Abstract
RNAi is a relatively new but powerful technology used for monitoring gene silencing in many species. The RNA corresponding to the gene of interest can be delivered into cells using various protocols, but the approach where both sense and antisense strands are located in the same transcript seems to be the most promising. However, the DNA sequencing step is a challenge due to the formation of a strong hairpin structure. In this work we present a very simple modification to the standard DNA sequencing protocol that allows sequencing through such difficult regions. The modification does not require any additional enzymatic or chemical manipulations. It simply adds a 5-min heat-denaturation step at 98°C, prior to addition of a Taq mix. The inclusion of this step is very effective in sequencing through many different categories of difficult templates. Other examples of the implementation of this step have been described previously and more detailed information on heat-denaturation of plasmid DNAs has been presented.
Keywords: Difficult templates, hairpins, repeats, siRNA, pDONR221
In recent years RNAi technology has emerged as a powerful tool to monitor gene silencing in a number of eukaryotic species including plants, fungi, insects, invertebrates, and mammalian systems.1–4 The sequence-specific degradation of mRNA is mediated through 21- and 22-nucleotide siRNAs cleaved by the ribonuclease III activity from longer dsRNAs.5
The chemically synthesized siRNA corresponding to a specific gene of interest can be transfected into cultured mammalian cells, resulting in gene silencing. The more elegant and efficient method to introduce siRNA into cells is to use specially engineered plasmids containing RNA polymerase III-specific promoters and terminators that direct the expression of short RNAs. Currently, two variants of siRNA-expressing vectors are used. In first variant, the sense and anti-sense transcripts that form a hairpin structure are independently produced within a host cell.6 In the second variant, both sense and anti-sense strands are separated by a short loop and expressed as a single transcript. This design results in the formation of a strong hairpin structure (19–29 bases long) that produces siRNA after intracellular processing.7,8 To achieve the desired gene silencing effect, the sequence of the siRNA and the targeted gene has to be strictly complementary.7,8 As genetic manipulations (and at the hairpin synthesis stage) can introduce point mutations, it is highly advisable to confirm the DNA sequence of the construct prior to carrying out planned experiments. The presence of a strong hairpin structure prevents getting clear data from such fragments using standard sequencing protocols. Recently, Ducat et. al.9 described a method, using restriction cleavage in the region close to a loop between the inverted repeats, that allows sequencing of plasmids with siRNA. Although effective, this method has at least two obvious limitations. First, the desired restriction site has to be present in the specific location (this may be introduced at the design stage). Second, an additional step (restriction digest) needs to be part of a sequencing protocol, which adds labor, cost and time to the entire process. This methodology may not be feasible when a large number of siRNA containing plasmids needs to be analyzed.
In this article we describe a very simple modification that allows sequencing through hairpin structures without the need for additional enzymatic steps. The other class of plasmid DNAs that contain inverted repeat structures potentially difficult to sequence is the Gateway cloning system (Invitrogen, Carlsbad, CA).10,11 Recently, Esposito et al.12 reported a method of sequencing DNA fragments cloned into pDONR223 Gateway vector. To obtain good quality data (especially for short inserts of less than 500 bases) they introduced a blocking oligonucleotide that prevents formation of a 15-base hairpin impeding clean read-through during sequencing. In this article, it is demonstrated that the same simple modification used to sequence plasmids with siRNAs allows sequencing of short and long DNA fragments cloned into pDONR221 (which is the same as pDONR223 except for a different resistance marker, kanamycin, rather than spectinomycin in pDONR223; Invitrogen).
MATERIALS AND METHODS
DNA sequencing was carried out as follows: An aliquot (0.1–0.25 μg) of plasmid DNA was combined with 1 μL of 5–10 μM primer and the volume was adjusted to 7 μL with 10 mM Tris/0.01 mM EDTA, pH 8.0. This mixture was heat-denatured for 5 min at 98°C in a thermocycler (PTC-225, MJ Research, Waltham, MA), placed on ice and 2 μL of the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit mix was added (v. 3.0, 3.1 or dGTP 3.0). The final dilution factor of all dye terminators was fourfold in 10 μL reaction volumes. All dye-terminator reagents were purchased from Applied Biosystems (Foster City, CA). The amplification reactions were performed in a PTC-225 thermocycler for 40 cycles (96°C for 10 sec, 50°C for 5 sec, and 60°C for 2 min). Following the cycling step, 20 μL of water was added to reactions and the excess dye was removed by gel filtration on a 96-well filter plate filled with Sephadex G-50 beads. The filter plate (cat. no. MAHV N45 50) was from Millipore (Bedford, MA) and Sephadex G50 was from Amersham Biosciences (Piscatawy, NJ). The samples were then heat-denatured for 2 min at 90–95°C and electrophoresed on an ABI3100 or ABI3700 genetic analyzer (Applied Biosystems) under conditions recommended by the manufacturer.
In addition, we evaluated the effect of a number of reagents on the read length. Each component from the set of Sequence Enhancer Rx (Invitrogen, Carlsbad, CA) and betaine (Sigma-Aldrich, St. Louis, MO) was separately included in the sequencing mix with or without a heat-denaturation step. Only reagent C had some effect (5.9% ± 5.5% increase in Q > 20 read length, n = 20). Sequences were viewed and edited using Sequencher 4.1.2 (Gene Codes, Ann Arbor, MI).
To evaluate the effectiveness of this sequencing approach, we tested 20 plasmid vectors (pENTR11, Invitrogen) containing different 29 bp hairpins, with 4 mismatches. The mismatches are located at different positions of stem part of hairpins. These hairpins contained from 8 to 20 G-C pairs and the GC content varied from 46% to 67%. The data for 7 representative 29-bp hairpins with greater than 13 G-C pairs are summarized in Table 1. The total number of G-C pairs, their distribution, and the position of mismatches as well as the overall GC content did not have any significant effect on the quality of the sequencing data and read lengths, expressed in terms of Q ≥ 20,13 providing that the heat-denaturation step is part of the protocol. For this set BigDye 3.0, 3.1 or dGTP 3.0 produced similar results. The plasmid DNA used for this part of work was prepared using Qiagen’s (Valencia, CA) EndoFree plasmid purification kit. Similar approach, heat-denaturation, and modified chemistry, can be used to effectively sequence through another class of hairpin structures, as indicated in Figure 1. In some cases the combination of BD3.1 and dGTP chemistries may be needed to resolve compression issues.
TABLE 1.
Sequencing Through Hairpin Structures
| BigDye‰ Terminator Mix (Q ≥ 20 Phred Scores) | |||||||||||
| BD V3.0 Treatment | BD dGTPV3.0 Treatment | BD V3.1 Treatment | |||||||||
| Hairpin Type (#) | No of GC Pairs | Overall GC Content (%) | No HD | +HD | +HD+C | No HD | +HD | +HD+C | No HD | +HD | +HD+C |
| 1 | 20 | 67 | 366 | 543 | 551 | 336 | 527 | 555 | 493 | 605 | 622 |
| 2 | 19 | 64 | 196 | 573 | 589 | 345 | 452 | 460 | 448 | 590 | 596 |
| 3 | 17 | 60 | 435 | 597 | 574 | 442 | 515 | 555 | 453 | 599 | 599 |
| 4 | 16 | 58 | 285 | 542 | 549 | 440 | 537 | 548 | NT | NT | NT |
| 5 | 15 | 55 | 288 | 512 | 605 | 333 | 520 | 551 | NT | NT | NT |
| 6 | 14 | 53 | 352 | 506 | 523 | 437 | 489 | 511 | NT | NT | NT |
| 7 | 13 | 50 | 471 | 600 | 607 | 361 | 488 | 554 | NT | NT | NT |
NT, not tested. No/+ HD, no heat-denaturation/ + 5 min heat denaturation at 98° C with reagent C prior to adding appropriate Taq mix.
Three different chemistries were evaluated for the effectiveness of reading through various hairpin structures. Only representative data for 29 bp hairpins are shown. The remaining 13 hairpins gave similar results. The BigDye 3.1 chemistry was tested only on three 29 bp hairpins, as it did not offer any advantages over BigDye 3.0 chemistry. No improvement was observed when reagent C was part of a standard sequencing protocol (not shown). All sequencing reactions for this set were run on an ABI3100 DNA sequencer equipped with a 50-cm capillary array.
The overall GC content refers to the hairpin part only.
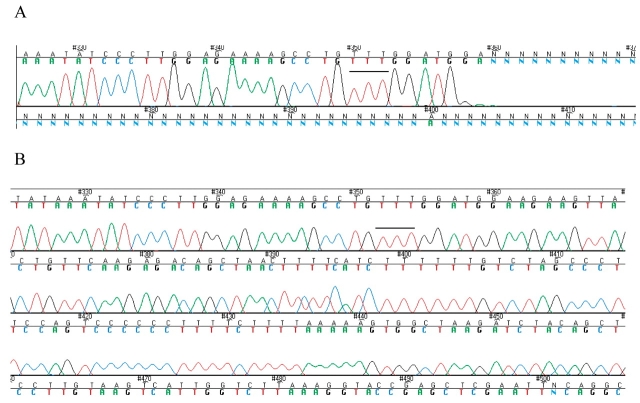
FIGURE 1.
Sequencing through a 19 bp perfect hairpin. A: Sequence trace obtained using BigDye V3.0 dye-terminator. B: Sequencing trace obtained with dGTP BigDye V3.0 in the presence of reagent C. The black horizontal line over three Ts indicates the same position in the sequence. The scale in both panels is different to show extended data.
Using exactly the same protocol we were able to sequence through a 630-bp insert in pDONR221 Gateway vector and in vector only. Following the standard molecular biology procedures14 these two plasmid DNAs were prepared using GeneElute plasmid purification kit (Sigma-Aldrich). The data for this part are presented in Table 2. Adding the heat-denaturation step extended the read length by about 200–600 bases with Q ≥ 20, depending on the direction of the primer relative to the hairpin. In pDONR221 only, sequencing with M13R or M13F primers with the standard protocol produced very short reads, confirming the data obtained by Esposito.12 The presence of other reagents (data for reagent C only is shown here) in the sequencing mix only slightly increased the read length. It seems that the 15-bp hairpin structure in this vector is not abolished under standard sequencing conditions, and that the prolonged heat denaturation prior to adding Taq mix is needed to obtain a clean read through.
TABLE 2.
Sequencing Through an Inverted Repeat in pDONR221 Vector
| BigDye‰ Terminator Mix Q ≥ 20 phred scores | ||||||||||
| BD V3.0 Treatment | dGTPV3.0 Treatment | BD V3.1 Treatment | ||||||||
| DNA Type | Primer Type | No HD | +HD | +HD+C | No HD | +HD | +HD+C | No HD | +HD | +HD+C |
| pDONR | M13F | 310 | 513 | 694 | 511 | 705 | 765 | 482 | 618 | 592 |
| 221 only | M13R | 81 | 703 | 734 | 584 | 805 | 771 | 261 | 681 | 684 |
| pDONR | M13F | 778 | 780 | 762 | 784 | 797 | 793 | 800 | 828 | 820 |
| 221+630 | M13R | 805 | 793 | 825 | 800 | 794 | 814 | 820 | 823 | 820 |
| bp insert | ||||||||||
Three different chemistries with forward (M13 F) and reverse (M13 R) primers were used to sequence through an inverted repeat in pDONR221 vector only and in the vector containing a 630 bp insert. Other abbreviations are as in the description to Table 1. All sequencing reactions for this set were run on an ABI 3700 DNA sequencer.
CONCLUSION
The controlled heat denaturation of all plasmid DNAs is part of a standard protocol in our sequencing laboratory, and since its introduction a few years ago the success rate and the trace quality has increased significantly.15,16 In our experience, this heat-induced plasmid denaturation is the single most effective step when sequencing many different types of difficult templates. The addition of other reagents and/or using alternative chemistries often improves the quality of sequencing data.16
Acknowledgments
I would like to thank Ms. Lora Haines for suggestions and critical reading of this manuscript and Ms. Katarzyna Bajson for skillful technical support. I would also like to thank Hemchand Sookdeo and Charlie Richard for continued encouragement and support throughout this work.
REFERENCES
- 1.Hammond SM, Caudy AA, Hannon GJ. Posttranscriptional gene silencing by double-stranded RNA. Nat Rev Genet 2001;2:110–119. [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. RNAi and double-stranded RNA. Genes & Dev 1999;13:139–141. [PubMed] [Google Scholar]
- 3.Hannon GJ. RNA interference. Nature 2002;418:244–251. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 2002;26:199–213. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001;409:363–366. [DOI] [PubMed] [Google Scholar]
- 6.Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol 2002;20:497–500. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002;296:550–553. [DOI] [PubMed] [Google Scholar]
- 8.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev 2002;16:948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducat DC, Herrera FJ, Triezenberg SJ. Overcoming obstacles in DNA sequencing of expression plasmids for short interfering RNAs. BioTechniques 2003;34:1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res 2000; 10:1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheo D, Brasch MA, Temple GF, Hartley JL, Byrd D, 2001. Use of multiple recombination sites with unique specificity in recombinational cloning. International patent application no. WO 01/42509 A1.
- 12.Esposito D, Gillette W, Hartley JL. Blocking oligonucleotides improve sequencing through inverted repeats. BioTechniques 2003;35:914–920. [DOI] [PubMed] [Google Scholar]
- 13.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998;8:175–185. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Russell DW. Molecular Cloning, 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2001.
- 15.Kieleczawa J. Controlled heat-denaturation of DNA plasmids. In Kieleczawa J (ed): DNA Sequencing: Optimizing the Process and Analysis. Sudbury, MA: Jones and Bartlett, 2005:1–10.
- 16.Kieleczawa J. Sequencing of difficult DNA templates. In Kieleczawa J. (ed): DNA Sequencing: Optimizing the Process and Analysis. Sudbury, MA: Jones and Bartlett, 2005:27–34.