Abstract
Ubiquitin is a member of the family of low-molecular-weight heat shock proteins that serve a vital role in physiological and pathological protein turnover. It appears to be one of the proteins involved in cell alterations during aging, degenerative disorders, and age-related cognitive decline. It is not known exactly how ubiquitin alterations are related to aging disorders; however, it is possible that ubiquitin is one of the target proteins for free-radical attack. In vivo, the free radical superoxide reacts with nitric oxide to form peroxynitrite, a powerful oxidant. Peroxynitrite may react directly with proteins, lipids, and other molecules to cause damage, with ubiquitin being a possible target. In vitro reaction of peroxynitrite with ubiquitin produces two modified forms of the protein, one oxidized at methionine and the other nitrated at tyrosine, which were characterized by electrospray ionization time-of-flight mass spectrometry. The exact location of the nitrated tyrosine residue was determined by in-source collision-induced dissociation using electrospray ionization time-of-flight mass spectrometry.
Keywords: Oxidative damage, aging, ubiquitin, 3-nitrotyrosine, ESI, TOF
Alzheimer’s disease (AD) is chronic common, progressive neurodegenerative disorder. Alzheimer (1907) originally described it in a 51-year old woman.1 Recently, oxidative stress has been proposed as a pathologic mechanism in Alzheimer’s disease.2,3 One mechanism of oxidative damage is the nitration of tyrosine residues in proteins, mediated by peroxynitrite breakdown to form 3-nitrotyrosine.4–7 Peroxynitrite can be generated by direct reaction of the two free radicals superoxide anion and nitric oxide under pathological circumstances.8–10 In turn, peroxynitrite can react with a wide range of biomolecules, resulting in peroxidation, oxidation, and/or nitration of proteins, lipids, and other molecules, and as a consequence cause their inactivation.11–16
Exactly how ubiquitin alterations are related to aging is not clear. Ubiquitin is a member of the family of low-molecular-weight, heat shock proteins that serve a vital role in physiological and pathological protein turnover.17–18 Ubiquitin appears to be one of the proteins involved in cell alterations during aging, degenerative disorders, and age-related cognitive decline, such as AD.19–21 Furthermore, researchers have shown evidence supporting a direct link between oxidative damage to the neuronal ubiquitination/deubiquitination machinery and the pathogenesis of sporadic AD.22 Several groups also reported that oxidative stress is associated with the alteration of ubiquitin.23–25 Since oxidative stress has been implicated in the pathogenesis of AD and other neurodegenerative disorders,26–29 it is useful to determine whether products of ubiquitin oxidation (e.g., by peroxynitrite) may be characterized by modern analytical techniques.
MATERIALS AND METHODS
Sample Preparation
Peroxynitrite (Calbiochem, La Jolla, CA) was obtained as an aqueous solution (170–200 mM) and stored frozen at −20°C until use. Ubiquitin (Sigma-Aldrich, St. Louis, MO) was stored at 4°C until use. Peroxynitrite was added (six aliquots of 2 μL each) to a stirred solution at 37°C of ubiquitin (5 mg) in Tris buffer (50 mM, 1 mL).30 A portion of the resulting solution was diluted 100-fold with 0.1% TFA and analyzed. Another portion of the product solution was denatured, digested using a trypsin digestion protocol, and analyzed. In a similar fashion, ubiquitin was analyzed in its native form and as a trypsin digest.
Instrumentation
All samples were analyzed using an LC/MSD TOF system (Agilent Technologies, Inc., Palo Alto, CA) with capillary pump, autosampler, column compartment, DAD, and TOF mass spectrometer.
LC Conditions
Chromatographic separation was achieved using a Zorbax Poroshell 300 SB-C18 column, 1.0 × 75 mm, 5-μm particle size (Agilent Technologies part # 661750-902) using a gradient (A = 0.1% formic acid in water, B = 0.1% formic acid in acetonitrile, 20% B to 100% B in 5.5 min) at 0.5 mL/min flow rate for intact protein analysis. The column temperature was set at 75°C. For protein digest analysis, the column was a Zorbax Extend-C18, 2.1 × 150 mm, 5-μm particle size (Agilent Technologies, part # 773700-902) at room temperature. The following gradient was used with flow rate at 0.5 mL/min: 0 min, 3% B; 5 min, 3% B; 5.01 min, 15% B; 25 min, 60% B; 30 min, 60% B; 45 min, 80% B; 50 min, 3% B; stop time 51 min, post time 4 min (A = 0.1% formic acid, B = 0.1% formic acid/methanol).
TOF Conditions
The TOF system used a combination of a dual-nebulizer ESI source and an automated calibrant delivery system to continuously introduce low-level reference masses to achieve accurate mass assignments. For intact protein analysis, the drying gas flow was set to 12 L/min, with gas temperature at 350°C; the nebulizer was set to 45 psig and capillary voltage was −4000 V; a fragmentor setting of 225 V was used with skimmer 60 V; the mass range was set to 300–3500 m/z with transients/scan equal to 10,000; and internal reference mass correction was used. For protein digests, the drying gas flow was set to 6 L/min, with gas temperature at 325°C; the nebulizer was set to 40 psig and capillary voltage to −4000 V; fragmentor voltage was cycled between 135, 225, and 375 V, with the skimmer at 60 V; the mass range was set to 100–3000 m/z, with the transient/scan equal to 8836; and internal reference mass correction was used.
Data Processing
Intact protein molecular weights were determined by deconvolution (Agilent Protein confirmation software). The protein digest data were searched using PMF search (Agilent Spectrum Mill MS Proteomics Workbench software) against the NCBInr database, including the variable modifications N-terminal pyroglutamate, oxidized methionine, and nitrotyrosine (user-defined). The peptide mass tolerance error was set to 5 ppm.
RESULTS AND DISCUSSION
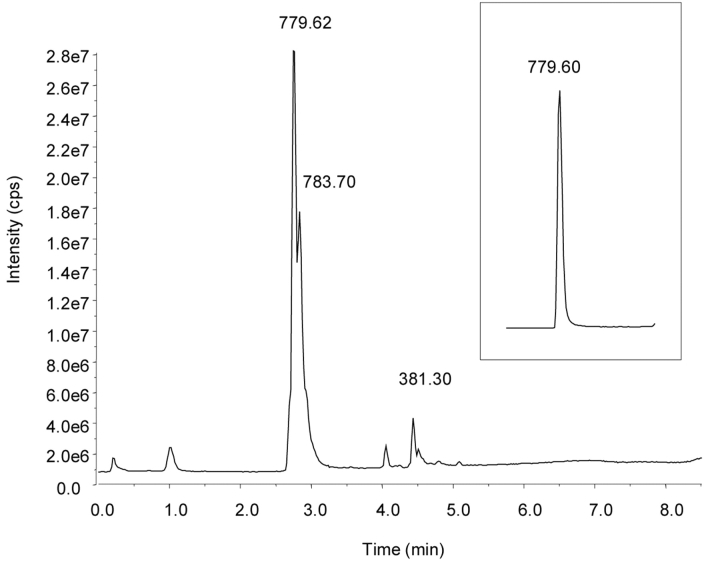
The results of these experiments provide definitive evidence of ubiquitin oxidation and nitration after peroxynitrite treatment, starting at the intact protein level and focusing more specifically down to the peptide and amino acid levels. The total ion chromatograms of peroxynitrite-treated ubiquitin and the native protein are shown in Figure 1. Manual deconvolution of the averaged spectra (Figure 2) indicated the presence of at least two additional proteins with the addition of 16 and 45 Da, consistent with an oxidized and nitrated species, respectively. These modified proteins essentially coeluted with the unreacted protein under the chromatographic conditions. The calculated molecular weight for all proteins yielded a value within 0.3 Da (0.003%) of the expected value.
FIGURE 1.
Total ion chromatograms of the reaction products of peroxynitrite and ubiquitin and for native ubiquitin (inset). The numbers over the peaks indicate the m/z value of the most abundant ion.
FIGURE 2.
Deconvoluted spectrum of the averaged spectrum of the reaction products of peroxynitrite and ubiquitin, showing the presence of an oxidized species (error 0.1 Da) and a nitrated species (error 0.3 Da).
Liquid chromatography mass spectrometry (LC/MS) analysis of the native and modified ubiquitin digests (Figure 3) indicates that many of the peptides were the same in the two samples, as evidenced by the peaks with the same retention time. However, two peaks at retention time 7.7 min and 11.9 min in the analysis of the native sample decreased in amount in the treated sample, and two new peaks formed at retention time 7.1 min and 11.2 min for the treated sample. PMF searching of this data (Figure 4) yielded excellent correlation of the experimental peptide molecular weights with the theoretical digest values, with an average mass error of less than 2 ppm for each sample. Sequence coverage was 68% and 73% for the native and peroxynitrite-treated ubiquitin, respectively, and ubiquitin was the only protein retrieved from the database. Comparing the results, modifications were found on two of the peptides, consistent with oxidation at methionine and nitration at tyrosine. These peptides corresponded to the new peaks in the nitrated sample, and the unmodified forms of these two peptides corresponded to the peaks that decreased in size between the two samples. The significant shifts in retention times for these modified peptides were consistent with those reported in the literature.30,31
FIGURE 3.
Total ion chromatograms of trypsin digests of native ubiquitin (top) and modified ubiquitin (bottom). The numbers over the peaks indicate the mass of the most intense ion of the peak. The unique peaks in each chromatogram are labeled with the identity from the PMF search.
FIGURE 4.
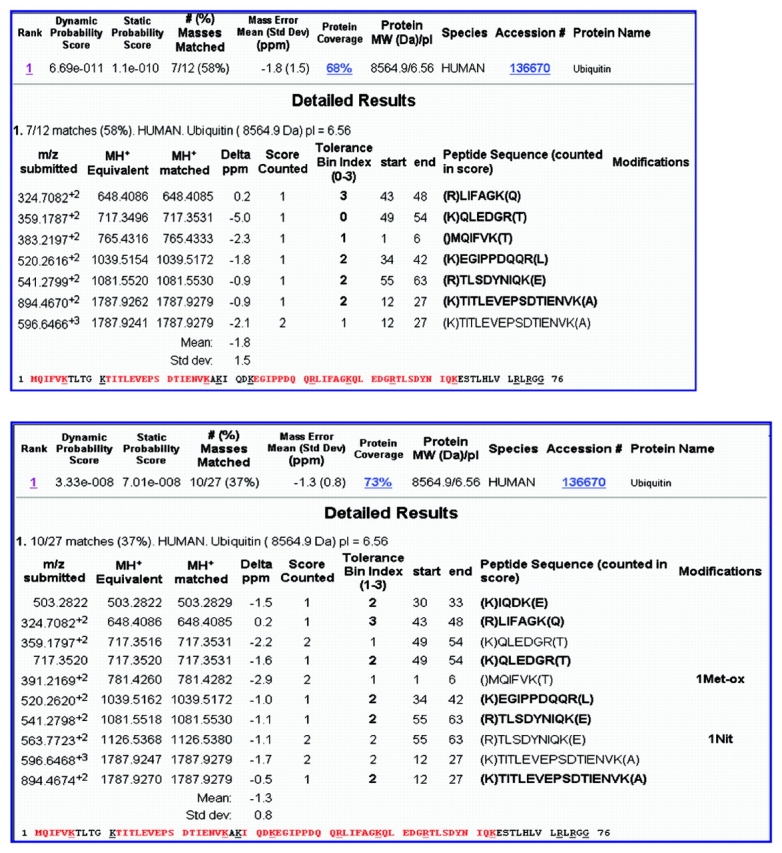
PMF search results of trypsin digests of native ubiquitin (top) and modified ubiquitin (bottom). The modifications were identified as oxidation of methionine and nitration of tyrosine (bottom). Sequence coverage was 68% and 73%, respectively.
The digest of the nitrated protein was also analyzed using programmable in-source CID (135, 225, 375 V fragmentor voltage). For the 225-V CID spectrum of the peptide identified as TLSD(nitroY)NIQK, several a-, b-, and y-series ions were identified (Figure 5; co-eluting compounds were also present). By comparing the measured m/z values with values from in silico fragmentation, the locus of nitration was unambiguously assigned to tyrosine, as all y-ions y5 and higher were shifted in m/z value by +45.0 Da compared to the theoretical collision-induced dissociation (CID) fragmentation of the peptide TLSDYNIQK (Table 1). Portions of the native and modified digest samples were also analyzed by ion trap MS/MS (Figure 6) and confirmed this assignment. This finding is also consistent with other work indicating that tyrosine residues are nitrated by peroxynitrite to form an end product 3-nitrotyrosine.8,9,16
FIGURE 5.
In-source CID-TOF MS of the peptide TLSD(nitroY)NIQK. Several diagnostic ions are labeled here and listed in Table 1. The larger abundance peaks are due to an unidentified coeluting compound.
TABLE 1.
Theoretical m/z Values for CID Fragments from Peptides TLSDYNIQK and TLSD(nitroY)NIQK Compared with the Experimentally Determined Values from a Spectrum Obtained at 225V Fragmentor Setting
| Ion | Native, Expected m/z | Nitrated, Expected m/z | Measured m/z |
| a2 | 187.14 | 187.14 | 187.1479 |
| b2 | 215.14 | 215.14 | 215.1430 |
| b3 | 302.17 | 302.17 | 302.1752 |
| b8 | 935.45 | 980.51 | n/a |
| y4 | 502.30 | 502.30 | 502.3049 |
| y5 | 665.36 | 710.36 | 710.3552 |
| y6 | 780.39 | 825.39 | 825.3843 |
| y7 | 867.42 | 912.42 | 912.4090 |
| y8 | 980.51 | 1025.51 | 1025.4915 |
| [M+2H]2+ | 541.28 | 563.77 | 563.7742 |
| [M+H]+ | 1081.55 | 1126.55 | 1126.5483 |
All ions y5 and higher for the nitrated peptide are shifted by +45.0 Da relative to the corresponding peptide from native ubiquitin, thus confirming nitration at tyrosine.
FIGURE 6.
Ion trap MS/MS spectra of the peptides TLSDYNIQK and TLSD(nitroY)NIQK, exhibiting the same CID fragment ions as for Figure 5 from the TOF instrument.
CONCLUSIONS
The reaction of peroxynitrite and ubiquitin in vitro produced at least two modified forms of the protein, one oxidized and one nitrated. The modified proteins were directly characterized by ESI-TOF, and their trypsin-digestion products were analyzed by ESI-TOF and PMF. The accurate mass capability of the ESI-TOF instrument combined with narrow PMF search criteria yielded unambiguous assignments for the modifications, one being oxidation at methionine, and the other nitration at tyrosine. The exact locus of the nitrotyrosine residue was pinpointed using ESI-TOF with in-source CID, even in the presence of coeluting potential interferences, and was confirmed using ion trap MS/MS.
Acknowledgments
The authors would like to thank Dr. Dana Hoch, MD, for the discussion of ubiquitin down-regulation and age-related cognitive decline.
REFERENCES
- 1.Fernandez-Vizarra P, Fernandez AP, Castro-Blanco S, Serrano J, Bentura ML, Martinez-Murillo R, et al. Expression of nitric oxide system in clinically evaluated cases of Alzheimer’s disease. Neurobiol Dis 2004;15: 287–305. [DOI] [PubMed] [Google Scholar]
- 2.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann NY Acad Sci 2001;928:22–38. [DOI] [PubMed] [Google Scholar]
- 3.Lahiri DK, Chen D, Ge, YW, Farlow M, Kotwal G, Kanthaswamy A, et al. Does nitric oxide synthase contribute to the pathogenesis of Alzheimer’s disease? Ann NY Acad Sci 2003;1010:639–642. [DOI] [PubMed] [Google Scholar]
- 4.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, et al. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch Biochem Biophys 1992;298:431–437. [DOI] [PubMed] [Google Scholar]
- 5.Pryor WA, Jin X, Squadrito GL. One- and two-electron oxidations of methionine by peroxynitrite. Proc Natl Acad Sci USA 1994;91:11,173–11,177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Good PF, Werner P, Hsu A, Olanow CW, Perl DP. Evidence for neuronal oxidative damage in Alzheimer’s disease. Am J Pathol 1996;49:21–28. [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeiffer S, Mayer B. Lack of tyrosine nitration by peroxynitrite generated at physiological pH. J Biol Chem 1998;273:27,280–27,285. [DOI] [PubMed] [Google Scholar]
- 8.Beckman JS, Chen J, Ischiropoulos H, Crow JP. Oxidative chemistry of peroxynitrite. Methods Enzymol 1994;233:229–240. [DOI] [PubMed] [Google Scholar]
- 9.Yi D, Smythe GA, Blount BC, Duncan MW. Peroxynitrite-mediated nitration of peptides: characterization of the products by electrospray and combined gas chromatography–mass spectrometry. Arch Biochem Biophys 1997;344:253–259. [DOI] [PubMed] [Google Scholar]
- 10.Beal MF. Oxidatively modified proteins in aging and disease. Free Radic Biol Med 2002;32:797–803. [DOI] [PubMed] [Google Scholar]
- 11.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite-induced membrane lipid peroxidation: The cytotoxic potential of superoxide and nitric oxide. Arch Biochem Biophys 1991;288:481–487. [DOI] [PubMed] [Google Scholar]
- 12.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem 1991;266:4244–4250. [PubMed] [Google Scholar]
- 13.Hu P, Ischiropoulos H, Beckman JS, Matalon S. Peroxynitrite inhibition of oxygen consumption and sodium transport in alveolar type II cells. Am J Physiol 1994;266:L628–L634. [DOI] [PubMed] [Google Scholar]
- 14.Tien M, Berlett BS, Levine RL, Chock PB, Stadtman ER. Peroxynitrite-mediated modification of proteins at physiological carbon dioxide concentration: pH dependence of carbonyl formation, tyrosine nitration, and methionine oxidation. Proc Natl Acad Sci USA 1999;96:7809–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi D, Ingelse BA, Duncan MW, Smythe GA. Quantification of 3-nitrotyrosine in biological tissues and fluids: Generating valid results by eliminating artifactual formation. J Am Soc Mass Spectrom 2000;11:578–586. [DOI] [PubMed] [Google Scholar]
- 16.Sarver A, Scheffler NK, Sheltlar MD, Gibson BW. Analysis of peptides and proteins containing nitrotyrosine by matrix-assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom 2001;12:439–448. [DOI] [PubMed] [Google Scholar]
- 17.Ciechanover A, Orian A, Schwarz AL. The ubiquitin-mediated proteolytic pathway: Mode of action and clinical implications. J Cell Biochem 2000;77(Suppl 34):40–51. [DOI] [PubMed] [Google Scholar]
- 18.Wang D-S, Bennett DA, Mufson EJ, Mattila P, Cochran E, Dickson DW. Contribution of changes in ubiquitin and myelin basic protein to age-related cognitive decline. Neurosci Res 2004;48:93–100. [DOI] [PubMed] [Google Scholar]
- 19.Shang F, Gong X, Palmer HJ, Nowell Jr. TR, Taylor A. Age-related decline in ubiquitin conjugation in response to oxidative stress in the lens. Exp Eye Res 1997;64:21–30. [DOI] [PubMed] [Google Scholar]
- 20.Bulteau AL, Szweda LI, Friguet B. Age-dependent declines in proteasome activity in the heart. Arch Biochem Biophys 2002;397:298–304. [DOI] [PubMed] [Google Scholar]
- 21.Konishi Y, Beach T, Sue LI, Hampel H, Lindholm K, Shen Y. The temporal localization of frame-shift ubiquitin-B and amyloid precursor protein, and complement proteins in the brain of non-demented control patients with increasing Alzheimer’s disease pathology. Neurosci Lett 2003;348:46–50. [DOI] [PubMed] [Google Scholar]
- 22.Choi J, Levey A, Weintraub ST, Weintraub ST, Rees HD, Gearing M, et al. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem 2004;279:13,256–13,264. [DOI] [PubMed] [Google Scholar]
- 23.Shang F, Taylor A. Oxidative stress and recovery from oxidative stress are associated with altered ubiquitin conjugating and proteolytic activities in bovine lens epithelial cells. Biochem J 1995;307:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang F, Gong X, Taylor A. Activity of ubiquitin-dependent pathway in response to oxidative stress. Ubiquitin-activating enzyme is transiently up-regulated. J Biol Chem 1997;272:23,086–23,093. [DOI] [PubMed] [Google Scholar]
- 25.Ramanathan M, Hassanain M, Levitt M, Seth A, Tolman JS, Fried VA, et al. Oxidative stress increases ubiquitin-protein conjugates in synaptosomes. Neuroreport 1999;10:3797–3802. [DOI] [PubMed] [Google Scholar]
- 26.Bowling AC, Beal MF. Bioenergetic and oxidative stress in neurodegenerative diseases. Life Sci 1995;56:1151–1171. [DOI] [PubMed] [Google Scholar]
- 27.Morrison BM, Hof PR, Morrison JH. Determinants of neuronal vulnerability in neurodegenerative diseases. Ann Neurol 1998;44:S32–S44. [DOI] [PubMed] [Google Scholar]
- 28.Multhaup G, Ruppert T, Schlicksupp A, Hesse L, Beher D, Masters CL, et al. Reactive oxygen species and Alzheimer’s disease. Biochem Pharmacol 1997;54:533–539. [DOI] [PubMed] [Google Scholar]
- 29.Christen Y. Oxidative stress and Alzheimer disease. Am J Clin Nutr 2000;71(Suppl):621S–629S. [DOI] [PubMed] [Google Scholar]
- 30.Sokolovsky M, Riordan JF, Vallee BL. Tetranitromethane. A reagent for the nitration of tyrosyl residues in proteins. Biochemistry 1966;5:3582–3589. [DOI] [PubMed] [Google Scholar]
- 31.Gevaert K, Van Damme J, Goethals M, Thomas GR, Hoorelbeke B, Demol H, et al. Chromatographic isolation of methionine-containing peptides for gel-free proteome analysis. Mol Cell Proteomics 2002;1:896–903. [DOI] [PubMed] [Google Scholar]