Abstract
We developed the SNPlex Genotyping System to address the need for accurate genotyping data, high sample throughput, study design flexibility, and cost efficiency. The system uses oligonucleotide ligation/polymerase chain reaction and capillary electrophoresis to analyze bi-allelic single nucleotide polymorphism genotypes. It is well suited for single nucleotide polymorphism genotyping efforts in which throughput and cost efficiency are essential. The SNPlex Genotyping System offers a high degree of flexibility and scalability, allowing the selection of custom-defined sets of SNPs for medium- to high-throughput genotyping projects. It is therefore suitable for a broad range of study designs. In this article we describe the principle and applications of the SNPlex Genotyping System, as well as a set of single nucleotide polymorphism selection tools and validated assay resources that accelerate the assay design process. We developed the control pool, an oligonucleotide ligation probe set for training and quality-control purposes, which interrogates 48 SNPs simultaneously. We present performance data from this control pool obtained by testing genomic DNA samples from 44 individuals. in addition, we present data from a study that analyzed 521 SNPs in 92 individuals. Combined, both studies show the SNPlex Genotyping system to have a 99.32% overall call rate, 99.95% precision, and 99.84% concordance with genotypes analyzed by TaqMan probe–based assays. The SNPlex Genotyping System is an efficient and reliable tool for a broad range of genotyping applications, supported by applications for study design, data analysis, and data management.
Keywords: Single nucleotide polymorphism, genotyping, high throughput, pharmacogenomics
Differences between individual genomes provide a wealth of information regarding the elements responsible for phenotypic differences among individuals.1 Such differences, including single tandem repeats (STRs) and single nucleotide polymorphisms (SNPs), are important for studies investigating the genetic nature of complex diseases, drug responses, or quantitative traits, or for human identification.
SNPs are the most abundant markers in the human genome, and SNP genotyping, therefore, is a key technology for genome-wide analysis of genetic variation. Different genotyping applications require screening of different numbers of SNPs. The determination of a single SNP (or mutation) can be sufficient to screen for the presence of a Mendelian disease, such as cystic fibrosis.2 However, to evaluate whether mutations within a class of genes contribute to a disease, hundreds to thousands of SNPs must be studied in association studies. In addition to these quantitative aspects, a genotyping system must provide the tools required for efficient and flexible study designs, low sample consumption, reasonable assay duration, access to automation, and efficient data analysis and management, in addition to excellent assay performance (concordance, precision, and call rate).
Many SNP genotyping assays have been described in the literature.3–5 These approaches utilize stringent allele-specific hybridization alone, or hybridization coupled to an enzymatic step, such as 5′ nuclease, ligation, primer extension, or flap endonuclease. Several platforms have been designed for the analysis of SNP genotypes, including electrophoresis, fluorescent readout, oligonucleotide microarrays, mass spectrometry, and beads. Ligation reactions can detect SNPs with high specificity,6–7 and ligation reactions can be used for SNP detection prior to or after PCR.8 While other approaches have distinct individual advantages, we believe that the SNPlex Genotyping System meets all the criteria listed above, combining robust SNP detection with automated assay readout and data analysis. SNPlex system experiments are analyzed on industry-standard CE platforms and processed by a suite of supporting applications.
Assay Overview
The SNPlex Genotyping System consists of a set of pre-optimized, universal assay reagents that are utilized independently of the genotypes studied. The only SNP-specific components of the assay are the ligation probes that participate in the oligonucleotide ligation (OLA). Currently, up to 48 SNPs can be addressed simultaneously in one OLA reaction.
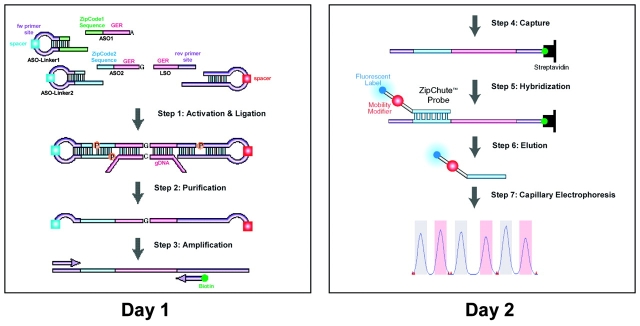
The assay workflow for the SNPlex Genotyping System involves the following seven steps, designed for easy automation, which can be completed within two days (Figure 1): (1) allele-specific OLA reaction; (2) purification of OLA reaction by exonucleolytic digestion of excess probes and linkers; (3) universal PCR reaction to amplify ligation products; (4) capturing of biotin-labeled PCR products in streptavidin coated microtiter plates; (5) binding of ZipChute probes to single-strand PCR products; (6) elution of hybridized ZipChute probes; and (7) detection by CE.
FIGURE 1.
A SNPlex Genotyping System assay protocol can be completed within two days. On the first day, the OLA reaction, exonuclease purification, and PCR amplification are performed. On the second day, the amplicons are immobilized on streptavidin-coated microtiter plates. ZipChute probes are hybridized to complementary ZipCode sequences, and non-hybridized ZipChute probes are washed away. The bound ZipChute probes are eluted and analyzed by CE.
Step one consists of the OLA reaction, during which allele-specific oligonucleotide (ASO) probes and locus-specific oligonucleotide (LSO) probes hybridize to the genomic target sequence. Typically, 37 ng of gDNA is used, resulting in the consumption of <1 ng of gDNA per genotype. These allele-specific and locus-specific probes ligate when they are hybridized to a perfectly matching sequence at the SNP site. Simultaneously, universal linkers are ligated to the distal termini of the ASO and LSO ligation probes. These linkers contain universal PCR primer–binding sequences as well as sequences complementary to ASO and LSO probes. A unique ZipCode sequence is attached at the 5′ end of the genomic equivalent sequence within each ASO. Consequently, by virtue of the ZipCode sequence, the OLA step encodes the genotype information of every SNP into unique ligation products. All probes are designed to function under the same hybridization conditions; therefore, no optimization of OLA reaction conditions is required.
In step two, unligated probes and linkers, as well as the genomic DNA, are removed by enzymatic digestion using exonuclease I and lambda exonuclease. This step is necessary to ensure the efficiency of the subsequent PCR reaction. Step three involves the simultaneous PCR amplification of purified ligation products with a single pair of PCR primers, one of which is biotinylated. Since we use a universal pair of PCR primers, no optimization of PCR reaction conditions is necessary. Next (step four), biotinylated amplicons are bound within wells of streptavidin-coated microtiter plates. Subsequently, the non-biotinylated strands are removed, leaving single-stranded amplicons bound to the microtiter plate.
In step five, fluorescently labeled universal Zip-Chute probes hybridize to the bound single-stranded amplicons. Each ZipChute probe contains a sequence complementary to the unique ZipCode sequence within each ASO; therefore, in order to analyze 48 SNPs, 96 different ZipCode sequences and 96 unique ZipChute probes are required. Each ZipChute probe further contains a mobility modifier, which assigns to each ZipChute probe a specific rate of mobility during CE. Finally, in steps six and seven, the specifically bound ZipChute probes are eluted into CE buffer and analyzed on an Applied Biosystems 3730/3730xl DNA Analyzer.
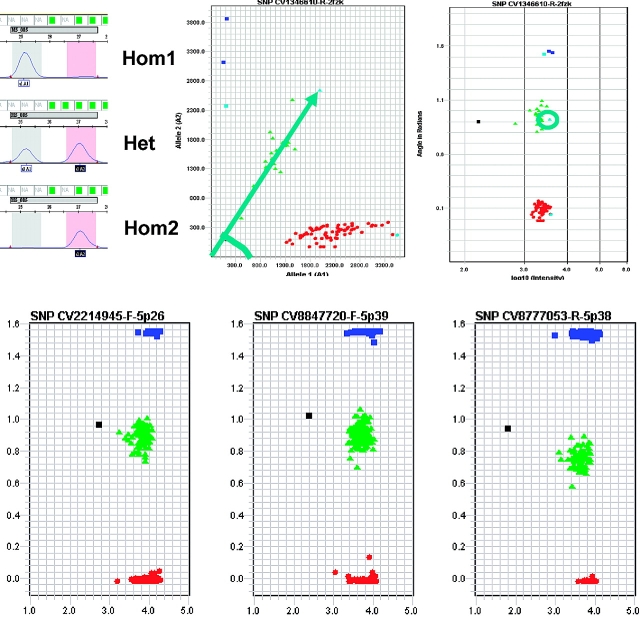
GeneMapper software is used for analyzing the raw CE data and calling SNP genotypes (Figure 2). Because one SNP is typically characterized by two possible alleles, two fluorescent peaks in a CE electropherogram represent the two alleles of a specific SNP. GeneMapper analysis software assigns individual genotypes, based on the intensity and location of peaks.
FIGURE 2.
GeneMapper software analyzes the electropherogram of a SNPlex system sample (A). Two neighboring peaks indicate the genotype of a particular SNP. The software creates for each SNP a genotype plot and uses a clustering algorithm to assign genotypes. SNP genotypes can be displayed as a Cartesian plot (middle ) in which the intensity of both peaks is measured on the x and y axis, or as a polar plot (right), in which the intensity of the peaks is measured on the x axis (length of arrow in Cartesian plot), and the ratio of both peak heights (angle of arrow in Cartesian plot) is measured on the y axis. The data point at the top of the arrow in the Cartesian plot is circled in the polar plot. B Cluster plots representing three SNPs from the control pool probe set, analyzing 44 genomic DNAs eight times on one 384-well plate. in each plot, 352 genotype calls are shown. The black square represents the position of a data point if no genomic DNA is used (non-template control).
In this paper we describe the performance of the SNPlex Genotyping System. We used a probe set, called the control pool, which interrogates 48 population-validated SNPs (Table 1). We tested this control pool against 44 genomic DNAs, which were each represented eight times on a 384-well plate. In addition, we describe the design and performance of 11 probe sets that analyzed 521 SNPs in 92 individuals. For both studies, we report the pass rate, call rate, precision, and concordance with data from TaqMan probe-based assays.
TABLE 1.
Composition of Control Pool
| SNP # | hCV | TSC | rs# | AFAm | Cauc | Chin | Japn |
| 1 | hCV2058031 | TSC0630913 | rs1425151 | 0.23 | 0.26 | 0.28 | 0.28 |
| 2 | hCV7547436 | TSC0691193 | rs1457947 | 0.39 | 0.42 | 0.49 | 0.44 |
| 3 | hCV1901045 | TSC0806211 | rs1323881 | 0.29 | 0.2 | 0.38 | 0.37 |
| 4 | hCV7536854 | TSC0556240 | rs1388276 | 0.46 | 0.33 | 0.34 | 0.45 |
| 5 | hCV2597763 | TSC0926934 | rs1980408 | 0.12 | 0.23 | 0.48 | 0.44 |
| 6 | hCV2059319 | 0.43 | 0.31 | 0.15 | 0.08 | ||
| 7 | hCV8929459 | TSC0339341 | rs1035089 | 0.16 | 0.42 | 0.38 | 0.38 |
| 8 | hCV2986015 | TSC0295775 | rs705681 | 0.21 | 0.45 | 0.22 | 0.22 |
| 9 | hCV8720462 | TSC0165692 | rs893613 | 0.3 | 0.48 | 0.41 | 0.4 |
| 10 | hCV349615 | TSC0314577 | rs992690 | 0.16 | 0.24 | 0.34 | 0.36 |
| 11 | hCV8879897 | 0.43 | 0.46 | 0.44 | 0.45 | ||
| 12 | hCV7505765 | TSC0265430 | rs961495 | 0.26 | 0.19 | 0.2 | 0.08 |
| 13 | hCV1637791 | TSC0016017 | rs729673 | 0.45 | 0.4 | 0.24 | 0.2 |
| 14 | hCV1691378 | TSC0265475 | rs1115261 | 0.48 | 0.46 | 0.18 | 0.28 |
| 15 | hCV3194437 | TSC0301076 | rs984071 | 0.2 | 0.29 | 0.17 | 0.17 |
| 16 | hCV7571632 | TSC0243654 | rs927221 | 0.16 | 0.15 | 0.28 | 0.45 |
| 17 | hCV7537265 | TSC0129483 | rs748573 | 0.32 | 0.22 | 0.33 | 0.33 |
| 18 | hCV7589926 | 0.3 | 0.11 | N/A | N/A | ||
| 19 | hCV8845932 | TSC0324505 | rs1156404 | 0.27 | 0.47 | 0.17 | 0.17 |
| 20 | hCV2179737 | 0.33 | 0.2 | 0.43 | 0.31 | ||
| 21 | hCV8792022 | TSC0213380 | rs879253 | 0.21 | 0.45 | 0.21 | 0.08 |
| 22 | hCV2489240 | TSC0098582 | rs1016146 | 0.12 | 0.35 | 0.48 | 0.27 |
| 23 | hCV2025116 | TSC0757622 | rs1507213 | 0.12 | 0.48 | 0.48 | 0.5 |
| 24 | hCV1653240 | TSC0136611 | rs751340 | 0.42 | 0.37 | 0.24 | 0.18 |
| 25 | hCV357822 | TSC0783613 | rs1520483 | 0.1 | 0.41 | 0.48 | 0.48 |
| 26 | hCV8686971 | TSC0465947 | rs1570903 | 0.23 | 0.44 | 0.45 | 0.26 |
| 27 | hCV706864 | TSC0071873 | rs288423 | 0.42 | 0.37 | 0.43 | 0.38 |
| 28 | hCV3017144 | TSC0084538 | rs1007106 | 0.19 | 0.34 | 0.45 | 0.45 |
| 29 | hCV9621778 | TSC0318440 | rs995178 | 0.41 | 0.49 | 0.49 | 0.45 |
| 30 | hCV8747570 | TSC0679949 | rs1597695 | 0.35 | 0.39 | 0.5 | 0.44 |
| 31 | hCV8862622 | TSC0825772 | rs1334334 | 0.15 | 0.24 | 0.27 | 0.17 |
| 32 | hCV8946637 | TSC0809047 | rs794108 | 0.46 | 0.38 | 0.29 | 0.37 |
| 33 | hCV1358402 | TSC0463216 | rs1569244 | 0.42 | 0.17 | 0.41 | 0.46 |
| 34 | hCV7500677 | TSC0296508 | rs238196 | 0.12 | 0.1 | 0.28 | 0.3 |
| 35 | hCV9589619 | TSC0984433 | rs1925643 | 0.42 | 0.3 | 0.42 | 0.42 |
| 36 | hCV8921382 | TSC0910879 | rs1713423 | 0.3 | 0.5 | 0.31 | 0.41 |
| 37 | hCV1688032 | 0.22 | 0.31 | 0.16 | 0.23 | ||
| 38 | hCV9636350 | 0.16 | 0.37 | 0.36 | 0.37 | ||
| 39 | hCV2962785 | 0.4 | 0.11 | 0.17 | 0.24 | ||
| 40 | hCV2780152 | TSC0851851 | rs1861606 | 0.49 | 0.31 | 0.29 | 0.36 |
| 41 | hCV2569743 | TSC0430769 | rs1548543 | 0.22 | 0.34 | 0.31 | 0.29 |
| 42 | hCV468629 | TSC0463040 | rs1569125 | 0.41 | 0.29 | 0.22 | 0.2 |
| 43 | hCV1534177 | 0.22 | 0.2 | 0.27 | 0.29 | ||
| 44 | hCV11164916 | 0.19 | 0.12 | N/A | N/A | ||
| 45 | hCV2214945 | TSC1086371 | rs220860 | 0.25 | 0.22 | 0.22 | 0.22 |
| 46 | hCV8847720 | TSC0695101 | rs1460239 | 0.29 | 0.4 | 0.19 | 0.29 |
| 47 | hCV8777053 | TSC0070807 | rs954779 | 0.28 | 0.18 | 0.32 | 0.3 |
| 48 | hCV7443819 | TSC0267953 | rs963014 | 0.48 | 0.4 | 0.08 | 0.11 |
For the control pool, SNPs were selected with minor allelic frequencies (MAF) above 0.1 for African-American (AfAm) and Caucasian (Cauc) populations. For Chinese (Chin) and Japanese (Japn) samples, at least 42 SNPs have a MAF above 0.1. SNPs were selected from the Celera Discovery System (CDS) data-base. The Control Pool SNP number and SNP IDs are displayed as well: Celera SNP ID (hCV); The SNP Consortium ID (TSC); dbSNP reference cluster ID (rs#).
RESULTS
We used the control pool, analyzing 48 SNPs, to test the performance of the SNPlex system. To design the control pool, we selected population-validated SNPs for which TaqMan probe-based assays were available. Each SNP has a minor allele frequency of at least 0.1 in one or more of the following populations: African-American, Caucasian, Japanese, and Chinese (Table 1). Using the control pool probe set, we then analyzed 44 gDNAs of Caucasian origin, each spotted eight times per 384-well plate, which equals 16,896 genotype calls per plate. The plates with sample DNA used in this study were pre-manufactured and contained 37 ng of dried-down fragmented gDNA.
SNPlex System Performance Evaluation I
To test the robustness of the SNPlex Genotyping System, three laboratories each tested three identical 384-well plates and analyzed them on three 3730xl DNA analyzers. For each plate, we determined the pass rate, call rate, precision, and the concordance with genotypes from TaqMan probe–based assays (Table 2). The pass rate is the percentage of SNPs that meet minimal quality requirements. GeneMapper analysis software uses empirical parameters, such as signal strength and cluster separation, to determine whether the plot characteristics of a SNP are within assay specifications. The call rate is the percentage of successful genotype calls per passing SNP; precision is the reproducibility of identical genotype calls within one plate; and concordance is the agreement between genotype calls measured with TaqMan probe–based assays and the SNPlex Genotyping System. Output tables for pass rate and call rate were calculated by GeneMapper software.
TABLE 2.
SNPlex System Performance Evaluation I
| Laboratory | CE | Pass Rate | Call Rate | Precision | Concordance |
| 1 | 3730_1 | 100 | 99.56 | 100 | 100 |
| 1 | 3730_1 | 100 | 99.75 | 100 | 100 |
| 1 | 3730_1 | 100 | 99.67 | 100 | 100 |
| Average 1 | 100 | 99.66 | 100 | 100 | |
| 2 | 3730_2 | 100 | 99.94 | 100 | 100 |
| 2 | 3730_2 | 100 | 99.92 | 99.99 | 100 |
| 2 | 3730_2 | 100 | 99.94 | 100 | 99.99 |
| Average 2 | 100 | 99.93 | 100 | 100 | |
| 3 | 3730_3 | 100 | 99.81 | 100 | 100 |
| 3 | 3730_3 | 100 | 99.31 | 99.83 | 100 |
| 3 | 3730_3 | 100 | 94.96 | 99.71 | 99.79 |
| Average 3 | 100 | 99.03 | 99.85 | 99.95 | |
| Average 1–3 | 100 | 99.21 | 99.95 | 99.98 |
Overview of performance data for nine 384-well plates, in which 48 SNPs were analyzed in 44 gDNAs. Each gDNA was spotted eight times on each 384-well plate, resulting in 16,896 genotype calls per plate. Three plates were each tested in a different laboratory and analyzed on three 3730xl DNA analyzers. all values represent percentages.
SNPlex System Performance Evaluation II
To analyze the performance of the SNPlex Genotyping System on a more representative and larger set of SNPs, we chose confirmed SNPs from human chromosome 21. The SNP set was selected from the dbSNP NCBI database. Initially, 2476 SNPs were selected from the first 10 MB of chromosome 21. The SNPs had to be bi-allelic markers, mapping to a unique region in the genome. SNPlex system probe sets were designed using our automated assay design pipeline. After screening the SNP sequences, the pipeline produced designs and multiplex pools containing 2243 SNPs, a successful design rate of 90.59%. Those SNPs predicted to have a low likelihood of providing valid data are rejected by the assay design pipeline. Pre-screening SNPs reduces the cost of synthesis and shortens analysis time. A subset of 521 SNPs in 11 multiplex pools was tested in triplicate, using 92 genomic DNAs from the Coriell Diversity Panel (Coriell Institute for Medical Research, Camden, NY). For this second data set, we calculated the assay pass rate, call rate, precision and concordance with genotype data from TaqMan assays (Table 3)
TABLE 3.
SNPlex System Performance Evaluation II
| Experiment | Pass Rate | Call Rate | Precision | Concordance |
| 1 | 92.52% | 99.48% | NA | 99.71% |
| 2 | 92.48% | 99.08% | NA | 99.66% |
| 3 | 94.22% | 99.74% | NA | 99.73% |
| Average | 93.07% | 99.43% | 99.94% | 99.70% |
Overview of performance data of 521 SNPs analyzed in triplicate in 92 genomic DNAs. Each percentage value is based on 47,932 genotype calls. The precision of genotype calls can be calculated only between different runs and is not applicable (NA) for individual runs.
The average pass rate for all 521 SNPs in the three assay replicas was 93.07%. The overall assay conversion rate (design rate × assay pass rate) therefore was 84.31%. The average precision of the genotype calls was 99.94%. Precision in this case means the reproducibility of identical genotype calls across different plates, prepared and analyzed at different times. Concordance with TaqMan assay results was 99.70%. The concordance calculation was based on TaqMan assay data available for 19,116 (40%) of the 47,932 tested genotypes.
DISCUSSION
We have presented a detailed overview of the SNPlex Genotyping System, a high-throughput SNP genotyping technology that provides high assay reproducibility and accuracy. The SNPlex Genotyping System is a novel platform that uses industry-standard CE instruments to enable medium- to high-throughput, cost-effective SNP genotyping projects in a multiplexed format. The flexibility and scalability of the SNPlex Genotyping System regarding study design and sample throughput make it well suited for applications in modern genetics research and pharmacogenetics. Additionally, the system’s online tools enable researchers to plan and manage customized SNP genotyping studies. The chemistry protocol can be completed in two days, using commonly available automation systems, which allows it to analyze up to 1.5 million genotypes in a five-day week.
The user friendliness of a genotyping platform is partly determined by the methodology of the genotyping assay. Of equal importance is whether it supports the whole process, from study design to data analysis and management. It is therefore important to consider the ease that a platform provides for the selection of genetic markers that can successfully convert to high-performing assays. Although SNPs are abundant in the human genome and in large databases of candidate genes,9–10 not all putative polymorphisms are suitable for the development of genotyping assays. It has been reported several times in the literature that only 50% of SNPs selected at random from dbSNPs typically yield working assays, and this results in significant delays and expense.11–13
SNPbrowser Software
To simplify the SNP genotyping selection process, we developed the freely available SNPbrowser software, a tool that assists in the knowledge-based selection of markers for association studies. SNPbrowser software is a stand-alone free application, based on the Windows operating system, which displays SNPs for the entire human genome, as well as the empirically observed patterns of linkage disequilibrium (LD). The SNPs displayed are those for which it is possible to design an in silico SNPlex system assay. The software wizards allow researchers to prioritize the selection of SNPs based on the patterns of linkage disequilibrium as observed by De La Vega et al.,14 and calculated from the HapMap data (www.hapmap.org), and by metrices, such as minor allelic frequencies and TaqMan assay validation. Currently, more than 150,000 SNPs are validated by TaqMan assays and can be designed as SNPlex system assays. In addition, more than 8 million SNPs in dbSNP have been pre-screened by the design pipeline, and the passing SNPs are available as SNPlex system assays. Wizards further help to supplement gaps with coding or pre-designed, functionally tested assays that help ensure the highest probability of success for an association study.
Additionally, wizards can generate lists of SNPs based on a number of study design approaches, including picket-fence distribution, pairwise r2, and haplotype r2. After SNP selection, SNPs can be submitted to the SNPlex system design pipeline for custom assay design.
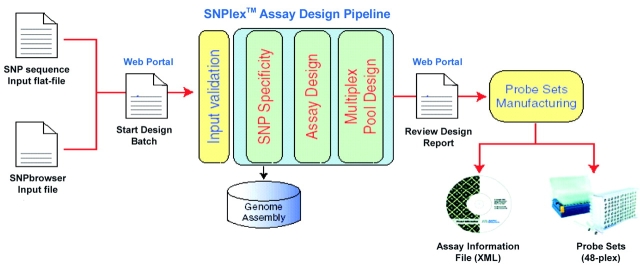
To facilitate the process, an assay design submission tool is available through the Applied Bio-systems website (www.appliedbiosystems.com). The SNPs can be submitted online as a list of identifiers (e.g., from SNPbrowser software), or as a flat file that contains the SNP identifier and sequence, including SNP alleles. After the submitted SNPs are verified for format, a design request is submitted to the automated assay design pipeline, which runs on a distributed computing environment at the back end (Figure 3). When the pool designs are available, they are sent to the researcher for review before the order is submitted.
FIGURE 3.
User-defined SNPlex system assays are designed by means of a proprietary assay design pipeline, which can be accessed through the Applied Biosystems website. Ligation probes for individual SNP assays are designed using thermodynamic rules to ensure annealing specificity and efficiency. each probe undergoes extensive genome screening and is filtered for secondary structure or probe-probe chimeras. After the SNP-specific probes are designed, stringent rules are used to determine optimal probe-pool composition. assay annotations and allele calling information are provided as assay information files (AIF) on CD-ROM.
Assay Design
Assays for the SNPlex Genotyping System are designed by an automated high-throughput pipeline (Figure 3). Assays can be designed for human as well as non-human SNPs. The multi-step pipeline combines SNP-specific assays into compatible multiplex pools to ensure robust assay performance. These steps include: (1) screening the SNP context sequences against the human genome to avoid designing assays for SNPs in repetitive or duplicated genomic regions that would lead to low specificity (SNP sequences can also be screened against the mouse and rat genome); (2) selecting the most suitable strand and probe sequences by applying rules that maximize assay and manufacturing success; (3) assigning ZipCode sequences to each SNP assay; and (4) separating the assays into compatible probe pools that are screened for probe-probe interactions, spurious ligation templates, and unintended probe combinations that may have a significant genomic target. During the screening step, SNP sequences are eliminated if they are redundant in the target genome, contain non-target polymorphisms near the target SNP, or contain sequence motifs that are incompatible with the assay. Because testing the uniqueness of SNP targets with the widely used BLAST algorithm15 is slow and lacks sensitivity, we developed a proprietary, fast, and sensitive SNP-centered genome-screening algorithm that uses empirically weighted match/mismatch scores around the SNP loci. SNP assays that pass the above-described screens are used for the composition of compatible probe pools. The resulting pool of OLA probes targets between 24 and 48 viable, mutually compatible SNPs.
Control Pool Performance
The SNPlex System control pool is a set of high-quality probes interrogating 48 population-validated SNPs. The set of human gDNAs (gDNA Panel), pre-distributed in a 384-well plate, together with the control pool probes, form a tool for training, quality control, and troubleshooting purposes.
The gDNA Panel is a collection of genomic DNAs from 44 Caucasians, approximately evenly distributed between male and female. With this DNA panel, 45 of 48 SNPs produce cluster plots with three clusters. The three SNPs with only two clusters have minor allelic frequencies of 11%, 15%, and 19% in Caucasian populations, which could explain the occurrence of only two clusters. Using the actual minor allelic frequencies, the predicted statistical number of SNPs with two clusters is 2–7 for 48 SNPs in 44 genomic DNA samples. The observed three SNPs with two clusters are, therefore, within the expected range. As almost all SNPs interrogated by the control pool have minor allelic frequencies higher than 0.1 in African-American, Caucasian, Chinese, and Japanese populations, the polar plots of SNPs should display three clusters independent of the ethnic origin of the sample collection. The control pool is therefore useful for testing customer-supplied gDNA samples as well.
The goal of studying control pool SNPs in a gDNA panel (SNPlex System Performance Evaluation I) was to measure the robustness of the SNPlex Genotyping System in three independent laboratory settings. Performance consistency is important for a high-throughput genotyping system, and it has a direct impact on the cost per genotype.
The control pool SNPs were selected for high performance; therefore, the overall assay pass rate of 100% was not surprising. From a user perspective, the more relevant data are the call rate, precision, and concordance of the genotype calls, and the way in which these values differ across the three laboratories. The call rate was consistently above 99.3%; one plate in one laboratory showed a call rate of 95%; however, concordance and precision were above our assay specifications of 99.5% and 99.7%, respectively.
The very high precision, as well as the concordance with TaqMan genotyping data, confirms the high performance of the SNPlex Genotyping System. Likewise, the low standard deviation of precision and concordance across the three laboratories demonstrates the quality of performance. Precision and concordance were calculated for passing genotype calls, and they are therefore not affected by the assay call rate.
Design Pipeline Evaluation
The goal of SNPlex System Performance Evaluation II was to test both the design and assay conversion rate in a more realistic setting. The study used 2476 confirmed SNPs from the first 10 MB of chromosome 21, chosen from the NCBI database and entered into the assay design pipeline. Additional selection criteria required these SNPs to involve only two possible polymorphic bases and to map to only one position in the human genome. A design could be made for 2243 of the 2476 submitted SNPs, which translates to a design rate of 90.6%. The design rate is very much influenced by the quality of sequences submitted (see discussion above). SNPs fail the design pipeline for the following reasons: (1) the SNP may be within a repeat sequence found many times in the genome; (2) the SNP sequence may contain a second SNP near the first; and (3) the SNP context sequence may contain one or more sequences of low complexity. SNP sets containing TaqMan assay–validated SNPs usually produce a design rate of greater than 90%. For customer-supplied SNP sets containing confirmed dbSNPs, the design rate is usually between 80% and 85%. The most common cause of failed assay designs are multiple genome hits and nucleotide compositions that conflict with the assay design rules. Another factor negatively impacting the design rate is a low number of submitted SNPs.
This paper describes the performance characteristics of the SNPlex Genotyping System. We have designed a set of probes that interrogates 48 SNPs (the control pool), and a 384-well plate that analyzes 44 genomic DNAs (the gDNA panel). These optimized components enable the user of the SNPlex Genotyping System to measure and validate assay performance, and to identify components that impact assay function. To test the performance of the system, we processed nine 384-well plates in three laboratories and analyzed the results on three CE instruments.
In a second experiment, we analyzed SNPs from 10 MB of chromosome 21 in 92 samples and measured the assay design efficiency and the performance characteristics. The SNPlex Genotyping System showed a consistent high performance in both experiments. Upcoming development opportunities include fixed sets of SNP assays for applications such as linkage mapping, or nsSNP sets for direct association studies for human as well as non-human organisms.
Acknowledgments
The authors wish to acknowledge the following groups at Applied Biosystems for their support in the development of the advances described here: Advanced Development and Manufacturing, Analysis Software R&D, Bioinformatics R&D, Genomic Applications R&D, Genotyping Applications Marketing, Global Oligo Operations, Pilot Operations Lab, and Product Test. Thanks are due to Josh Goldsmith and Dale Baskin for comments on the manuscript. Special thanks to Amy C. Kivett, Rizza A. Padilla, and Nitesh R. Patel for performing SNPlex system experiments. We also want to acknowledge the work of Shirley J. Johnson, Ernest J. Friedlander, Dominika Maglasang, Allison Holt, and Amanda Bach, who contributed during the early stages of the project.
The genomic DNA samples analyzed by Control Pool probes contain DNA samples supplied by the European Collection of Cell Cultures (ECACC). Purchase of the plate conveys the right to use such DNA samples as internal controls in connection with use of the SNPlex Genotyping System, but does not convey any right to replicate or redistribute such DNA samples.
REFERENCES
- 1.Venter C, Adams MD, Myers EW, et al. The sequence of the human genome genome. Science 2001;291(5507): 1304–1351. [DOI] [PubMed] [Google Scholar]
- 2.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 1989;245(4922):1066–1073. [DOI] [PubMed] [Google Scholar]
- 3.Jannetto PJ, Laleli-Sahin E, Wong SH. Pharmacogenomic genotyping methodologies. Clin Chem Lab Med 2004;42(11):1256–1264. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Sullivan PF. Single nucleotide polymorphism genotyping: Biochemistry, protocol, cost and throughput. Pharmacogenomics J 2003;3(2):77–96. [DOI] [PubMed] [Google Scholar]
- 5.Kwok, PY. Methods for genotyping single nucleotide polymorphisms. Ann Rev Genomics Hum Genetics 2001;(2):235–258. [DOI] [PubMed]
- 6.Khanna M, Cao W, Zirvi M, Paty P, Barany F. Ligase detection reaction for identification of low abundance mutations. Clin Biochem 1999;32(4):287–290. [DOI] [PubMed] [Google Scholar]
- 7.Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, Barany F. Universal DNA microarray method for multiplex detection of low abundance point mutations. J Mol Biol 1999;292, 251–262. [DOI] [PubMed] [Google Scholar]
- 8.Eggerding FA, Iovannisci DM, Brinson E, Grossman P, Winn-Deen ES. Fluorescence-based oligonucleotide ligation assay for analysis of cystic fibrosis transmembrane conductance regulator gene mutations. Hum Mutat 1995;5(2):153–165 [DOI] [PubMed] [Google Scholar]
- 9.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res 2001;29(1):308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerlavage A, Bonazzi V, di Tommaso M, Lawrence C, Li P, Mayberry F, et al. The Celera Discovery System. Nucleic Acids Res 2002;30(1):129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marth G, Yeh R, Minton M et al., Nat Genet. 2001;27(4):371–372. [DOI] [PubMed] [Google Scholar]
- 12.Marsh S, Kwok P, McLeod HL, SNP databases and pharmacogenetics: Great start, but a long way to go. Hum Mutat. 2002;20(3):174–179. [DOI] [PubMed] [Google Scholar]
- 13.Reich DE, Gabriel SB, Altshuler D, Nat Genet 2004;33(4):457–458 [DOI] [PubMed] [Google Scholar]
- 14.De La Vega FM, Isaac H, Collins A, Scafe CR, Halldorsson BV, Su X, et al. The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern. Genome Res 2005;15:454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool. J Mol Biol 1990:215(3):403–410. [DOI] [PubMed] [Google Scholar]