amino acid sequence information simultaneously. In order to elucidate peptide sequence information for the glycopeptide isoforms, additional MS3 analysis selecting core peptide ions based on MS/MS product ion was performed with higher collision energy introduced (50 eV in this work). A selected MS3 spectrum of m/z 475 ion from MS/MS of m/z 1170.4 ion is shown in Figure 4. The MS3 spectrum file was searched using Mascot to determine the amino acid sequence. The search result shows that the top hit sequence is NLTK, which matches the amino acid sequence of the RNase B and fits the NXS/T sequence motif for N-linked glycosylation. Therefore, the asparagine 34 residue in RNase B is identified as the glycosylation site. The molecular weight of the pattern ions identified in the precursor ion scan and subsequent fragmentation information together with the molecular weight of the amino acid sequence confirms the presence of five different high- mannose-type glycopeptides in RNase B as Man5–9GlcNAc2-NLTK.
FIGURE 4.
MS3 mass spectrum derived by collision-induced fragmentation of (M + 2H)2+ at m/z 1170.4 → m/z 475.4 for the identification of the amino acid sequence of the RNase B glycopeptide. “→ “ Indicates that the MS/MS product ion at m/z 475.4 from the first precursor ion at m/z 1170.4 is used as the second precursor ion for MS3 analysis.
Identification of a Trace Amount of a New Glycoform in RNase B Protein
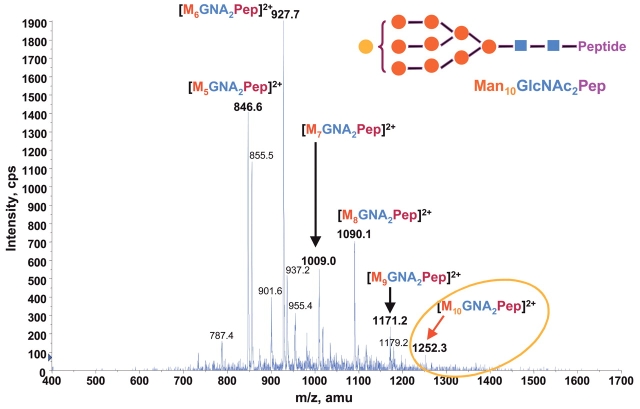
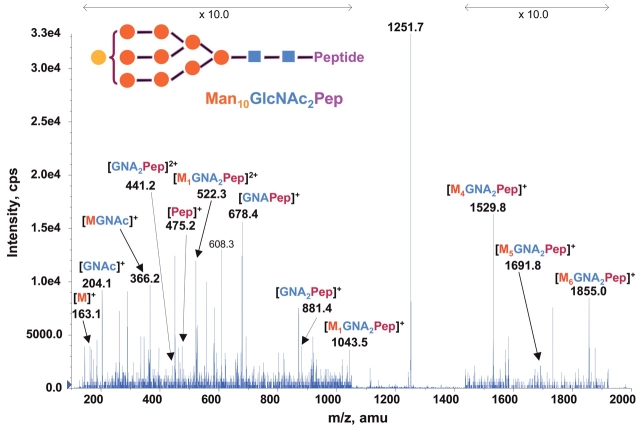
Through methylation analysis, sequential exoglycosidase digestion followed by nuclear magnetic resonance and liquid chromatography–tandem mass spectrometry analyses, the structures of RNase B oligosaccharides have previously been shown to be N-linked high-mannose type. It contains two GlcNAc residues and five to nine mannose (Man5–9GlcNAc2) whose branch pattern has been determined.15,30n The relative molar proportions of five glycoform pools from Man5 to Man9 were determined to be 57, 31, 4, 7, and 1%, respectively.30 During our chip-based nanoelectrospray ionization precursor ion scanning analysis on Y1+ ion, at m/z 678.4 for RNase B digestion with increased concentration at 1 pmol/μL, we unexpectedly found an additional glycoform ion at m/z 1252.3. As shown in Figure 5, the precursor ion scanning spectrum reveals six glycoform ions with mass differences of 81 Da. Although the intensity of the new potential glycoform ion at m/z 1252.3 is relatively low, it is still observable with a signal/noise ratio above 4. The survey scan spectrum on the same sample showed the doubly charged ion at m/z 1251.7 which is exactly 162 Da more than that of the identified Man9GlcNAc2 glycoform, suggesting that a trace amount of a glycoform containing the tenth hexose residue may exist. The subsequent product ion scan on the doubly charged ion at m/z 1251.7 provides additional convincing evidence for this finding. Figure 6 shows the product ion spectrum of m/z 1251.7 containing many high m/z Y- type fragment ions and low m/z B ions as found for other glycoform ions (Fig. 3). The results confirm that there is a trace amount of an additional Hex10GlcNAc2 glycoform in RNase B protein. It should be noted that the fragmentation of the m/z 1251.7 ion is not nearly as good as the other glycoform ions at collision energy of 28 eV (Figs. 3 and 6). The reason for this is not known. Increasing the energy to 70 eV did slightly improve fragmentation (data not shown). We also tried an MS3 analysis of the core peptide at m/z 475 from the MS/MS product ion scan of the new glycoform ion (m/z 1251.7). No meaningful MS3 spectrum was obtained, as apparently the intensity of the selected m/z 475 ion is too low to be detected. Further treatment with mannosidase for the RNase B digest followed by MS/MS analysis will determine whether the new hexose residue is mannose. Since RNase B oligosaccharides are proven to be N-linked high-man-nose type, it is highly likely that the RNase B has a trace amount of Man10GlcNAc2 glycoform in addition to the five previously reported Man5–9 GlcNAc2 glycoforms.
FIGURE 5.
Precursor ion scan mass spectrum (m/z 400–1700) of bovine RNase B tryptic digest at a concentration of 1 pmol/μL for the larger oxonium Y1+ ion at m/z 678.4. The orange circle denotes a new glycopeptide isoform containing Hex10GlcNAc2 oligosaccharide detected.
FIGURE 6.
Enhanced product ion mass spectrum (m/z 100–2000) derived by collision-induced dissociation of the (M + 2H)2+ at m/z = 1251.7 precursor ion of the RNase B tryptic digest at a concentration of 1 pmol/μL.
The Benefits of Chip-Based Infusion NanoESI/4000 Q Trap for Glycosylation Studies
In glycosylation studies, identification and determination of glycosylation site, peptide sequence, and carbohydrate structure are essential requiring multiple approaches with different techniques. This includes peptide separation, mass spectrometry analysis of individual glycoform ions, and various steps of exoglycosidase digestion or chemical modification followed by repeat analyses of the treated samples. Therefore, the analysis procedures can be both time and sample consuming. Chip-based infusion nanoelectrospray ionization analysis has been reported for improved ionization efficiency and low flow rates providing extended analysis time for completing tandem MS (MSn) analyses on oligosaccharides.21,32 The system provides long-lasting, stable nanoESI signal, allowing us to optimize various MS scan conditions for highly sensitive detection and sequencing of minor glycopeptide components in complex samples. In this work, we took advantage of the system’s ability to extend analysis time by acquiring data for more than 40 min. During this time we sequentially acquired precursor ion scans on three different ions, MS/MS product ion scans of five precursor ions and MS3 scans of two ions in a single analysis and consumed only 6 μL of samples. Using the chip-based system coupled with 4000 Q Trap mass spectrometer, we were able to characterize the glycopeptide isoforms of RNase B digest at 100 fmol/μL. The high-sensitivity performance provided by the system also allows us to identify a trace amount of a new glycopeptide isoform of RNase B digest without LC separation of the samples.
CONCLUSION
In this paper we report using chip-based nanoelectro-spray ionization coupled to a quadrupole linear ion trap mass spectrometer operated in precursor ion scanning mode for rapid and sensitive glycopeptide detection and characterization of bovine RNase B. The automated chip-based nanoelectrospray ionization platform coupled to the 4000 Q Trap offers a powerful analytical combination providing extended analysis time with limited sample consumption for highly sensitive precursor ion scanning and MS/MS and MS3 product ion scans in a single analysis. This approach requires minimum sample handing for direct analysis of unseparated glycopeptide mixtures. A precursor ion scanning approach for the glycopeptide-based Y1 ion is evaluated and used to obtain the glycosylation site profiles for a complex glycoprotein digest. The combined triple quadrupole and linear ion trap scan features offered by the 4000 Q Trap are demonstrated to be a valuable system for glycosylation studies.
Acknowledgments
We would like to thank Larry Klecha for the initial installation of the NanoMate for the 4000 Q Trap. We also thank Professors Jack Henion, and Kelvin Lee and Dr. Colleen Van Pelt for reviewing this manuscript and for providing helpful comments.
REFERENCES
- 1.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1999;1473:4–8. [DOI] [PubMed] [Google Scholar]
- 2.Maguire TM, Thakore J, Dinan TG, Hopwood S, Breen KC. Plasma sialyltransferase levels in psychiatric disorders as a possible indicator of HPA axis function. Biol Psychiatry 1997;41:1131–1136. [DOI] [PubMed] [Google Scholar]
- 3.Dwek RA. Glycobiology: More functions for oligosaccharides. Science 1995;269:1234–1235. [DOI] [PubMed] [Google Scholar]
- 4.Turner GA. Haptoglobin. A potential reporter molecule for glycosylation changes in disease. Adv Exp Med Biol 1995;376:231–238. [PubMed] [Google Scholar]
- 5.Dwek RA. Glycobiology: Toward Understanding the Function of Sugars. Chem Rev 1996;96:683–720. [DOI] [PubMed] [Google Scholar]
- 6.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science 2001;291:2364–2369. [DOI] [PubMed] [Google Scholar]
- 7.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem 1985;54:631–664. [DOI] [PubMed] [Google Scholar]
- 8.Paulson JC. Glycoproteins: What are the sugar chains for? Trends Biochem Sci 1989;14:272–276. [DOI] [PubMed] [Google Scholar]
- 9.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science 2001;291:2351–2356. [DOI] [PubMed] [Google Scholar]
- 10.Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom Rev 2004;23:161–227. [DOI] [PubMed] [Google Scholar]
- 11.Di Jeso B, Liguoro D, Ferranti P, et al. Modulation of the carbohydrate moiety of thyroglobulin by thyrotropin and calcium in Fisher rat thyroid line-5 cells. J Biol Chem 1992;267:1938–1944. [PubMed] [Google Scholar]
- 12.Okafo G, Burrow L, Carr SA, Roberts GD, Johnson W, Camilleri P. A coordinated high-performance liquid chromatographic, capillary electrophoretic, and mass spectrometric approach for the analysis of oligosaccharide mixtures derivatized with 2-aminoacridone. Anal Chem 1996;68:4424–4430. [DOI] [PubMed] [Google Scholar]
- 13.Chaimbault P, Petritis K, Elfakir C, Dreux M. Ion-pair chromatography on a porous graphitic carbon stationary phase for the analysis of twenty underivatized protein amino acids. J Chromatogr A 2000;870:245–254. [DOI] [PubMed] [Google Scholar]
- 14.Kelly JF, Locke SJ, Ramaley L, Thibault P. Development of electrophoretic conditions for the characterization of protein glycoforms by capillary electrophoresis-electrospray mass spectrometry. J Chromatogr A 1996; 720:409–427. [DOI] [PubMed] [Google Scholar]
- 15.Liu T, Li JD, Zeng R, Shao XX, Wang KY, Xia QC. Capillary electrophoresis-electrospray mass spectrometry for the characterization of high-mannose-type N-glycosylation and differential oxidation in glycoproteins by charge reversal and protease/glycosidase digestion. Anal Chem 2001;73:5875–5885. [DOI] [PubMed] [Google Scholar]
- 16.Chelius D, Wu SL, Bondarenko PV. Identification of N-linked oligosaccharides of rat insulin-like growth factor binding protein-4. Growth Horm IGF Res 2002;12:169–177. [DOI] [PubMed] [Google Scholar]
- 17.Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal Chem 1996;68:1–8. [DOI] [PubMed] [Google Scholar]
- 18.Bahr U, Pfenninger A, Karas M, Stahl B. High-sensitivity analysis of neutral underivatized oligosaccharides by nanoelectrospray mass spectrometry. Anal Chem 1997;69:4530–4535. [DOI] [PubMed] [Google Scholar]
- 19.Creaser CS, Reynolds JC, Harvey DJ. Structural analysis of oligosaccharides by atmospheric pressure matrix-assisted laser desorption/ionisation quadrupole ion trap mass spectrometry. Rapid Commun Mass Spectrom 2002; 16:176–184. [DOI] [PubMed] [Google Scholar]
- 20.Sheeley DM, Reinhold VN. Structural characterization of carbohydrate sequence, linkage, and branching in a quadrupole ion trap mass spectrometer: Neutral oligosaccharides and N-linked glycans. Anal Chem 1998;70:3053–3059. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Chelius D. Characterization of protein glycosylation using chip-based infusion nanoelectrospray linear ion trap tandem mass spectrometry. J Biomol Tech 2004;15:120–133. [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie MA, Gill AC, Deery MJ, Lilley K. Precursor ion scanning for detection and structural characterization of heterogeneous glycopeptide mixtures. J Am Soc Mass Spectrom 2002;13:1065–1077. [DOI] [PubMed] [Google Scholar]
- 23.Jebanathirajah J, Steen H, Roepstorff P. Using optimized collision energies and high resolution, high accuracy fragment ion selection to improve glycopeptide detection by precursor ion scanning. J Am Soc Mass Spectrom 2003;14:777–784. [DOI] [PubMed] [Google Scholar]
- 24.Sandra K, Devreese B, Van Beeumen J, Stals I, Claeyssens M. The Q-trap mass spectrometer, a novel tool in the study of protein glycosylation. J Am Soc Mass Spectrom 2004;15:413–423. [DOI] [PubMed] [Google Scholar]
- 25.Schultz GA, Corso TN, Prosser SJ, Zhang S. A fully integrated monolithic microchip electrospray device for mass spectrometry. Anal Chem 2000;72:4058–4063. [DOI] [PubMed] [Google Scholar]
- 26.Van Pelt CK, Zhang S, Henion JD. Characterization of a fully automated nanoelectrospray system with mass spectrometric detection for proteomic analyses. J Biomol Tech 2002;13:72–84. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S, Van Pelt CK. Chip-based nanoelectrospray mass spectrometry for protein characterization. Expert Rev Proteomics 2004;1:449–468. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Van Pelt CK, Henion JD. Automated chip-based nanoelectrospray-mass spectrometry for rapid identification of proteins separated by two-dimensional gel electrophoresis. Electrophoresis 2003;24:3620–3632. [DOI] [PubMed] [Google Scholar]
- 29.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB/MS spectra of glyco-conjugates. Glycoconjugates 1988;5:397–409. [Google Scholar]
- 30.Fu D, Chen L, O’Neill RA. A detailed structural characterization of ribonuclease B oligosaccharides by 1H NMR spectroscopy and mass spectrometry. Carbohydr Res 1994;261:173–186. [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki N, Ohta M, Hyuga S, Hashimoto O, Hayakawa T. Analysis of carbohydrate heterogeneity in a glycoprotein using liquid chromatography/mass spectrometry and liquid chromatography with tandem mass spectrometry. Anal Biochem 1999;269:297–303. [DOI] [PubMed] [Google Scholar]
- 32.Zamfir A, Vakhrushev S, Sterling A, Niebel HJ, Allen M, Peter-Katalinic J. Fully automated chip-based mass spectrometry for complex carbohydrate system analysis. Anal Chem 2004;76:2046–2054. [DOI] [PubMed] [Google Scholar]