Abstract
RNA isolation from yeast is complicated by the need to initially break the cell wall. While this can be accomplished by glass bead disruption or enzyme treatment, these approaches result in DNA contamination and/or the need for incubation periods. We have developed a protocol for the isolation of RNA samples from yeast that minimizes degradation by RNases and incorporates two purification steps: acid phenol extraction and binding to a silica matrix. The procedure requires no precipitation steps, facilitating automation, and can be completed in less than 90 min. The RNA quality is ideal for microarray analysis.
Keywords: RNA isolation, yeast, Saccharomyces cerevisiae, silica matrix, microarray analysis
Microarray approaches to study gene expression patterns require highly purified RNA preparations, free of DNA, that can be efficiently converted to fluorescently tagged molecules.1 Degradation of RNA during and after the purification is a significant concern. Silica-based matrices allow for rapid and efficient purification of RNA in the presence of chaotropic ions that immediately denature RNases.2,3 These chaotropic reagents effectively lyse mammalian cells; however, yeast cell walls are not easily broken. Prior incubation of the yeast cells with lyticase aids in cell-wall disruption, but given that the average in vivo half-life for yeast mRNA is approximately 10 min,4,5 this incubation period will result in dramatic changes to the RNA profile. We have thus developed an RNA isolation protocol that takes advantage of the purification provided by the silica-based isolation matrices but does not require a preincubation step to digest the yeast cell wall.
METHODS AND RESULTS
We have adapted the protocol from the NucleoSpin RNA II kit (Macherey-Nagel, Germany) to allow rapid cell lysis and increased RNA purification. Saccharomyces cerevisiae are grown in rich media to exponential phase (A600 nm approximately 2.0). The culture is placed on ice for 5 min. Five milliliters of yeast cells (approximately 108) are harvested by centrifugation at 1000 g and washed with ice-cold water. The cell pellet is resuspended in 300 μL of RA1 buffer, provided by the manufacturer, and 0.3 g of 500-μm acid-washed glass beads (BioSpec Products, Inc., Bartlesville, OK) is added. The cells are vortexed at high speed for 2 min. During this step, the glass beads break the cells and, although heating occurs, RNases are efficiently denatured by the guanidine contained in the RA1 buffer. Glass beads and cell debris are pelleted by centifugation at 16,000 g for 5 min at 4°C. The aqueous phase is then vortexed with 500 μL of acid phenol (Fisher Scientific) for 10 sec, and incubated on ice for 5 min. Two hundred microliters of chloroform (EM Science, Gibbstown, WV) is added and the aqueous phase collected after centrifugation at 16,000 g for 5 min at 4°C. The chloroform extraction is then repeated. The acid phenol extraction further ensures denaturation of RNases and eliminates DNA, which remains in the phenol phase.3
To complete the purification of the RNA, 300 μL of RNase-free 70% ethanol is added to the aqueous phase. This solution can then be applied directly to the silica matrix in the NucleoSpin RNA II kit. Further steps proceed as described by the manufacturer (Macherey-Nagel, Easton, PA). The matrix is desalted, and then DNA is removed by treatment with DNase. The matrix is washed and RNA eluted with 40–60 μL of water.
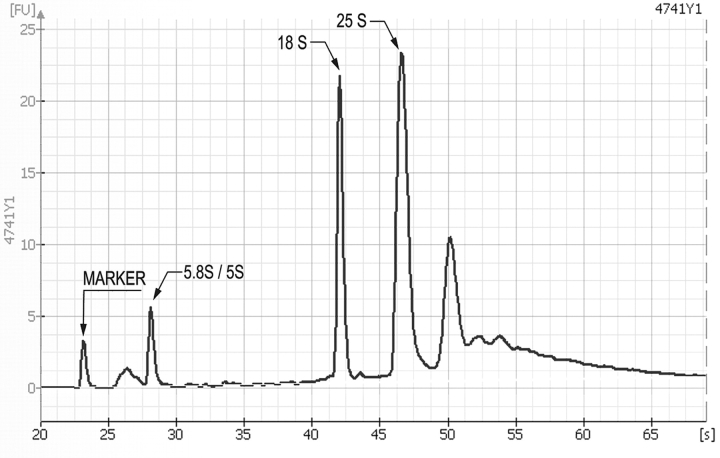
Using this procedure, we have isolated 25–30 μg of RNA from 108 cells. The A260/A280 ratio for six samples averaged 2.0 ± 0.1, indicating that the samples were free of protein. Samples were also analyzed on an Agilent 2100 Bioanalyzer (Figure 1). Based upon a rRNA [25S:18S] ratio of 1.9, we conclude that the RNA was intact. In addition, the RNA was found to be fully stable after storage at −70°C for 3 wks, after which it was efficiently labeled and used in microarray analyses.
FIGURE 1.
Electropherogram of an RNA sample purified from BY4741. The sample was analyzed on an Agilent 2100 Bioanalyzer, using an RNA 6000 Nano kit. The chip was first primed with gel matrix containing RNA dye. Each sample well was filled with marker before the sample was loaded. RNA migration through the capillary was detected by fluorescence of the RNA dye. The 5s/5.8s, 18s, and 25s peaks are visible at 27, 42, and 46 sec, respectively. rRNA[25s:18s] ratio is 1.9. The RNA was analyzed at Robarts Research Institute, London, ON, Canada.
DISCUSSION
This approach for purification of RNA from Saccharomyces cerevisiae has several features that make it advantageous for microarray analysis. The cells are rapidly broken by glass beads, and RNases are quickly inactivated in guanidine. The extraction with acid phenol aids in the rapid elimination of protein and DNA, though incubation with hot phenol is not required as it is in other protocols.3,6 Use of the silica matrix allows for additional purification and concentration of the RNA sample under conditions in which any RNase is inactive. The need for reagents such as diethylpyrocarbonate to inactivate RNases is unnecessary, allowing for maximal probe synthesis in subsequent steps. Finally, the protocol is rapid and allows efficient handling of multiple samples.
In developing this protocol, we tried changing the order of the steps. In initial trials, phenol extraction preceded guanidine extraction. While efficient cell lysis did occur, binding to the silica matrix was seriously compromised.
Acknowledgments
We thank Julie Genereaux for technical assistance, Megan Davey for comments on the manuscript, Cristina Dumitrescu for assistance with graphics, David Carter for RNA analysis, and Shaukat Rangwala (MOgene) for advice leading to microarray analyses. This work was supported by a Canadian Institutes of Health Research grant to CJB.
REFERENCES
- 1.Gasch AP. Yeast genomic expression studies using DNA microarrays. Methods Enzymol 2002;350:393–414. [DOI] [PubMed] [Google Scholar]
- 2.Krawetz SA, States JC, Dixon GH. Isolation and fractionation of total nucleic acids from tissues and cells. J Biochem Biophys Methods 1986;12:29–36. [DOI] [PubMed] [Google Scholar]
- 3.Wise JA. Preparation and analysis of low molecular weight RNAs and small ribonucleoproteins. Methods Enzymol 1991;194:405–415. [DOI] [PubMed] [Google Scholar]
- 4.Koch H, Friesen JD. Individual messenger RNA half-lives in Saccharomyces cerevisiae. Mol Gen Genet 1979;170:129–135. [DOI] [PubMed] [Google Scholar]
- 5.Santiago TC, Purvis IJ, Bettany AJ, Brown AJ. The relationship between mRNA stability and length in Saccharomyces cerevisiae. Nucleic Acids Res 1986;14:8347–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol 1991;194:398–405. [DOI] [PubMed] [Google Scholar]