Abstract
Aggrecanase activities of ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) proteinases were measured with a recombinant aggrecan fragment and two monoclonal antibodies. Recombinant human aggrecan interglobular domain was first incubated in the presence of ADAMTS enzymes. The aggrecan peptide with the N-terminal sequence ARGSVIL released upon hydrolysis was then quantified in an enzyme-linked immunosorbent assay (ELISA) with an anti-neoepitope antibody specific for the N-terminal ARGSVIL sequence and a second anti-aggrecan peptide antibody. For higher sensitivity of the assay, P1-P5 residues of the aggrecanase site within the aggrecan sub-strate were changed by in vitro mutagenesis. Specific activities of recombinant truncated ADAMTS1 and ADAMTS4 estimated with authentic aggrecan interglobular domain amounted to 2.4 ± 0.4 and 21.7 ± 9.5 nmoles hydrolyzed substrate/min·mg, respectively. The values were 10.3 ± 5.1 and 151.5 ± 93.5 nmoles/min·mg for hydrolysis of the modified substrate. The aggrecanase activity assay can be used for (1) kinetic characterization of aggrecanase activities of human and animal ADAMTS, (2) screening of inhibitors for aggrecan hydrolyzing ADAMTS, and (3) estimation of aggrecanase activities in biological samples.
Keywords: ADAMTS, aggrecanase, enzymatic assay, inhibitor screening, drug discovery
Aggrecan is a large proteoglycan present in cartilage, tendons, aorta wall, vertebrate discs, and the perineuronal net.1,2 The core protein folds into three globular domains: G1, G2, and G3. The first two domains are connected by a short 128–amino acid polypeptide referred to as the aggrecan interglobular domain (IGD), while the second and the third domain are separated by a 1491–amino acid sequence carrying a great number of sulfated glucosaminoglycan chains.3,4 In cartilage, aggrecan forms aggregates with link protein and hyaluronan. The aggregates, with their high negative charge, attract water molecules and endow cartilage with resistability to compression and deformation.
Excessive degradation of aggrecan is an early event in cartilage destruction.5,6 Aggrecan loss in osteoarthritis and other joint diseases is mainly due to the activity of glutamyl-specific multidomain metalloproteinases of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family. Among 19 human ADAMTS, six enzymes—ADAMTS1,7 ADAMTS4,8 ADAMTS5,9 ADAMTS8,10 ADAMTS9,11 and ADAMTS1512—hydrolyze aggrecan. A more detailed characterization of aggrecan degradation by ADAMTS4 and ADAMTS5 revealed five cleavage sites in vitro and in vivo.8,9 Four sites are located in the chondroitin sulfate–rich region between aggrecan globular domains G2 and G3 (sites E1480–G1481, E1667–G1668, E1771–A1772, E1871–L1872), and one site is situated in the interglobular domain between G1 and G2 (E373–A374). ADAMTS1, ADAMTS8, ADAMTS9, and ADAMTS15 likewise hydrolyze aggrecan at several sites, including the unique site within the interglobular domain.
Two other large proteoglycans, versican and brevican, are also hydrolyzed by enzymes of the ADAMTS family. Versican is degraded by ADAMTS1, ADAMTS4,13 and ADAMTS9,9 and brevican is cleaved by ADAMTS4.14
Quantitative estimation of aggrecanase activity of ADAMTS has been hampered by the fact that short peptides are not readily hydrolyzed by the enzymes. In addition to specific cleavage sites, secondary interaction sites have to be present in the substrates for effective hydrolysis.15 Purified aggrecan or aggrecan fragments were therefore preferred as substrates, whereby hydrolysis products of native aggrecan had first to be deglycosylated. They were then separated by electrophoresis, blotted, and quantified with neoepitope antibodies.16 Sequences from aggrecan IGD were used in two different methods. In the first method, the aggrecan-IGD sequence was fused to human IgG1 constant region. Cleavage of this substrate by aggrecanase generated the N-terminal sequence ARGSVIL. The ARGSVIL polypeptide was quantified in a sandwich ELISA with an anti-neoepitope antibody (antibody BC-3) and an antibody against human IgG1 constant region.17 A disadvantage of the fusion substrate is that it includes a rather large part of the human IgG1 sequence, which excludes estimation of aggrecanase activity in human body fluids. In the second method, a biotinylated synthetic polypeptide consisting of aggrecan residues Q354–L394 was used as substrate. The polypeptide was bound to streptavidin-coated microtiter plates. Nascent ARGSVIL sequences arising from hydrolysis by ADAMTS4 were targeted again with the anti-neoepitope antibody BC-3 and then quantified with a secondary peroxidase (POD)-conjugated antibody. As indicated by the authors, the reaction had to be carried out with the substrate pre-fixed to streptavidin plates. The reported Km value was an unfavorably high 0.48 mM.18
The present work describes an improved method for aggrecanase activity determination. The samples to be investigated are first incubated with recombinant aggrecan IGD. After hydrolysis, the larger poly-peptide is quantified with two novel monoclonal antibodies—an anti-ARGSVIL neoepitope antibody and an aggrecan sequence antibody. Further improvement is achieved with an engineered aggrecan IGD, which is cleaved more effectively by aggrecanases than authentic aggrecan IGD. The higher sensitivity allows measurement of aggrecanase activity in biological samples.
MATERIALS AND METHODS
Materials
ADAMTS1 cDNA was amplified from human brain cDNA (BD Biosciences Clontech). cDNA for ADAMTS4 was obtained from Kazusa DNA Research Institute (Chiba, Japan), and cDNA for ADAMTS5 was kindly provided by Dr. Kern (Sanofi Aventis). BaculoGold linearized baculovirus DNA and pVL1392 transfer vector were from BD Biosciences Pharmingen. Restriction enzymes, T4 DNA ligase, and Vent DNA polymerase were from New England Biolabs. Ex-Cell 401 insect serum-free medium was purchased from JRH Bosciences. POD-labeled goat anti-mouse IgG was obtained from Dianova GmbH. The ECL-detection kit was from Amersham Life Sciences, and Talon metal affinity resin was from BD Biosciences Clontech. MMP-14 catalytic domain and MMP-13 were from Invitek GmbH. Other chemicals were from Sigma.
Human osteoarthritic synovial fluid (SF) was obtained from the Orthopädische Klinik Berlin-Buch. Samples were centrifuged to remove cells and stored frozen at −80°C until use.
Expression and Purification of Truncated ADAMTS1, ADAMTS4, and ADAMTS5
For expression of truncated ADAMTS1, PCR was performed with forward primer 5′-AACTGCAGAT-GGGGAACGCGGAGCGG GCTC containing a Pst I site and reverse primer 5′-GC TCTAGATTAATGATGAT-GATGATGATGATTGCTTGGACAGTCCTC AAGGTTA containing an Xba I site. The resulting cDNA of 1.9 kb codes for ADAMTS1 residues 1–617 followed by a C-terminal His tag. By analogy, cDNA for ADAMTS4 was amplified with primers 5′-ATAGAGCGGCCGCATGTCCCAGACAGGCTCGCATCC and 5′-CCGCTCTAGAATTAGTGATGATGATGATGATGGGCTGAGC-CAGTTGGG CA, and cDNA for truncated ADAMTS5 was amplified with the primer pair 5′-ATAGAGCGGCCGCATGCTGCTCGGGTGGGC and 5′-AAGCTCTAGATTAATGATGATGATGATGATGACCATTGGGT-GGGCAGGGC.
Primers for ADAMTS4 contain a Not I site and an Xba I site, respectively. The cDNA amplified with these oligonucleotides codes for ADAMTS4 residues 1–579 followed by a C-terminal His tag. Primers for ADAMTS5 likewise contain a Not I site and an Xba I site. The amplified cDNA codes for ADAMTS5 residues 1–625 and a His tag.
The three cDNAs were cut with respective restriction enzymes and then ligated into the baculovirus tranfer vector pVL1392 to obtain plasmids pVL-ADAMTS1trunc, pVL-ADAMTS4trunc, and pVL-ADAMT-S5trunc. Recombinant baculoviruses were generated by cotransfection of tranfer plasmids with BaculoGoldTMDNA according to the instructions of the manufacturer. Isolated viruses were amplified and used to produce recombinant proteins. Sf9 insect cells were infected with the viruses in serum-free Ex-Cell 401 medium at an MOI equal to 5. After 3 d at 27°C, cultures were centrifuged 12 min at 6000 g. Proteins in the supernatants were precipitated with ammonium sulfate (476 g/L) in the cold. The precipitates were collected by centrifugation for 15 min at 10,000 g and solubilized in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM CaCl2, 0.05% Brij 35 (buffer A). Protein solutions were applied to Talon (3 mL Talon/L culture supernatant). After extensive washing with buffer A, His-tagged ADAMTS proteins were eluted from Talon with 50 mM imidazole in buffer A. N-terminal amino acid sequences of purified active ADAMTS1 and ADAMTS4 were confirmed by N-terminal sequencing.
Expression and Purification of Aggrecan IGD, Aggrecan IGD-s, and TVK Protein
cDNA for aggrecan IGD was amplified with primers 5′-CATACCATGGCCACCGCAGAAGACTTTGTG-GACATC and 5′-CCTCTCGAGGTGATGGTGATGGT-GATGTCCCCCTGGCAAATGCGGCTG. The amplified cDNA codes for aggrecan residues T331–G458. The forward primer introduces an Nco I site and the reverse primer prolongs aggrecan-IGD cDNA with 6 codons for His residues and an Xho I site. The cDNA was ligated into a modified pET19 vector (Novagen), and the vector was transfected into E. coli BL21(DE3). Recombinant bacteria were grown at 37°C to a density of OD600 = 1 and then induced with 1 mM IPTG for 3.5 h at 20°C. Aggrecan IGD was purified from IPTG-induced bacteria by Talon chromatography. Bacteria from a 2-L culture were homogenized in lysis buffer (1 mg/mL lysozyme, 1 mM PMSF, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 5% ethyleneglycol, 1 mM imidazole, 0.3 M NaCl, 50 mM Tris-HCl [pH 8.0]). The suspension was centrifuged for 20 min at 50,000 g and the clear supernatant was applied to Talon (1 mL Talon for lysates from a 2-L bacterial culture). After extensive washing with buffer A, aggrecan IGD was eluted with buffer A containing 100 mM imidazole. Aggre-can IGD was further fractionated by gel filtration on a Hiload 16/60 Superdex 75 prep grade column equilibrated in buffer A. Two fractions of different elution volume were separated and denoted aggrecan IGD1 and aggrecan IGD2.
In addition to aggrecan IGD, two variants were produced: Aggrecan IGD-s and TVK protein. In aggrecan IGD-s, the amino acid sequence around the aggrecana-se site was changed from PRNITEGE ARGSVIL to PTS-FKEEE ARGSVIL. In vitro mutagenesis was carried out by the method of overlap extension using oligonucleotides 5′-CCTACTAGTTTTAAGGAGGAAGAAGCCC-GAGGCAGCGTG and 5′-TTCTTCCTCCTTAAAAC-TAGTAGGCAGTGGCAGCTCCAT.
TVK protein consisted of aggrecan residues T381 . . . G458. This sequence represents the C-terminal part of aggrecan IGD downstream from the aggrecanase site. cDNA for TVK protein was amplified with forward primer 5′-CATACCATGGCCACCGTA-AAGCCCATCTTCGAGG and the same reverse primer as used for amplification of aggrecan IGD.
Recombinant expression and purification of aggrecan IGD-s and TVK protein on Talon were carried out as described above for aggrecan IGD. Aggrecan IGD-s was further fractionated on Superdex 75 again as for aggrecan IGD. Two distinct fractions of different elution volumes were designated aggrecan IGD-s1 and aggrecan IGD-s2.
Hydrolysis of Aggrecan IGD and Aggrecan IGD-s by ADAMTS1 and ADAMTS4
Recombinant aggrecan IGD (10 μM) or aggrecan IGD-s (10 μM) was incubated at 37°C in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM CaCl2 with 0.4 μM truncated ADAMTS1 or 0.1 μM truncated ADAMTS4. After defined time intervals, aliquots were withdrawn from the reaction mixtures and analyzed by SDS-PAGE.
Preparation of ARGSVIL Peptides
For preparation of ARGSVIL peptide 1 and ARGSVIL peptide 2, 150 μg/mL of aggrecan IGD1 and aggrecan IGD2, respectively, were incubated for 2 h at 37°C with 5 μg/mL truncated ADAMTS4 in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM CaCl2, 1 μM leupeptin, 1 μM pepstatin, 1 mM Pefabloc, 0.05 % Brij 35. Complete digestion of substrates was controlled by SDS-PAGE. Reactions were stopped and diluted with 30 mM Tris-HCl (pH 7.5), 144 mM NaCl, 1.8 mM CaCl2, 6 mM EDTA, 0.6 % BSA, 0.38 μM pepstatin, 0.38 μM leupeptin, 0.16 mM Pefabloc, 0.01 % Brij 35.
ARGSVIL peptide s1 and ARGSVIL peptide s2 were obtained analogously by digestion of aggrecan IGD-s1 and aggrecan IGD-s2 with truncated ADAMTS4.
Antibodies against Aggrecan Neoepitope ARGSVIL and Aggrecan-IGD Sequence
For quantitative determination of the larger C-terminal polypeptide produced from aggrecan IGD by aggrecanases, two monoclonal antibodies were produced: (1) an antibody recognizing the N-terminal sequence ARGSVIL and (2) an antibody binding to the aggrecan-IGD sequence C-terminal from the aggrecanase site.
The first antibody was raised against the synthetic peptide ARGSVILTGC coupled via the Cys residue to KLH (keyhole limpet hemocyanin). To obtain the second antibody, TVK protein was used as antigen. Immunizations with both antigens, cell fusions, and hybridoma selections were carried out in accordance with standard protocols. Several antibody-producing hybridomas were obtained with both antigens. Among the anti-N-terminal ARGSVIL antibodies, one antibody of immunoglobulin subtype IgG1 performed well in Western blots and ELISA with ADAMTS4-digested aggrecan IGD. A dissociation constant of Kdiss = 10−8 M was determined for the complex of the antibody with ARGSVIL peptide in a competition ELISA.
Different antibodies against TVK protein were then tested for their ability to bind simultaneously with the selected ARGSVIL antibody to ARGSVIL peptides. The best-performing antibody was selected and labeled with horseradish peroxidase according to a standard procedure.
Western Blots
Proteins were separated in 12% polyacrylamide gels and then electrophoretically transferred to nitrocellulose membranes. The membranes were blocked with 0.36% gelatin in 50 mM Tris-HCl (pH 7.4), 200 mM NaCl, 0.1% Tween-20. They were then incubated for 1.5 h with mouse monoclonal antibody against the N-terminal sequence ARGSVIL (0.5 μg antibody/mL 1 % BSA, 20 mM Na phosphate [pH 7.4], 150 mM NaCl, 0.004% Tween-80). Blots were washed and subsequently incubated with peroxidase-labeled goat anti-mouse IgG. Peroxidase activity was detected with the ECL system of Amersham Biosciences.
ELISA for ARGSVIL Peptides
ARGSVIL peptides were estimated with anti-ARGSVIL antibody and POD-labeled aggrecan sequence antibody. Microtiter plate wells were coated overnight with 100 μL solution of 4 μg anti-ARGSVIL antibody/mL 15 mM Na2CO3, 35 mM NaHCO3. Unspecific binding sites were blocked for 1 h at room temperature with 1% BSA, 0.15 M NaCl, 20 mM Na-phosphate buffer (pH 7.2). Plates were then washed two times with wash buffer (0.15 M NaCl, 0.05% Tween-20, 20 mM Na-phosphate [pH 7.2]). The ELISA was started by pipetting 100-μL aliquots of ARGSVIL peptides into wells. After a 1.5-h incubation, plates were washed three times with wash buffer. Aliquots of 100 μL peroxidase-labeled aggrecan sequence antibody were then added and plates were incubated again for 1.5 h. Thereafter, plates were washed five times. Peroxidase reactions were initiated by addition of 100 μL TMB solution. The reactions were allowed to proceed for 30 min. They were stopped with 100 μL 0.25 M H2SO4. Absorbance was read at λ = 450 nm with a reference filter at λ = 620 nm.
Aggrecanase Activity Assay
Various concentrations of truncated ADAMTS1, ADAMTS4, or ADAMTS5 were incubated for 15 min at 37°C with 0.1 μM aggrecan IGD in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM CaCl2, 1 μM leupeptin, 1 μM pepstatin, 1 mM Pefabloc, 10 μM ZnCl2, 0.05% Brij 35. The total reaction volumes were 100 μL. Reactions were stopped by addition of 150 μL 10 mM EDTA, 20 mM Na-phosphate (pH 7.2), 150 mM NaCl, 1% BSA. The mixtures were subsequently analyzed with the ELISA described above. Calibration curves for standard ARGSVIL peptide were run in parallel, and the amounts of ARGSVIL peptide produced in hydrolytic reactions were calculated from the calibration curves.
Sensitive Aggrecanase Assay
In the sensitive aggrecanase activity assay, 0.1 μM aggrecan IGD-s was used as substrate. Other conditions were as described above for the standard assay.
RESULTS
Expression and Purification of Truncated ADAMTS1, ADAMTS4, and ADAMTS5, and of Aggrecan Interglobular Domain
Truncated ADAMTS1, ADAMTS4, and ADAMTS5 were expressed in Sf9 insect cells. The secreted enzymes each consisted of a catalytic, a disintegrin, and a thrombospondin type-1 domain followed by a His tag. The precise amino acid sequences of truncated enzymes were ADAMTS1 (F253–N617)–6His, ADAMTS4 (F 213–A573)–6His, and ADAMTS5 (S262–G625)–6His, with numbers in parentheses referring to amino acid residues in full-length enzymes. The three sequences have calculated Mrs of 41,017 Da, 40,366 Da, and 40,804 Da, respectively.
Recombinant ADAMTS1 and ADAMTS4 were purified from insect cell-culture supernatants by Talon chromatography. As judged by SDS-PAGE, the final preparations contained greater than 90% truncated ADAMTS1 and ADAMTS4, respectively (Figure 1). ADAMTS5 was expressed at lower levels than ADAMTS1 and ADAMTS4. The enzyme could not be enriched in a single chromatography step to the same extend as ADAMTS1 and ADAMTS4. Fractions eluted from Talon contained only about 20–30% ADAMTS5 protein as judged from SDS-PAGE (not shown).
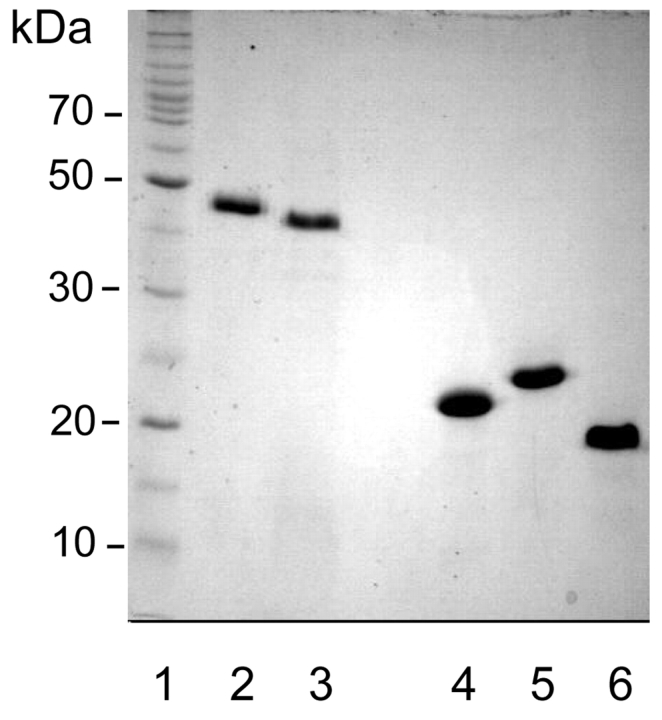
FIGURE 1.
SDS-PAGE of recombinant ADAMTS and aggrecan fragments. Two-microgram quantities of purified protein preparations were separated in 12% acrylamide gels and stained with Coomassie Brilliant Blue. 1 = marker proteins, 2 = truncated ADAMTS1, 3 = truncated ADAMTS4, 4 = aggrecan IGD, 5 = aggrecan IGD-s, 6 = TVK protein.
Recombinant aggrecan IGD and a variant with an engineered aggrecanase site, aggrecan IGD-s, were expressed in E. coli and purified from E. coli lysates. The calculated Mrs of the two proteins are 15,493 Da and 15,544 Da. However, in SDS-PAGE the fragments migrated with Mrs of about 21 kDa and 23 kDa when compared to globular marker proteins (Figure 1). Slow migration in electrophoresis is most probably due to the extended structures of the two proteins.
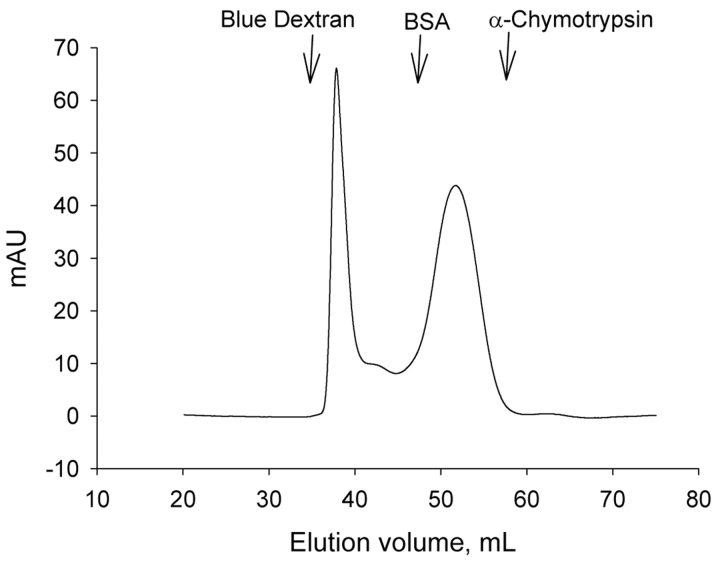
When purified aggrecan IGD was subjected to gel filtration on Superdex 75, the protein was separated into two fractions (Figure 2). One fraction appeared in the flow-through eluate, while the other fraction eluted like a globular protein of Mr 40 kDa. The different forms of aggrecan IGD contained in the two fractions were designated aggrecan IGD1 and aggrecan IGD2, respectively. When separated fractions were re-chromatographed on Superdex 75, each eluted in the same volume as before, with little indication of a second fraction. This experiment indicated that there is no rapid interconversion between the two forms of aggrecan IGD. Since the two fractions showed identical behavior in SDS-PAGE, it is assumed that aggrecan IGD in the flow-through eluate is an aggregate that is dissociated by SDS, and aggrecan IGD in the second fraction is a monomer form. As will be outlined below, aggrecan IGD1 and aggrecan IGD2 are hydrolyzed with comparable rates by aggrecanases. However, they behave differently in a sandwich ELISA with monoclonal antibodies.
FIGURE 2.
Gel filtration of aggrecan IGD on HiLoad 16/60 Superdex 75 pg. A solution of 4 mg aggrecan IGD/mL was loaded onto a HiLoad 16/60 Superdex 75 pg column preequilibrated in 50 mM Tris-HCL (ph 7.5), 150 mM NaCL, 5 mM CaCl2. Chromatography was carried out with an Äkta purifyer and the flow rate was 1 mL/min. Molecular-weight markers were Blue Dextran, bovine serum albumin (Mr 67 kDa), and bovine α-chymotrypsin (25 kDa).
Purified aggrecan IGD-s was separated on Superdex 75 into two fractions in the same way as was aggrecan IGD (experiments not shown). The two forms of the modified substrate were named aggrecan IGD-s1 and aggrecan IGD-s2.
Throughout this paper, aggrecan IGD refers to the nonseparated mixture of aggrecan IGD1 and aggrecan IGD2, and aggrecan IGD-s indicates the mixture of aggrecan IGD-s1 and aggrecan IGD-s2.
Hydrolysis of Aggrecan IGD and Aggrecan IGD-s by ADAMTS1, ADAMTS4, and ADAMTS5
Aggrecan IGD, aggrecan IGD-s, and their constituent forms separated by gel filtration are hydrolyzed by truncated ADAMTS1, ADAMTS4, and ADAMTS5. Figure 3A demonstrates hydrolysis of aggrecan IGD1 and aggrecan IGD2 by ADAMTS4, and in Figure 3B hydrolysis of aggrecan IGD1 and hydrolysis of aggrecan IGD-s1 by the same enzyme are compared.
FIGURE 3.
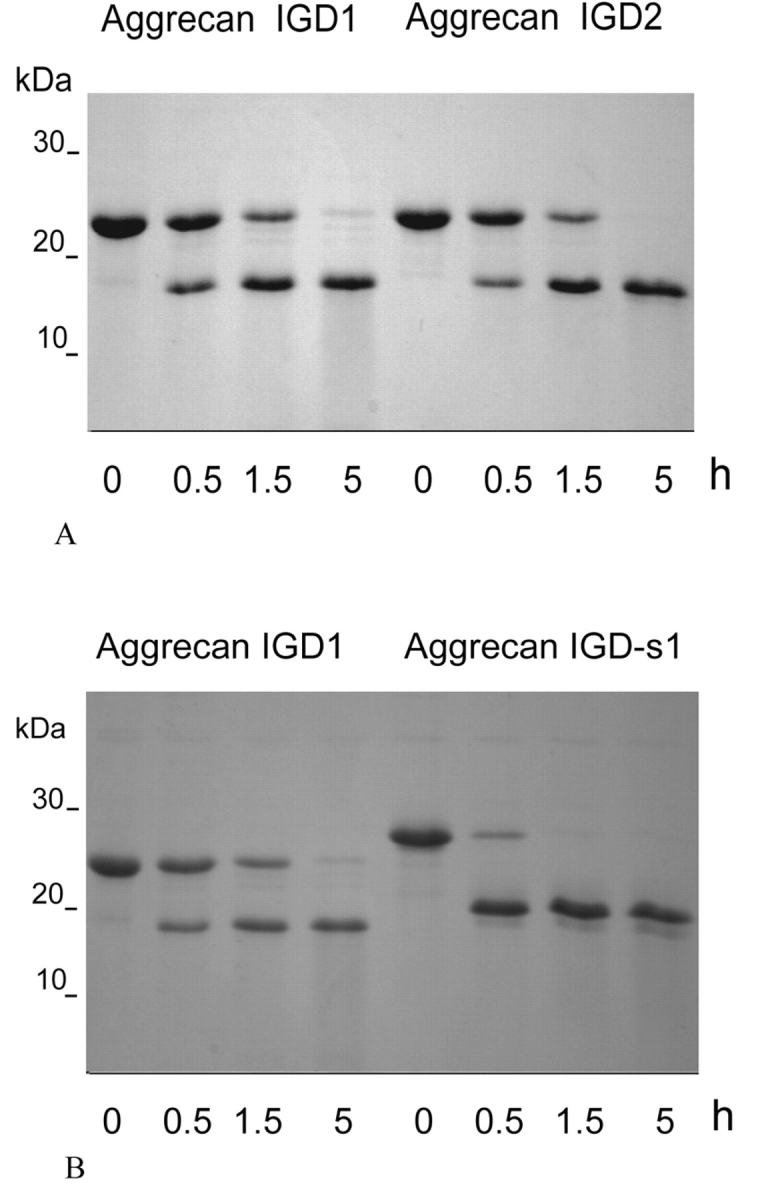
Proteolysis of recombinant aggrecan IGD and aggrecan IGD-s in the presence of truncated ADAMTS4. 10 μM aggrecan IGD1, aggrecan IGD2, or aggrecan IGD-s1 were incubated at 37°C with 0.1 μM truncated ADAMTS4 in 50 mM Tris-HCL (ph 7.5), 150 mM NaCl, 5 mM CaCl2. After various time intervals, 12-μL aliquots were withdrawn from the reaction mixtures and analyzed by SDS-PAGE. A: Proteolysis of 10 μM aggrecan IGD1 and aggrecan IGD2. B: Proteolysis of 10 μm aggrecan IGD1 and aggrecan IGD-s1.
One product of the hydrolysis reaction is a polypeptide of apparent Mr 16 kDa. N-terminal sequencing of the polypeptide confirmed the expected sequence ARGSVIL. The polypeptides released from aggrecan IGD and from aggrecan IGD-s consisted of the same amino acid sequence. However, for unambiguousness the polypeptide released from aggrecan IGD was named ARGSVIL peptide (subtypes ARGSVIL peptide-1 and ARGSVIL peptide-2) and the polypeptide produced from aggrecan IGD-s was designated ARGSVIL peptide-s (subtypes ARGSVIL peptide-s1 and ARGS-VIL peptide-s2).
It is evident from Figure 3B that truncated ADAMTS4 hydrolyzes aggrecan IGD-s1 more effectively than aggrecan IGD1. Under the specified conditions, hydrolysis of aggrecan IGD-s1 is nearly complete within 0.5 h, while less than half of aggrecan IGD1 is hydrolyzed within this time. Similarly, aggrecan IGD-s2 is degraded faster than aggrecan IGD2. Accelerated hydrolysis of aggrecan IGD-s as compared to aggrecan IGD was also observed in the presence of either truncated ADAMTS1 or truncated ADAMTS5 (experiments not shown).
ELISA for ARGSVIL Peptides
For quantification of ARGSVIL peptides, two monoclonal antibodies binding simultaneously to the peptides were produced. The first antibody specifically recognized the neoepitope N-terminal sequence ARGSVIL. This antibody was named anti-N-terminal ARGSVIL neoepitope antibody. The second antibody binds at some distance from the N-terminus, not interfering with binding of the neoepitope antibody. This antibody is designated aggrecan sequence antibody.
The neoepitope antibody should not exhibit significant affinity for ARGSVIL sequences within polypeptide chains if it is used to quantify ARGSVIL peptides in the presence of substrates aggrecan IGD or aggrecan IGD-s. The discriminating properties of the neoepitope antibody were proved in Western blots and in ELISA experiments.
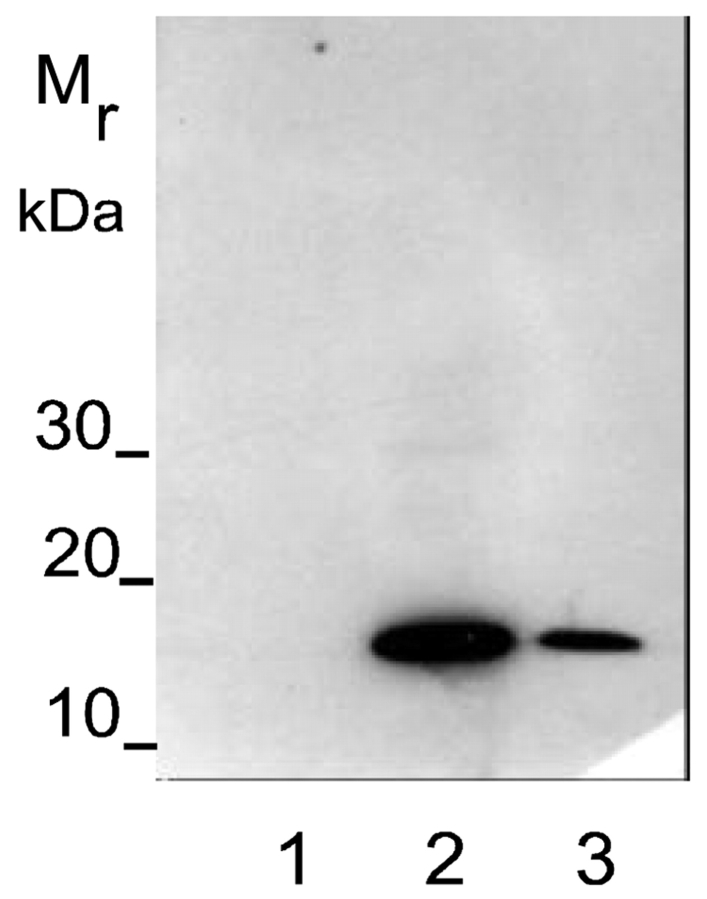
Figure 4 demonstrates heavy staining of 100 ng and 25 ng ARGSVIL peptide in a Western blot following incubation of the blot with neoepitope antibody and a POD-labeled secondary antibody. Under the same conditions, 100 ng aggrecan IGD were not stained at all.
FIGURE 4.
Western blot of recombinant aggrecan IGD and ARGSVIL peptide with anti-neoepitope antibody (anti-N-terminal-ARGSVIL antibody). Proteins were subjected to electrophoresis, blotted, and stained as described under Materials and Methods. Lane 1 = 100 ng aggrecan IGD, lane 2 = 100 ng ARGSVIL peptide, lane 3 = 25 ng ARGSVIL peptide.
ELISA experiments with the two separated forms of aggrecan IGD and corresponding ARGS-VIL peptides are demonstrated in Figure 5. Figure 5A gives data for substrates aggrecan IGD1 and aggrecan IGD2, while Figure 5B presents data for the hydrolysis products ARGSVIL peptide-1 and ARGS-VIL peptide-2.
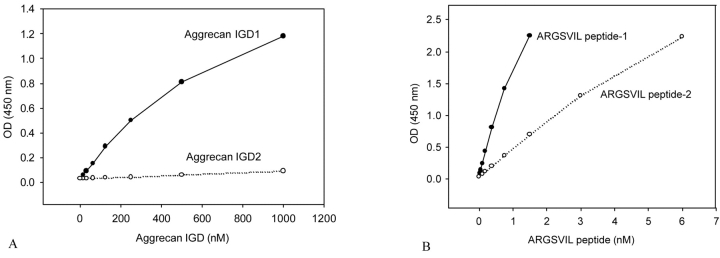
FIGURE 5.
ELISA for aggrecan IGD and ARGSVIL peptide. The ELISA was carried out with neoepitope and aggrecan sequence antibodies as described under Materials and Methods. A ELISA : for aggrecan IGD1 and aggrecan IGD2. B: ELISA for ARGSVIL peptide-1 (derived from aggrecan IGD1) and ARGSVIL peptide-2 (derived from aggrecan IGD2).
Three observations are readily apparent from the figures. First, the two forms, aggrecan IGD1 and aggrecan IGD2, behave differently in the ELISA. The same is true for the two ARGSVIL peptides. Second, ARGSVIL peptides give high absorbance values at concentrations below 5 nM, while aggrecan IGD substrates should be present in at least 1000-fold higher concentrations to give comparable absorbance values. Third, the greater difference in absorbance (about 10,000-fold) is observed for the pair aggrecan IGD2 and ARGSVIL peptide-2. Based on these data, aggrecan IGD2 was chosen as substrate in the aggrecanase activity assay, and the substrate concentration was fixed at 0.1 μM except for experiments in which the dependence of initial velocities of ADAMTS-catalyzed reactions on substrate concentrations was measured.
Aggrecanase Activity Assay
The aggrecanase activity assay was carried out in two steps. In the first step, aggrecan IGD2 was digested with aggrecanase to yield the product ARGSVIL peptide-2. The reaction was terminated with EDTA-containing buffer. In the second step, the concentration of ARGSVIL peptide-2 was determined by the ELISA method with neoepitope and sequence antibodies. For precise calculation of product, ARGSVIL peptide-2 standard concentrations of the peptide were run in parallel.
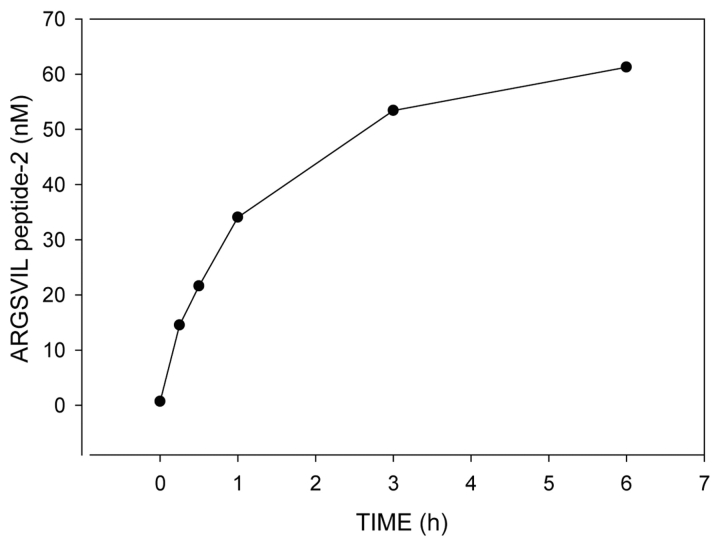
Figure 6 demonstrates a time course of the hydrolysis of 100 nM aggrecan IGD2 by 1.5 nM truncated ADAMTS4. Under the specified conditions, fast hydrolysis at the beginning of the reaction slows down rapidly, and about 6 h are required to cleave 60% of the substrate. As initial velocities should be measured in activity assays, all further reactions were carried out for 15 min only.
FIGURE 6.
Time course of aggrecan IGd2 hydrolysis by truncated ADAMTS4. Aggrecan IGd2 at a concentration of 0.1 μM was incubated at 37°C with 1.5 nM truncated ADAMTS4 in 50 mM Tris-HCL (pH 7.5), 150 mM NaCL, 5 mM CaCl2, 1 μM leupeptin, 1 μM pepstatin, 1 mM Pefabloc, 0.05 % Brij 35. At various time intervals, aliquots of the reaction mixture were withdrawn and analyzed for the hydrolysis product ARGSVIL peptide-2 by the ELISA method.
Variation of the substrate concentration between 20 and 400 nM had little effect on initial hydrolysis rates, indicating that the Km value of ADAMTS4 for aggrecan IGD2 lies below 20 nM. A precise estimation of the value was not possible, as initial hydrolysis rates at substrate concentrations below 20 nM proved difficult to measure.
Variation of the concentration of truncated ADAMTS4 between 0.02 and 1.5 nM yielded a linear relationship between reaction velocity and enzyme concentration. The experimental data are shown in Figure 7. Figure 7A demonstrates the dependence of OD values on ADAMTS4 concentrations. Figure 7B gives the standard curve for ARGSVIL peptide-2. In Figure 7C, OD values for different ADAMTS4 concentrations are replaced by concentrations of ARGSVIL peptide-2 calculated from the ARGSVIL peptide-2 standard curve. ARGSVIL peptide-2 concentrations were multiplied by a factor of 2.5 to account for dilution of aggrecanase reactions with EDTA stop solution. The slope of the linear dependence of product formation on ADAMTS4 concentration defines the specific activity of truncated ADAMTS4 with respect to the substrate aggrecan IGD2. From data in Figure 7C, a value of 0.45 nM ARGSVIL peptide-2/min·mg was calculated. The mean value for five ADAMTS4 preparations was 0.88 ± 0.38 nM ARGSVIL peptide-2/min·mg. When related to mg enzyme, the value is 21.7 ± 9.5 nmoles ARGSVIL peptide-2/min-mg.
FIGURE 7.
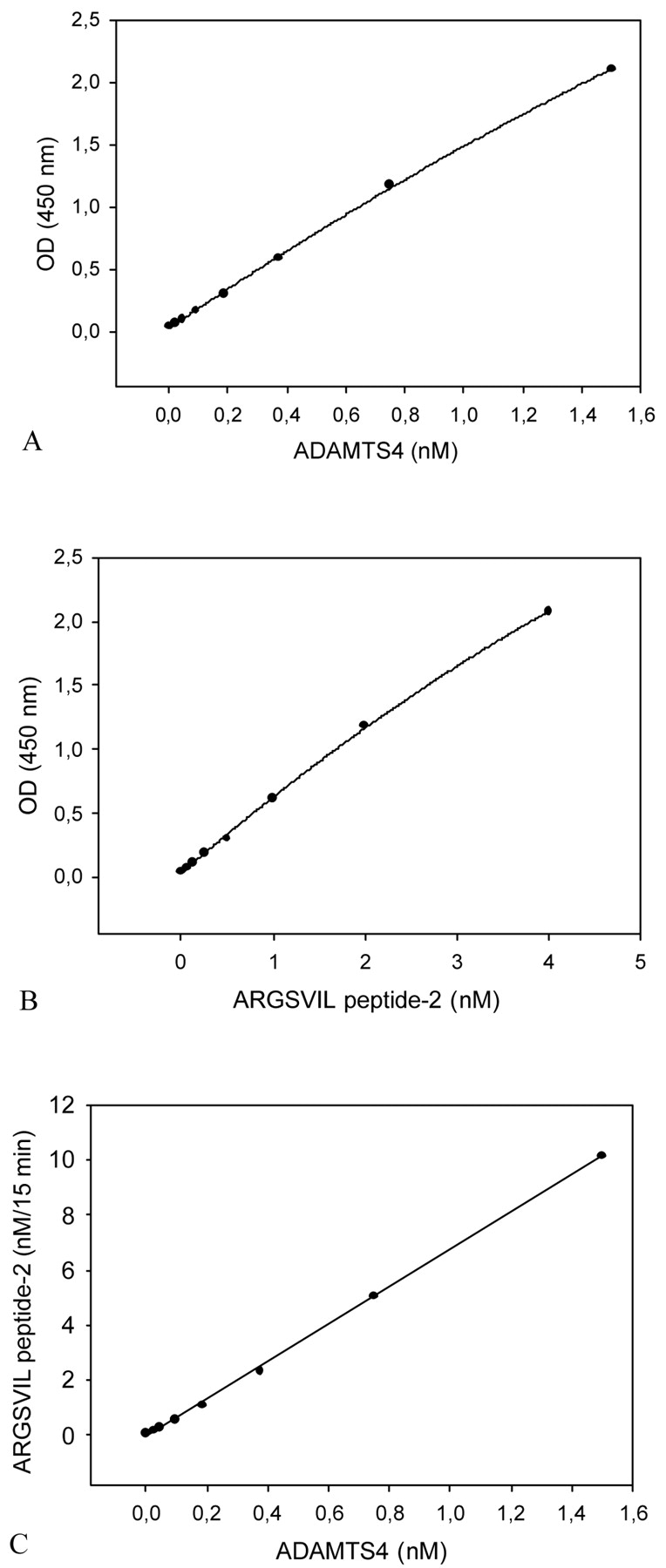
Aggrecanase activity assay: standard curves for truncated ADAMTS4 and for ARGSVIL peptide-2. The aggrecanase activity assay was carried out as described under Materials and Methods with 0.1 μM aggrecan-IGD2 and ADAMTS4 concentrations ranging from 0.024 to 1.5 nM. OD values measured in the ELISA were plotted against ADAMTS4 concentrations (A). Standard concentrations of ARGSVIL peptide-2 varying from 0.062 to 4 nM were determined in parallel (B). The standard curve for ARGSVIL peptide-2 was then used to calculate the amounts of ARGSVIL peptide-2 formed in ADAMTS4-catalyzed reactions (C). The slope of the linear dependence of formed product ARGSVIL peptide-2 versus ADAMTS4 concentrations defines the specific activity of ADAMTS4.
In similar experiments with three different preparations of truncated ADAMTS1, the following activity values were determined: 0.1 ± 0.02 nM ARGSVIL peptide-2/min·mg and 2.4 ± 0.4 nmoles ARGSVIL peptide-2/min-mg.
The aggrecanase activity assay measures also activity of truncated ADAMTS5. Specific activity values for this enzyme were not determined, however, as ADAMTS5 preparations were only partially purified.
The sensitivity of detection of ADAMTS4, defined as two standard deviations above the mean of calculated concentrations of 40 blank samples, was 0.012 nM ADAMTS4. Reproducibility of the aggrecanase activity assay was characterized as follows: The within-assay precision measured by assaying three different ADAMTS4 concentrations 16 times yielded CV% values of ≤9.9. The between-assay precision estimated in 14 repeated measurements amounted to CV% ≤ 13. The CV% values of the precision profile varied between 6 and 12.1.
Proteinases cleaving aggrecan IGD2 at sites other than the aggrecanase site were not detected with the assay. For example, peptides produced from aggrecan IGD by MMP-13 or MMP-14 catalytic domain gave no absorbance values above control levels in the ELISA.
Sensitive Aggrecanase Activity Assay
To increase the sensitivity of the assay, a mutated substrate that is cleaved by aggrecanases more effectively than authentic aggrecan IGD was produced. For in vitro mutagenesis, the limiting condition imposed by use of the anti-N-terminal ARGSVIL antibody for detection of hydrolysis product had to be taken into account: Only P residues of the cleavage site could be changed. As a rational choice, P residues in aggrecan IGD were replaced by P residues from a preferential aggrecanase site in the chondroitin sulfate–rich region of aggrecan. The site PTTFKEEE1667–1668GLGS is hydrolyzed by ADAMTS4 much more effectively than the site NITEGE373–374ARGSVIL.8 P residues from the former site were therefore grafted into the aggrecanase site of recombinant aggrecan IGD to obtain the substrate aggrecan IGD-s and the subforms aggrecan IGD-s1 and aggrecan IGD-s2. When aggrecan IGD-s1 or aggrecan IGDs2 were used as substrate instead of aggrecan IGD1 or aggrecan IGD2, much lower concentrations of truncated ADAMTS1 and ADAMTS4 could be detected in the aggrecanase activity assay. The highest sensitivity was achieved with aggrecan IGD-s1. Figure 8 demonstrates data obtained with aggrecan IGD-s1 and truncated ADAMTS4. For calibration, ARGSVIL peptide-s1 was used. The standard curve for ADAMTS4 extended from 3.1 to 100 pM, and the sensitivity of detection of ADAMTS4 was 2 pM (Figure 8A). This is a considerable increase in sensitivity compared to the standard activity assay (compare data in Figure 7, which were obtained with the same ADAMTS4 preparation). Specific activity values calculated for ADAMTS4 from data in Figure 8C amounted to 2.4 nM ARGSVIL peptide-s1/min·mg or 60 nmoles ARGSVIL peptide-s1/min-mg. For five different ADAMTS4 preparations, a mean value of 151.5 ± 93.5 nmoles ARGSVIL peptide-s1/min-mg was determined. The activity values measured with the substrate aggrecan IGD-s1 for three different ADAMTS1 preparations were 0.41 ± 0.21 nM ARGSVIL peptide-s1/min·mg or 10.3 ± 5.1 nmoles ARGSVIL peptide-s1/min-mg. When compared with data of the standard assay, the values demonstrate a four- to sevenfold faster hydrolysis by ADAMTS1 and ADAMTS4 of the mutated substrate than of the substrate with authentic aggrecan sequence. Due to its higher sensitivity for detection of aggrecanase activity, the assay with the substrate aggrecan IGD-s1 was named sensitive aggrecanase assay.
FIGURE 8.
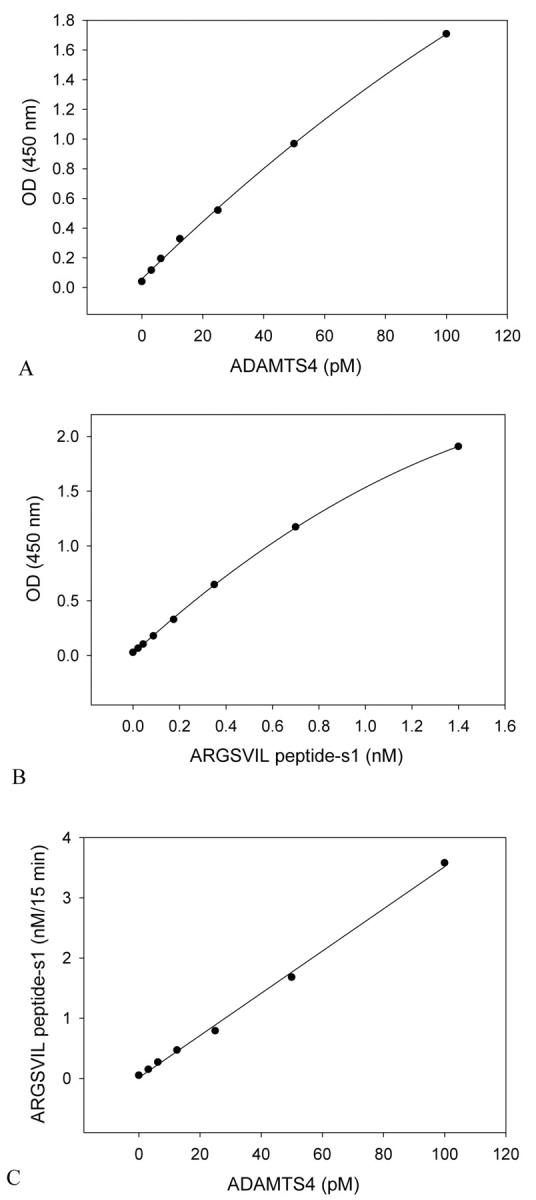
Sensitive aggrecanase activity assay: standard curves for truncated ADAMTS4 and for ARGSVIL peptide-s1. The sensitive aggrecanase activity assay was performed with 0.1 μM aggrecan IGD-s1 and ADAMTS4 concentrations ranging from 3.13 to 100 pM. OD values measured in proteolytic reaction mixtures were plotted against ADAMTS4 concentrations (A). A series of ARGSVIL peptide-s1 concentrations varying from 0.022 to 1.4 nM was measured on the same microtiter plate (B). The peptide standard curve was used to calculate amounts of peptide produced in ADAMTS4-catalyzed reactions. The slope of the linear dependence of product ARGSVIL peptide-s1 vs ADAMTS4 concentrations (C) gives the specific activity of ADAMTS4 with respect to the substrate aggrecan IGD-s1.
CV% values for within-assay and between-assay precisions of the sensitive assay were determined as ≤11.5 and ≤12.9, respectively. The CV% values of the precision profile varied between 4.9 and 22%. The sensitive aggrecanase activity assay, like the standard assay, does not measure activities of matrix metalloproteinases MMP-13 and MMP-14.
Measurement of Aggrecanase Activity in the Presence of Synovial Fluid
The activity assays were used to measure aggrecanase activity in diluted samples of synovial fluid. Two sets of experiments were carried out: (1) the activity of recombinant truncated ADAMTS4 was measured in the absence and in the presence of various concentrations of synovial fluid; (2) the endogenous aggrecanase activity of synovial fluid was assessed.
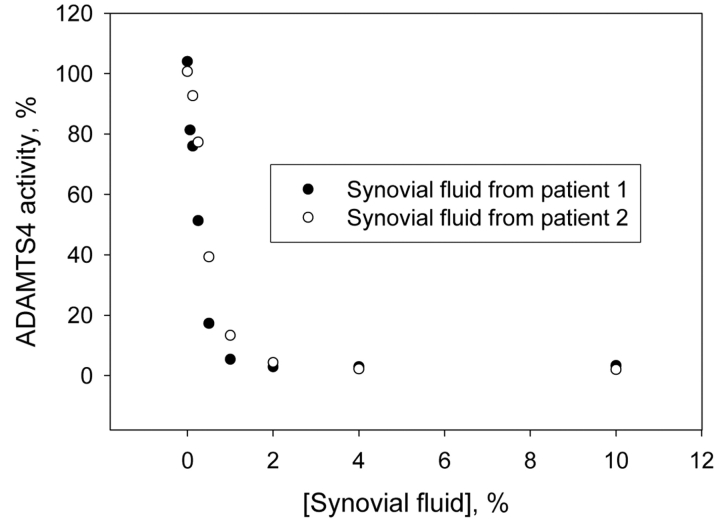
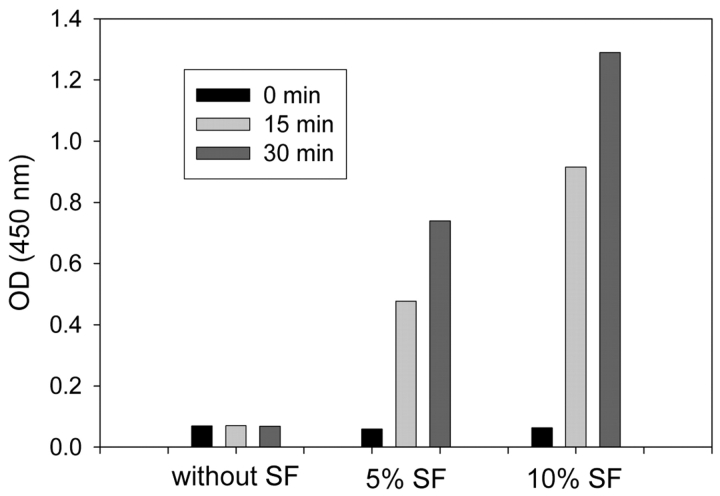
When synovial fluid was added to recombinant ADAMTS4, the enzyme was strongly inhibited. Half-maximal inhibition of 1.5 nM ADAMTS4 occurred at 0.5–0.7% synovial fluid in the assay mixture, and more than 90% of ADAMTS4 activity was suppressed in the presence of 2–4% synovial fluid (Figure 9). A synovial fluid concentration of 10% was only a little more inhibitory, and with some samples, 10% synovial fluid caused less inhibition than 2 or 4% concentrations. Examination of the latter samples with the sensitive aggrecanase activity assay revealed a time- and concentration-dependent endogenous aggrecanase activity (Figure 10). In repeated measurements of a selected synovial fluid sample, an activity value of 0.73 ± 0.04 nmoles hydrolyzed aggrecan IGD-s1/15 min was estimated for 10% synovial fluid diluted in reaction buffer.
FIGURE 9.
Inhibition of recombinant ADAMTS4 by synovial fluid. 0.1 μM aggrecan IGD2 was digested with 1.5 nM ADAMTS4 for 15 min at 37°C in the absence and in the presence of various concentrations of synovial fluid from two patients. Other conditions were as described under Materials and Methods for the aggrecanase activity assay. The activity measured in the absence of synovial fluid was set at 100%.
FIGURE 10.
Hydrolysis of aggrecan IGD-s1 in the presence of synovial fluid (SF). 0.25 μM aggrecan-IGd-s1 was incubated at 37°C in 50 mM Tris-HCL (pH 7.5), 150 mM NaCL, 5 mM CaCl2, 1 μM leupeptin, 1 μM pepstatin, 1 mM Pefabloc, 10 μM ZnCl2, 0.05% Brij 35 without or with 5% and 10% synovial fluid. After various time intervals, aliquots of the reaction mixtures were analyzed by the ELISA method. The concentration of ARGSVIL peptide-s1 produced in the presence of 10% synovial fluid within 15 min was estimated in 12 repetitive experiments as 0.71 ± 0.07 nM.
DISCUSSION
The aggrecanase activity assay described here allows reliable and sensitive measurement of peptide-bond hydrolysis at the aggrecanase site in aggrecan IGD. Hydrolysis reactions can be carried out under variable conditions, and aggrecanase concentrations can be varied over several orders of magnitude. Substrate cleavages at sites other than the aggrecanase site are not measured.
Essential components of the aggrecanase activity assay are: (a) recombinant aggrecan IGD, (b) a polypeptide standard representing the C-terminal fragment of aggrecan IGD beginning with the N-terminal sequence ARGSVIL, (c) an anti-neoepitope monoclonal antibody with high affinity for the ARGSVIL sequence at peptide N-termini and low affinity for the same sequence within continuous peptide chains, and (d) a second monoclonal antibody to the C-terminal fragment of aggrecan IGD, which can bind to this fragment together with the anti-neoepitope antibody.
Upon purification of recombinant aggrecan IGD, two forms of the polypeptide, aggrecan IGD1 and aggrecan IGD2, were separated. The two forms migrated identically in SDS-PAGE, and they were hydrolyzed at comparable rates by ADAMTS1 or ADAMTS4. However, the two forms differed in their hydrodynamic properties, as revealed by gel filtration, and they behaved differently in the ELISA with aggrecan neoepitope and aggrecan sequence antibodies. Aggrecan IGD1 gave stronger signals in the ELISA than aggrecan IGD2. Likewise, the ELISA calibration curves for the C-terminal fragments ARGSVIL peptide-1 and ARGSVIL peptide-2 were also different. OD values measured for 0.1–3 nM ARGSVIL peptide-1 were higher than OD values measured for corresponding concentrations of ARGSVIL peptide-2. As various preparations of aggrecan IGD contained variable amounts of aggrecan IGD1 and aggrecan IGD2, it was essential that the two forms be separated, so that an isolated homogeneous form was used as substrate and the corresponding ARGSVIL peptide was used as standard.
A possible explanation for different behavior of aggrecan IGD1 and aggrecan IGD2 in the ELISA is as follows: If aggrecan IGD1 is an aggregate and aggrecan IGD2 is a monomer, the aggregate, after binding to the neoepitope antibody on the microplate, may bind several sequence antibodies, while the monomer will bind only one sequence antibody. The same hypothesis applies to the derived C-terminal ARGSVIL peptides.
The performance of the aggrecanase activity assay depends—apart from the substrate—largely on properties of the anti-neoepitope antibody. The novel anti-ARGSVIL antibody shows a strong preference for the ARGSVIL sequence at the N-terminus of aggrecan fragments as compared to intrachain location of the sequence. ELISA signals arising from amino acids residues ARGSVIL in the uncleaved substrate can therefore be neglected. The neoepitope antibody proved also very useful for Western blot detection of ARGSVIL peptides and of aggrecan fragments in synovial fluid (data not shown).
The aggrecanase activity assay has been used to measure specific activities of truncated ADAMTS1 and ADAMTS4. Activity values of 2.4 and 21.7 nmoles ARGSVIL peptide/min-mg were estimated for the two enzymes, respectively. When other aggrecan-hydrolyzing enzymes, ADAMTS5, ADAMTS8, ADAMTS9, and ADAMTS15, are examined, all six ADAMTS can be classified in accordance with their relative hydrolytic efficiencies towards the E373–A374 bond in aggrecan IGD.
The assay will also prove useful to compare the activities of full-length and truncated ADAMTS. As repeatedly reported, full-length ADAMTS4 hydrolyzes aggrecan preferentially at sites in the chondroitin sulfate–rich region between globular domains G2 and G3.15,19 The enzyme has little activity against the E373–A374 bond located between domains G1 and G2. By contrast, ADAMTS4 mutants with deleted C-terminal region exhibited a greatly increased activity against the E373–A374 bond. Likewise, truncated forms of ADAMTS4 predominating after Il-1 treatment in chondrocyte cell cultures are highly active against the site in aggrecan IGD.20 The aggrecanase activity assay is a convenient tool to quantify activity changes occurring upon conversion of full-length to truncated forms of ADAMTS4 and possibly other aggrecan-hydrolyzing ADAMTS.
Another application of the aggrecanase activity assay is the screening of inhibitors as potential therapeutic agents to treat osteoarthritis and other diseases with excessive loss of aggrecan from tissues. The activity assay can easily be adapted for high-throughput screening of aggrecanase inhibitors in homogeneous phase. For example, aggrecan neoepitope and aggrecan sequence antibodies can be labeled with donor and acceptor fluorophores, respectively. Binding of both antibodies to the hydrolysis product, ARGSVIL peptide, can then be monitored by fluorescence resonance energy transfer. In an alternative format, fluorophores can be coupled to the neoepitope antibody and the C-terminal sequence of the substrate aggrecan IGD.
A more challenging task is the measurement of aggrecan hydrolyzing activities in biological samples. Enzyme activities in serum, synovial fluid, and other body fluids are complex parameters. In the case of aggrecanases, different enzymes either in full-length or truncated forms may contribute to overall activity, and inhibitors as TIMP-320,21 and α2-macroglubulin22 counterbalance the enzymes. In addition, a variety of proteins in biological fluids can be substrates for aggrecanases and compete with exogenously added substrate. When recombinant aggrecan IGD was incubated with chondrocyte cell-culture medium or synovial fluid, little product ARGSVIL peptide was found in the ELISA with aggrecan neoepitope and sequence antibodies. To increase the sensitivity of the assay, the aggrecanase site in aggrecan IGD was changed by in vitro mutagenesis. The rational approach was based on data demonstrating graded susceptibilities towards ADAMTS4 of different sites in isolated aggrecan.8 In the present work, P1–P5 residues of the site PTTFKEEE1667–1668GLGS were placed into recombinant aggrecan IGD. As a result, about four- to sevenfold faster hydrolysis of the modified substrate by ADAMTS1 and ADAMTS4 was observed. This experiment demonstrated that P1–P5 residues contribute significantly to more effective cleavage by ADAMTS4 of aggrecan site E1667–1668G as compared to other sites. More important, the modified substrate, aggrecan IGD-s, allowed measurement of much lower aggrecanase activity than the substrate with authentic aggrecan sequence. The detection limit for truncated ADAMTS4 was pushed down to 2 pM. Using aggrecan IGD-s1 as substrate aggrecanase activity of chondrocyte culture supernatants could easily be detected after 15-min incubations (data not shown). A time- and concentration-dependent cleavage of aggrecan IGD-s1 was also observed in the presence of some but not all human synovial fluid samples. In future studies, individual aggrecan-hydrolyzing ADAMTS will first be separated from synovial fluid with specific antibodies before their activity is analyzed with the aggrecanase activity assay.
Acknowledgments
The authors thank Thorsten Stroh for labeling aggrecan sequence antibody with peroxidase and Monique Oswald for excellent technical assistance. The work was supported by the German Ministry of Research and Technology (grant 01GG9830/0).
REFERENCES
- 1.Knudsen CB, Knudsen W. Cartilage proteoglycans. Sem Cell Developm Biol 2001;12:69–78. [DOI] [PubMed] [Google Scholar]
- 2.Matthews RT, Kelly GM, Zerillo CA, Gray G, Tiemeyer M, Hockfield S. Aggrecan glycoforms contribute to the molecular heterogeneity of perineuronal nets. J Neuroscience 2002;22:7536–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doege KJ, Sasaki M, Kimura T, Yamada Y. Complete coding sequence and deduced primary structure of the human cartilage large aggregating proteoglycan aggrecan. J Biol Chem 1991;266:894–902. [PubMed] [Google Scholar]
- 4.Watanabe H, Yamada Y, Kimata K. Roles of aggrecan, a large chondroitin sulfate proteoglycan, in cartilage structure and function. J Biochem (Tokyo) 1998;124:687–693. [DOI] [PubMed] [Google Scholar]
- 5.Lohmander LS, Neame PJ, Sandy JS. The structure of aggrecan fragments in human synovial fluid. Arthritis Rheum 1993;36:1214–1222. [DOI] [PubMed] [Google Scholar]
- 6.Malfait AM, Liu RQ, Ijiri K, Komiya S, Tortorella MD. Inhibition of ADAMTS4 and ADAMTS5 prevents aggrecan degradation in osteoarthritic cartilage. J Biol Chem 2002;277:22,201–22,208. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Manzaneque JC, Westling J, Thai SN, Luque A, Knauper V, Murphy G, et al. ADAMTS1 cleaves aggrecan at multiple sites and is differently inhibited by metalloproteinase inhibitors. Biochem Biophys Res Commun 2002;293:501–508. [DOI] [PubMed] [Google Scholar]
- 8.Tortorella MD, Pratta M, Liu RQ, Austin J, Ross OH, Abbaszade I, et al. Sites of aggrecan cleavage by recombinant human aggrecanase 1 (ADAMTS4). J Biol Chem 2000;275:18,566–18,573. [DOI] [PubMed] [Google Scholar]
- 9.Tortorella MD, Liu, RQ, Burns T, Newton RC, Arner E. Characterization of human aggrecanase 2 (ADAMTS5): Substrate specificity studies and comparison with aggrecanase 1 (ADAMTS4). Matrix Biol 2002;21:499–511. [DOI] [PubMed] [Google Scholar]
- 10.Collins-Racie LA, Flannery CR, Zeng W. ADAMTS8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol 2004;23:219–230. [DOI] [PubMed] [Google Scholar]
- 11.Sommerville RP, Longpre JM, Jungers KA, Engle JM, Ross M, Evanko S, et al. Characterization of ADAMTS9 and ADAMTS20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J Biol Chem 2003;278:9503–9513. [DOI] [PubMed] [Google Scholar]
- 12.Yamaji N, Nishimura K, Abe K, Ohara O, Nagase T, Nomura N. Novel metalloprotease having aggrecanase activity. European Patent 2000;00974894.8.
- 13.Sandy JD, Westling J, Denegy RD, Iruela-Arispe ML, Verscharen C, Rodriguez-Mazaneque JC, et al. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441–Ala442 bond, a site that is cleaved by ADAMTS1 and ADAMTS4. J Biol Chem 2001;276: 13,372–13,378. [DOI] [PubMed] [Google Scholar]
- 14.Matthews RT, Gary SC, Zerillo C, Pratta M, Solomon K, Arner EC, et al. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J Biol Chem 2000;275:22,695–22,703. [DOI] [PubMed] [Google Scholar]
- 15.Körber C, Büttner FH, Kern C, Schmiedeknecht G, Bartnik E. Truncation of the amino-terminus of the recombinant aggecan rAgg1mut leads to reduced cleavage at the aggrecanase site. Efficient aggrecanase catabolism may depend on multiple substrate interactions. Matrix Biol 2000;19:533–543. [DOI] [PubMed] [Google Scholar]
- 16.Gao G, Westling J, Thompson VP, Howell TD, Gottschall PE, Sandy JD. Activation of the proteolytic activity of ADAMTS4 (aggrecanase 1) by C-terminal truncation. J Biol Chem 2002;277:11,034–11,041. [DOI] [PubMed] [Google Scholar]
- 17.Hughes CE, Büttner FH, Eidenmüller B, Caterson B, Bartnik E. Utilization of a recombinant substrate rAgg1 to study the biochemical properties of aggrecanase in cell culture systems. J Biol Chem 1997;272:20,269–20,274. [DOI] [PubMed] [Google Scholar]
- 18.Miller JA, Liu RQ, Davis GL, Pratta MA, Trzaskos JM, Copeland RA. A microplate assay specific for aggrecanase. Analyt Biochem 2003;314:260–265. [DOI] [PubMed] [Google Scholar]
- 19.Kashiwagi M, Enghild JJ, Gendron C, Hughes C, Caterson B, Itoh Y, et al. Altered proteolytic activities of ADAMTS4 expressed by C-terminal truncation. J Biol Chem 2004;279:10,109–10,119. [DOI] [PubMed] [Google Scholar]
- 20.Pratta MA, Scherle PA, Yang G, Liu RQ, Newton RC. Induction of aggrecanase 1 (ADAMTS4) by interleukin-1 occurs through activation of constitutively produced protein. Arthritis Rheum 2003;48:119–133. [DOI] [PubMed] [Google Scholar]
- 21.Kashiwagi M, Tortorella MD, Nagase H, Brew K. TIMP 3 is a potent inhibitor of aggrecanase 1 (ADAMTS4) and aggrecanase 2 (ADAMTS5). J Biol Chem 2001;276: 12,501–12,504. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto G, Aoki T, Nakamura H, Tanzawa K, Okada Y. Inhibition of ADAMTS4 (aggrecanase 1) by tissue inhibitors of metalloproteinases (TIMP-1, -2, -3 and-4). FEBS Lett 2001;494:192–195. [DOI] [PubMed] [Google Scholar]
- 23.Tortorella MD, Arner EC, Hills R, Easton A, Korte-Sarfaty J, Fok K, et al. α2-Macroglobulin is a novel substrate for ADAMTS4 and ADAMTS5 and represents an endogenous inhibitor of these enzymes. J Biol Chem 2004;279:17,554–17,561. [DOI] [PubMed] [Google Scholar]