Abstract
Separation and enrichment of organelles from complex biological mixtures are important for proteomic analysis. Two widely used current standard techniques to isolate individual organelles include differential and density-gradient centrifugation. Although these techniques have proven useful for processing small volumes of sample, multiple rounds of centrifugation are required when performing a large-scale purification. In this report, we have introduced a novel technique: continuous-flow ultracentrifugation using a sucrose gradient to separate, accumulate, and highly enrich bovine heart mitochondria in one step. To demonstrate the advantage of the technique, mitochondrial proteins from two different bovine hearts (3–8 mo and 18–30 mo old) were examined. For each age group, 100 g of bovine heart tissue were homogenized by a blending procedure. After removal of the nuclei, the entire remaining homogenate was loaded onto a proteomics continuous-flow ultracentrifuge to separate and enrich the organelles. Fractions were collected and mitochondria-enriched fractions were identified by Western blot analysis. To study the protein profile changes with aging in the mitochondrial proteome, the mitochondria-enriched fractions were applied to two-dimensional gel electrophoresis. The resulting two-dimensional PAGE gels were subsequently analyzed by image analysis software to identify proteins unique to each age group and proteins with at least twofold differences in protein expression. These proteins were then digested with trypsin and identified by mass spectrometer. Significant differences in the protein profiles of the two differently aged mitochondria preparations were found. The continuous-flow ultracentrifugation technique was demonstrated to be a powerful tool for separation and enrichment of organelles and their sub-types.
Keywords: Proteomics, continuous-flow ultracentrifugation, sub-cellular fractionation, organelle separation, organelle enrichment, mitochondria, mitochondria enrichment, aging
Mitochondria are one of the most complex and most important organelles in a eukaryotic cell. Mitochondrial functions include processing bioenergetic metabolism, regulating cell death, modulating ionic homeostasis, oxidizing carbohydrates and fatty aids, and participating in many other catabolic and anabolic pathways. Consequently, mitochondrial mutations and dysfunction have been associated with many diseases, including neurodegenerative diseases such as Alzheimer’s and Parkinson’s,1 heart diseases, cancer, and various neuromuscular syndromes.2–5 There is increasing evidence that mitochondria play an important role in cell life and the process of aging.6–8 Therefore, analysis of age-related changes in subcellular status could lead to new insights into age-related functional changes and improve our understanding of the aging process.
Mitochondria, like other organelles, have been shown to vary with tissue type, animal age,9–10 disease,11 and function within the same cell type.12–13 Recent developments in proteomic techniques open the path towards a deeper exploration and better understanding of mitochondrial variations in structure and function. Until now, the most widely used proteomic approaches include two-dimensional electrophoresis (2DE) and multidimensional protein identification technology (MudPIT) combined with mass spectrometry analysis (MS).20–23 However, the ability to study the mitochondrial proteome and other complex biological mixtures appears to require better separation, particularly for bottom-up proteomic approaches. Therefore, subcellular fractionation and organelle enrichment play an important role in reducing the total cell lysate complexity and increasing the proteome coverage. Other pre-fractionation techniques have been reported and have proven useful prior to 2DE and MS analyses.24
The traditional methods used for isolation of mitochondria generally include one or both of the following steps: (1) differential centrifugation, which separates particles on the basis of size and yields several crude fractions. These fractions can be pelleted sequentially according to the gravitational (g) force of the centrifuge: ~1000g containing nuclei, heavy mitochondria, and plasma membrane sheet; ~5000 g containing light mitochondria, lysosomes, peroxisomes, and intact Golgi; ~100,000 g containing Golgi, endoplasmic reticulum (ER), microsomes, and endosomes. (2) Density-gradient centrifugation, which separates particles on the basis of density and size, yields a purer organelle fraction.14–17 Several density media, including sucrose, nycodenz, percoll, and ficoll 400, have been used for density-gradient centrifugation.18–19 However, most subcellular fractionation procedures have disadvantages, such as being labor intensive and time consuming. Here we present a technique that separates organelles and their subtypes, including mitochondrial subtypes, by sucrose density using proteomic continuous-flow ultracentrifugation (pCFU). At the same time, the organelles are effectively enriched at their buoyant densities, resulting in a more efficient proteomic analysis. Unlike classical methods, pCFU is not limited by sample volume and thus can be used to achieve significant accumulations of low-abundance components. In this report, we compare the mitochondria proteomic profiles of bovine hearts of two different ages using pCFU as the separation and enrichment tool.
MATERIALS AND METHODS
Chemicals and Animal Tissues
Bovine hearts were purchased from Animal Technologies (Tyler, Texas). Young bovine hearts were of ages 3–8 mo and adult bovine hearts were ages 18–24 mo. Immediately after sacrificing the animals, the organs were harvested and the hearts were flash frozen in liquid nitrogen. Sucrose was from Invitrogen (Carslbad, CA). Trypsin (mass spectrometry grade) was from Promega Biochemicals (Madison, WI). Protease inhibitor cocktail tablets were obtained from Roche Molecular Biochemicals (Indianapolis, IN). Magnesium chloride (MgCl2), HEPES, PBS buffer, ammonium bicarbonate (AB), acetonitrile (ACN), iodoacetomide (IA), Tris (2-carboxyethyl) phosphine hydrochloride (TCEP-Cl), acetic acid, methanol, Tris buffer, sodium chloride (NaCl), and Tween-20 were purchased from Sigma-Aldrich Chemicals (St. Louis, MO).
Tissue Homogenization
Flash-frozen bovine hearts were thawed at 4°C in 1X PBS. After removal of the connective tissue and blood vessels, the myocardium was minced. Approximately, 100 g of the minced heart tissue was randomly chosen and weighed. The weighed heart tissue was added to a pre-chilled 1-L commercial Waring blender. To the blender, 500 mL of homogenization buffer (20 mM HEPES, 5 mM MgCl2, 500 mM sucrose, protease inhibitor cocktail, pH 7.2) was added and the mixture was blended for 30 sec on high and 30 sec on low. The blended homogenate was then filtered through four layers of cheesecloth. The filtrate was centrifuged at 750 g for 10 min to remove nuclei. The resulting supernatants were saved. The pellets were weighed and resuspended in equal amounts of homogenization buffer. The suspension was reblended and recentrifuged using the same conditions. The resulting supernatant and the previous supernatant were combined, and the pellets were discarded. The clarified homogenate was diluted twofold using dilution buffer (20 mM HEPES, 5 mM MgCl2, pH 7.2) to bring the sucrose concentration to 250 mM. The homogenate was stored on ice for further separation.
Continuous-Flow Ultracentrifugation
The clarified bovine heart homogenate was separated and fractionated using a pCFU (Alfa Wassermann, West Caldwell, NJ). A PKII rotor (800 mL) was loaded with sucrose flow buffer (20 mM HEPES, 5 mM MgCl2, 250 mM sucrose, pH 7.2) using a peristaltic pump (Cole Parmer, Vernon, IL). Air bubbles were removed by accelerating the rotor to 20K rpm and simultaneously pumping flow buffer up through the bottom of the rotor. The rotor was then brought to rest and 400 mL of gradient buffer (60% sucrose w/v in dilution buffer) was pumped through the bottom of the rotor at 10 mL/min. The rotor was slowly ramped to 3500 rpm to establish a linear sucrose gradient. The centrifuge was accelerated to 20K rpm, and the bovine heart homogenate was loaded onto the rotor using the peristaltic pump. Eluent collected from the top of the rotor was reloaded at 35K rpm to allow any remaining light particles in the homogenate to enter into the sucrose gradient. The sample was banded for an additional 2 h at 35K rpm. After that time, the rotor was brought to rest and 35-mL fractions were collected from the bottom of the rotor. One milliliter of each fraction was aliquoted and stored overnight at 4°C. The remaining fractionated material was frozen at −70°C.
Density-Gradient Calculation
The refractive index of each fraction was measured to verify the linearity of the sucrose gradient. Refractive indices were measured on a Milton Roy Refractometer (Ivyland, PA). Sucrose percentages and densities were calculated using data from the International Critical Tables.36
Protein Assay
The protein concentration of each fraction from the pCFU separation was determined using the bicinchoninic acid assay kit from Pierce (Rockford, IL) following the manufacturer’s procedure.
SDS-PAGE and Western Blotting
SDS-PAGE samples were prepared as 1–2 mg/mL solutions. The protein samples were run on Criterion 4–20% polyacrylamide gels from Bio-Rad (Hercules, CA) in duplicate. One SDS-PAGE gel was stained with BioSafe Coomassie (Bio-Rad). The other SDS-PAGE gel was transferred onto a 0.2-μm PVDF membrane using the Bio-Rad Criterion blotting system for Western analysis. The blot was blocked with 5% nonfat milk overnight. To detect fractions enriched with mitochondria, blots were incubated with rabbit anti-Tom20 antibody from Santa Cruz Biotechnology (Santa Cruz, CA). Blots were washed three times with wash buffer (25 mM Tris, 125 mM NaCl, 0.1% Tween-20, pH 8.0) followed by incubation with goat anti-rabbit secondary antibody conjugated to HRP from Jackson Immunochemicals (West Grove, PA). Blots were washed three times with wash buffer and detected using Bio-Rad Immun-Star HRP substrate. The membranes were imaged using Kodak Biomax Light autoradiography film. Net band intensities were calculated using Nonlinear Dynamics (Durham, NC) Phoretix 1D expression software.
Two-Dimensional Gel Electrophoresis
Samples of approximatly 500 to 1000 μL from the fractions enriched with mitochondria were dialyzed overnight using Gebaflex tubes (Geba, Kfar-Hanagid, Israel) to remove sucrose. The dialyzed samples were then concentrated using Millipore (Bedford, MA) Bio-max centrifugal spin columns to give a sample volume of about 20 μL. The resulting samples were diluted into Bio-Rad 2D sample buffer with the final volume 85 μL. Eleven-centimeter IPG strips with pH ranges of 3–10, 5–8, and 7–10 (Bio-Ra) were used, and the strips were rehydrated at room temperature overnight. Rehydrated IPG strips were focused on a Bio-Rad Protean IEF System according to manufacturer’s instructions and then reduced and alkylated prior to running in the second dimension on 8–16% SDS-PAGE gels (Bio-Rad). Gels were later fixed in 40% MeOH:10% HOAc for 1 h and stained using Bio-Rad BioSafe Coomassie. Gels were destained as necessary and scanned as TIFF format images using an UMAX PowerLook 1120 scanner (UMAX Technologies, Dallas, TX). Images were analyzed using Nonlinear Dynamics Phoretix 2D Expression software. Samples were run in triplicate.
In-Gel Tryptic Digestion
Spots of interest were excised and destained thoroughly, followed by reduction with 20 mM TCEP-HCl in 25 mM AB (pH 8.0). The gel pieces were alkylated with 40 mM IA in 25 mM AB (pH 8.0). After washing in 25mM AB and 25 mM AB/50% ACN, the gel pieces were digested with 40 mM AB, 0.02 μg/μL trypsin (pH 8.0) for 16 h at 37°C.
Nano-LC-ESI-MS/MS
Peptide samples resulting from the in-gel digest were analyzed on a Thermo Electron LCQ Deca XP MAX ion-trap mass spectrometer with an NSI source (San Jose, CA). Samples were eluted using a linear gradient of 5% solvent B (0.1% formic acid in acetonitrile) to 65% solvent B for 30 min with a flow rate of 200 nL/min. The mass spectrometer was operated in a data-dependent MS/MS mode using a normalized collision energy of 35%. The capillary temperature of the ion source was set at 180°C. Protein identification was achieved using Thermo Electron BioWork 3.1 software with the most current NCBI non-redundant protein database. A minimum of two unique peptides found with a good Xcorr (+1 CS ≥ 1.5; +2 CS ≥ 2.0; +3 CS ≥ 2.5) were required for an identity assignment.
RESULTS AND DISCUSSION
Introduction of Continuous-Flow Ultracentrifugation using pCFU
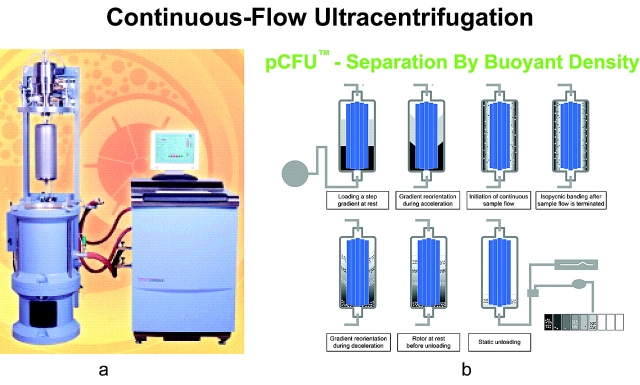
Continuous-flow ultracentrifugation technology was developed by Dr. Norman Anderson and coworkers in the 1960s29 and first commercialized by Alfa Wassermann (West Caldwell, NJ). Continuous-flow ultracentrifugation has been used for separation, isolation, clarification, and concentration of a wide variety of cellular components and macromolecules by their buoyant densities, using a density gradient as the separation medium.33–35 The separation mechanism is as follows (Figure 1): (1) material is loaded into the rotor at rest. (2) As the rotor is gradually accelerated, the gradient reorients itself vertically along the wall and forms a linear gradient. (3) Sample fluid is pumped into the bottom of the rotor using continuous flow. (4) The sample particles sediment radially into the gradient with increasing density. They band isopycnically in the cylindrical zones where the gradient density equals the particle’s buoyant density. (5) At the end of the run, the rotor is decelerated and (6) the gradient reorients to its original position without disturbing the particle bands. (7) The banded particles are now ready to be collected with the rotor at rest.
FIGURE 1.
a: PKII Continuous-flow Ultracentrifuge (CFU). b: Process of gradient formation and sample separation in the vertical rotor of the CFU.
pCFU provides a novel tool for sample preparation in proteomic analyses. Based on the principle of separation by buoyant density, combined with continuous flow, pCFU can be used for sample enrichment and accumulation, separation of organelles and their subtypes, separation of lipoproteins and other subcellular particles, and isolation of low-abundance components. Above all, pCFU provides separation without loss of biological information.
Mitochondria Separation, Enrichment, and Heterogeneity
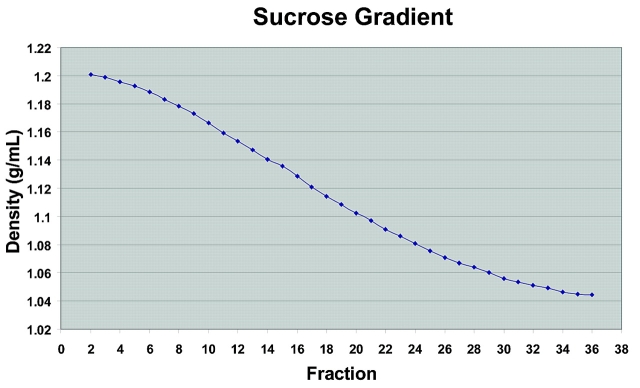
Sucrose is the typical and most frequently used medium for isolation of organelles, as it does not influence molecule-molecule interaction.25 In this application, we chose 8–60% sucrose as our separation gradient. In order to protect organelles and their integrity, 500 mM sucrose was used in the homogenization buffer. Figure 2 shows the formation of the linear gradient. Typically, a slightly lower percentage of sucrose at the high end (~55%, d = 1.2 g/mL) and a slightly higher percentage of sucrose (~12%, d = 1.04 g/mL) at the low end are observed, possibly due to the dead volume of the instrument. The sucrose linear gradient is formed as soon as the centrifuge is ramped up to 3500 rpm. Our instrument has demonstrated high consistency of linear density-gradient formation.
FIGURE 2.
Density profile of fractions from sucrose-density centrifugation of bovine heart. The sucrose density was calculated from refractive indices measured on a Milton Roy refractometer.
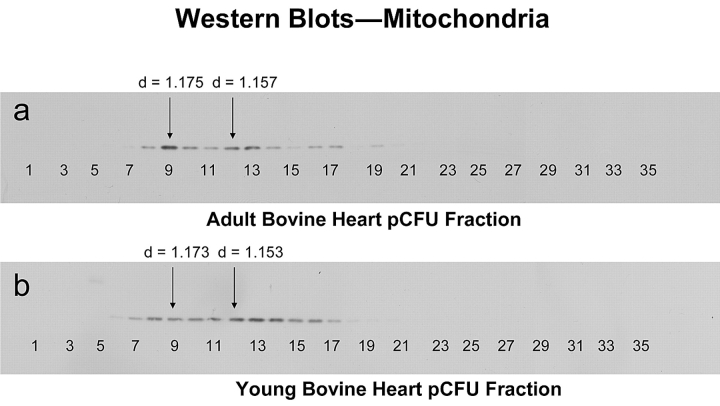
Mitochondria are double membrane–bound organelles with particle dimensions of 0.5–2 μm. Their density in sucrose gradients is about 1.17–1.21 g/mL. Mitochondrial proteins are approximately 16% of total cellular proteins. As described at the beginning of this article, the most widely used methods for isolation mitochondria until now were differential and density-gradient centrifugation. These methods are very labor intensive, requiring multiple runs, and separation of mitochondria sub-types is almost impossible. Using our pCFU technology, mitochondria and mitochondria sub-types were separated and highly enriched in a single step. The enrichment and relative quantitation of mitochondria in each fraction were assessed by Western blotting. Figure 3 shows the Western blot results of the mitochondrial distribution from adult and young bovine hearts. A total of 35 fractions were collected from each of the pCFU runs. Based on Western blot results, there were fourteen fractions from adult bovine heart and twelve fractions from young bovine heart containing mitochondria. Mitochondria were enriched three- to fourfold compared to the clarified homogenate before separation on pCFU. Similar mitochondria enrichment was also obtained from our previously reported study on rat liver mitochondria using pCFU.26 We observed that the relative intensities of the Western blot bands varied in both adult and young. In the adult Western blot analysis, the most intense band was fraction 9 (sucrose density = 1.175) and the second most intense band was fraction 12 (sucrose density = 1.157); in the young Western blot analysis, the most intense peak was fraction 13 (sucrose density = 1.147) and the next most intense peak was fraction 12 (sucrose density = 1.153). These Western blot results clearly illustrate the heterogeneity of mitochondria, which is consistent with the literature.27,28 This application demonstrated that pCFU has the ability to achieve both fractionation and enrichment of subcellular components at the level of organelle sub-types within the same process, which cannot be achieved using traditional methods.
FIGURE 3.
Western Blot analysis of equal protein amounts collected following continuous-flow ultracentrifugation of bovine heart homogenates (a) 18- to 30-month-old adult bovine heart (b) 3- to 8-month-old youth bovine heart. Samples were processed as described in Materials and Methods. Fractions selected for 2DE are marked with the arrows (fractions 9 and 12 in adult and 9 and 12 in the young bovine heart). d refers to sucrose density of the fraction.
Differential 2DE Analysis and Protein Identification by Nano-LC-ESI-MS/MS
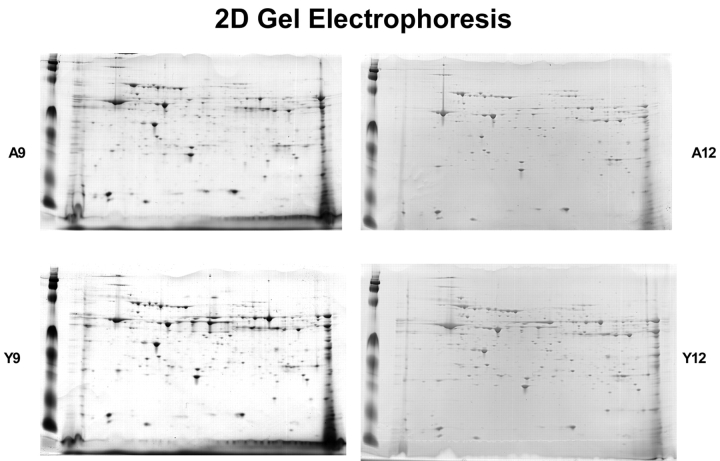
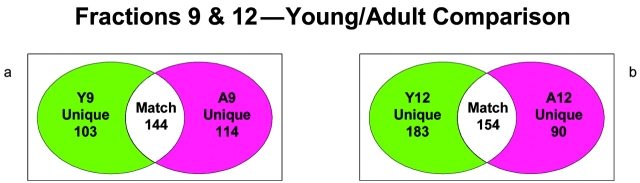
In order to examine age-related changes in the bovine heart mitochondrial proteome, two mitochondria-enriched fractions, A9 and A12 from adult bovine heart and Y9 and y12 from young bovine heart, were selected for two-dimensional gel electrophoresis analyses. A9 and Y9 have very similar sucrose densities, as do A12 and Y12. IPG strips with pH ranges of 3–10, 5–8, and 7–10 were used, and samples were run in triplicate. Figure 4 shows one group of 2D gels from adult fractions 9 and 12 (A9 and A12) and the corresponding young fractions 9 and 12 (Y9 and Y12). After analysis of 2D gel images, there were 144 matching spots in fraction 9 and 153 matching spots in fraction 12 (Figure 5) when comparing the two age groups. In fraction A9, there were 114 unique spots, and there were 90 unique spots in fraction A12. In fraction Y9, 103 unique spots were found, while 190 unique spots were found in Y12. The 2D gel image analysis results show a significant difference in the mitochondrial protein mapping profiles between the two age groups.
FIGURE 4.
2D gel electrophoresis of protein mixtures arising from fractions A9 (adult fraction 9), A12, Y9 (young fraction 9) and Y12 (see above). Samples were processed as described in Materials and Methods. Isoelectric focusing on these samples was performed using the broad pH range 3–10 and narrow range 5–8 and 7–10 strips. Above shown gels are 5–8.
FIGURE 5.
Comparison of 2D gels of protein mixtures arising from fractions 9 (a) and fractions 12 (b) of homogenate separations from young and adult bovine hearts. Isoelectric focusing on these samples was performed using the narrow pH range 5–8 IPG strips.
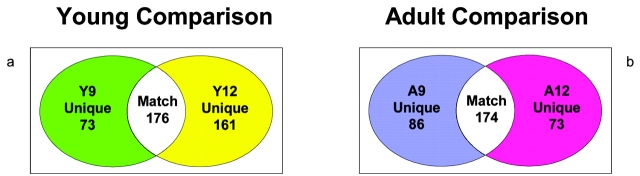
As previously discussed, pCFU achieves separation and enrichment at the organelle sub-type levels. Bovine heart mitochondria and sub-types were separated based on their shapes and densities as shown by Western blot analysis. To judge the heterogeneity of bovine heart mitochondria, two mitochondria-enriched fractions with similar sucrose densities from each age group were selected and analyzed by 2DE. Figure 6 illustrates the results of comparisons between and within age groups. Compared with the result from Figure 5, a higher number of matching spots were found within an age group than between age groups. These differences demonstrate the ability of pCFU to separate mitochondria with varied physical properties in one step.
FIGURE 6.
Numerical comparison of unique and matching gel spots from 2D gels of protein mixtures arising from fractions 9 and 12 of homogenate separations from young bovine heart (a) and from adult bovine heart (b). Isoelectric focusing on these samples was performed using the narrow pH range 5–8 IPG strips.
Preliminary comparisons of mitochondrial protein profiles between the adult and young bovine hearts were obtained by analyzing selected spots using nano-LC-ESI-MS/MS. The differences in protein expression levels were compared using a twofold cut-off value. Tables 1, 2, and 3 list the results of mass spectrometry analysis. Two selected fractions were resolved on pH 5–8 2D gels. From these 2D gels, 40 spots were selected for identification. Of these 40 spots, five unique proteins were identified in the adult that were not present in the young. Conversely, there were five unique proteins present in the young that were not in the adult. In addition, there were eight proteins found with at least a twofold difference in expression level between the adult and young bovine hearts. Among these identified proteins, there are important enzymes responsible for energy production and metabolism, a mitochondrial protein synthesis elongation factor, as well as proteins involved in the structural integrity of the organelles. Those oxidative modifications of the identified proteins could possibly contribute to the age-related degeneration and functional decline of heart proteins. Our results are consistent with the results of age-related increases in oxidative stress and free radical–dependent damage of proteins.30–32
TABLE 1.
Proteins Unique to Adult Bovine Heart Fractions 9 or 12
|
See Figure 5. Only selected 2D gel spots from pH 5–8 IPG strips were analyzed, using mass spectrometry as described in Materials and Methods.
TABLE 2.
Proteins Identified from 2D Gel Spots Unique to to Young Bovine Heart Fractions 9 or 12
|
See Figure 5. Only selected 2D gel spots from pH 5–8 IPG strips were analyzed, using mass spectrometry as described in Materials and Methods.
TABLE 3.
Proteins with Different Expression Levels in Fractions 9 or 12 that occurred in both Adult and Young Bovine Heart Fractions and showed at Least Twofold Increased Expression Levels
|
See Figure 5. Only selected 2D gel spots from pH 5–8 IPG strips were analyzed, using mass spectrometry as described in Materials and Methods.
CONCLUSION
It is known that mitochondria vary in structure and function. The current study has demonstrated a significant density-based heterogeneity in both young and adult bovine heart. pCFU resulted in separation of mitochondrial subtypes in both young and adult bovine heart. In addition, Western blot analysis indicates significant enrichment of total mitochondria vs. the clarified homogenate prior to separation. Comparisons of the 2D gel spots showed significant differences between adult and young mitochondrial subtypes within the same sucrose-density fraction. Also, significant differences were observed between subtypes within each age group. Matching spots showed significant percentages of unique proteins for each subtype in all comparisons. Protein identification data presented above represent only a small number of selected spots; it is thus expected that a significantly larger database can be generated through examination of all gel spots from all fractions at all pH ranges. The limitation of this technique is that endoplasmic reticulum and peroxisomal species may be present in some fractions due to their similar densities. This limitation is also present in traditional organelle separation methods.
REFERENCE
- 1.Chinnery PF, Howell, N, Andrews RM, Turnbull, DM. Clinical mitochondrial genetics. J Med Genet 1999;36:425–436. [PMC free article] [PubMed] [Google Scholar]
- 2.Hirano M, Davison M, DiMauro S. Mitochondria and heart. Curr Opin Cardiol 2001;16:201–210. [DOI] [PubMed] [Google Scholar]
- 3.Stadtman ER. Protein oxidation and aging. Science 1992;257:1220–1224. [DOI] [PubMed] [Google Scholar]
- 4.Behl C. Alzheimer’s disease and oxidative stress: Implications for novel therapeutic approaches. Prog Neurobiol 1999;57:301–323. [DOI] [PubMed] [Google Scholar]
- 5.Kovacic P, Jacintho JD. Mechanisms of carcinogenesis: focus on oxidative stress and electron transfer. Curr Med Chem 12001;8:773–796. [DOI] [PubMed] [Google Scholar]
- 6.Kalous M, Drahota Z. The role of mitochondria in aging. Physiol Res 1996;45:351–359. [PubMed] [Google Scholar]
- 7.Shigenaga M, Hagen T, Ames B. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA 1994;91:10,771–10,778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sastre J, Pallardo F, Garcia A, Vina J. Mitochondria, oxidative stress and aging. Free Rad Res 2000;32:189–198. [DOI] [PubMed] [Google Scholar]
- 9.Bakala H, Delaval E, Hamelin M, Bismuth J, Borot-Laloi C, Corman B. Changes in rat liver mitochondria with aging. Ion protease-like reactivity and N(epsilon)-carboxymethyllysine accumulation in the matrix. Eur J Biochem 2003;270:2295–2302. [DOI] [PubMed] [Google Scholar]
- 10.Cho YM, Bae SH, Choi BK, Song CW, Yoo JK, Paik YK. Differential expression of the liver proteome in senescence accelerated mice. Proteomics 2003;3:1883–1894. [DOI] [PubMed] [Google Scholar]
- 11.Modica-Napolitano JS, Singh KK. Mitochondria as targets for detection and treatment of cancer. Expert Rev Mol Med 2002;2002:1–19. [DOI] [PubMed] [Google Scholar]
- 12.Battersby BJ, Moyes CD. Influence of acclimation temperature on mitochondrial DNA, RNA, and enzymes in skeletal muscle. J Exp Biol 1998;201:2455–2460. [DOI] [PubMed] [Google Scholar]
- 13.Krysko DV, Roels F, Leybaert L, D’Herde K. Mitochondrial transmembrane potential changes support the concept of mitochondrial heterogeneity during apoptosis. J Histochem Cytochem 2001;49:1277–1284. [DOI] [PubMed] [Google Scholar]
- 14.Huber LA, Pfaller K, Vietor I. Organelle proteomics: Implications for subcellular fractionation in proteomics. Circ Res 2003;92:962–968. [DOI] [PubMed] [Google Scholar]
- 15.Fleischer S, Kervina M. Subcellular fractionation of rat liver. Methods Enzymol 1974;31:6–41. [DOI] [PubMed] [Google Scholar]
- 16.Palade GE. Cell fractionation: Important to cell-free systems development. Prog Clin Biol Res 1988;270: xix–xx. [PubMed] [Google Scholar]
- 17.Howell KE, Devaney E, Gruenberg J. Subcellular fractionation of tissue culture cells. Trends Biochem Sci 1989;14:44–47. [DOI] [PubMed] [Google Scholar]
- 18.Murayama K, Fujimura T, Morita M, Shindo N. Onestep subcellular fractionation of rat liver tissue using a nycodenz density gradient prepared by freezing-thawing and two-dimensional sodium dodecyl sulfate electrophoresis profiles of the main fraction of organelles. Electrophoresis 2001;22:2872–2880. [DOI] [PubMed] [Google Scholar]
- 19.Arnold R., Hrncirova P, Annaiah K, Novotny MV. Fast proteolytic digestion coupled with organelle enrichment for proteomic analysis of rat liver. J Proteome Res 2004;3:653–657. [DOI] [PubMed] [Google Scholar]
- 20.Chang J, Van Remmen H, Cornell J, Richardson A, Ward WF. Comparative proteomics: Characterization of a two-dimensional gel electrophoresis system to study the effect of aging on mitochondrial proteins Mech Ageing Dev 2003;124:33–41. [DOI] [PubMed] [Google Scholar]
- 21.Fountoulakis M, Berndt P, Langen H, Suter L. The rat liver mitochondrial proteins. Electrophoresis 2002;23: 311–328. [DOI] [PubMed] [Google Scholar]
- 22.Lescuyer P, Strub J, Luche S, Diemer H, Martinez P, Van Dorsselaer A, et al. Progress in the definition of a reference human mitochondrial proteome. Proteomics 2003;3:157–167. [DOI] [PubMed] [Google Scholar]
- 23.Journet A, Chapel A, Kieffer S, Roux F, Garin J. Proteomic analysis of human lysosomes: Application to monocytic and breast cancer cells. J Proteomics 2002;2: 1026–1040. [DOI] [PubMed] [Google Scholar]
- 24.Righetti P, Castagna A, Herbert B, Reymond F, Rossier JS. Prefractionation techniques in proteome analysis. Proteomics 2003;8:1397–1407. [DOI] [PubMed] [Google Scholar]
- 25.Rickwood, D Centrifugation: A practical approach. IRL Press, Oxford University 1992;77–138.
- 26.Drahos KL, Kiri AN, Lan W, Tran HC, McRorie DK, Horn MJ. Comparison of liver mitochondrial proteins from rats of different ages using a novel technique for fractionation and enrichment of organelles. A poster presented at the 44th annual meeting of the American Society for Cell biology, 2004, Washington, DC. Abstract number L304.
- 27.Gabaldon T, Huynen MA. Shaping the mitochondria proteome. Biochimica et Biophysica Acta 2004;1659: 212–220. [DOI] [PubMed] [Google Scholar]
- 28.Lopez M, Melov S, Applied proteomics: Mitochondrial proteins and effect on function Circ Res 2002;90:380–389. [DOI] [PubMed] [Google Scholar]
- 29.Anderson NG. Preparation zonal centrifugation. In Glick D (ed): Methods of Biochemical Analysis, 1st ed, vol XV. New York: Interscience Publishers, 1967:271–311. [DOI] [PubMed]
- 30.Judge S, Jang YM, Smith A, Hagen T, Leeuwenburgh C. Age-associated increases in oxidative stress and anti-oxidant enzyme activities in cardiac interfibrillar mitochondria: Implications for the mitochondrial theory of aging. FASEB J express article 10.1096/fj.04-2622fje, 2005. [DOI] [PubMed]
- 31.Barja G. Free radicals and aging. TRENDS in Neurosciences 2004;27:595–600. [DOI] [PubMed] [Google Scholar]
- 32.Kanski J, Behring A, Pelling J, Schoneich C. Proteomics identification of 3-nitrotyrosine-containing rat cardiac proteins: Effects of biological aging. Am J Physiol Heart Circ Physiol 2005;288:H371–H381. [DOI] [PubMed] [Google Scholar]
- 33.Higgins JA, Trout JM, Fayer R, Shelton D, Jenkins MC. Recovery and detection of Cryptosporidium parvum oocysts from water samples using continuous flow centrifugation. Water Res 2003;15:3551–3560. [DOI] [PubMed] [Google Scholar]
- 34.Caldwell SR, Varghese J, Puri NK. Large scale purification process for recombinant NS1-OspA as a candidate vaccine for Lyme disease. Bioseparation 1996;2:115–123. [PubMed] [Google Scholar]
- 35.Loewy, ZG. Separation and accumulation of subcellular components, and proteins derived thereform. International Application Published Under the Patent Cooperation Treaty (PCT). WO 2004; 083405.
- 36.Griffith OM. Techniques of Preparative, Zonal, and Continuous Flow Centrifugation, Beckman Instruments, Fullerton, CA, 1986:9.