Abstract
The broad dynamic range of protein abundances, which can vary from about 106 for cells to 1010 for tissues in complex proteomes, continues to challenge proteomics research. Proteome analysis, in particular organelle proteomics, using current approaches, requires extensive fractionation, separation, and enrichment. Over the years, organelle separation was achieved through the use of differential and density-gradient ultracentrifugation. However, the traditional fixed-volume process is a time-consuming and labor-intensive method, especially with large quantities of sample. Here, we present a novel tool for subcellular fractionation of biologically complex mixtures: continuous-flow ultracentrifugation of tissue homogenates to obtain both organelle separation and extensive organelle enrichment at the same time. In this study, rat liver tissues from two different age groups (3–8 wk and greater than 1 y old) were homogenized by blending. After removing nuclei, the resulting homogenates were further fractionated at the subcellular level by the use of sucrose gradient continuous-flow ultracentrifugation. Each organelle’s enriched fractions were identified by Western blot analysis. To study the possible effects of aging on the endoplasmic reticulum and Golgi apparatus, we compared the organelle protein profiles of the two groups of rat liver tissues using two-dimensional gel electrophoresis, digitized imaging of two-dimensional gel electrophoresis, and mass spectrometry. Significant differences in the protein profiles of both organelles were observed between the two groups of rat tissues. The technique described here for fractionation and enrichment of organelles demonstrated a useful tool for proteomics research, including identification of low-abundance proteins and post-translational modifications.
Keywords: Continuous-flow ultracentrifugation, Golgi apparatus, endoplasmic reticulum, organelle enrichment, organelle separation
With the completion of many genome sequencing projects, proteomics has emerged as a significant field of interest. Proteomics involves the analysis of protein mixtures in a cellular environment. Understanding the function and regulation of these proteins involves fractionation, quantitation, and characterization from complex protein mixtures.1–4 Alternative splicing of transcripts or post-translational modifications can increase the total number of different proteins compared to the encoding genes involved.
Proteomics has traditionally involved the use of two-dimensional gel electrophoresis to separate large sets of proteins, followed by mass spectrometry to identify the proteins corresponding to the individual gel spots. More recently, because of the complexity and heterogeneity of various proteomes, better methods of characterization are constantly being developed. One aspect is increased performance on two-dimensional gels through the use of larger gels and narrower pH ranges in the first dimension.5–6 In addition, preparative methods resulting in effective and simple sample fractionation procedures will help in the characterization prior to the isoelectric focusing step of two-dimensional gel analysis.7–9
The endoplasmic reticulum and the Golgi apparatus have long been studied for their involvement in protein synthesis, post-translational modification, and excretory processes. Understanding of the proteomes of these two organelles will help provide insight into the molecular changes within the organelles and their function.10–13 In order to study proteomic differences, laboratories first isolate the organelles, with every attempt to maintain reproducibility and native structure; the proteomes are then examined using two-dimensional gel electrophoresis followed by mass spectrometry of gel spots for protein identification. Recent research has focused on organelle enrichment and quantitative proteomic analysis. Here we present a technique that separates organelles and their subtypes by density using continuous-flow ultracentrifugation. When equilibrium is established, the organelles are effectively enriched at their buoyant densities, resulting in the increased concentration needed for sensitive proteomic analyses. Unlike classical methods, continuous-flow ultracentrifugation is not limited by sample volume and thus can be used to achieve significant accumulations of low-abundance proteins in individual organelles. This technique is used here to examine and compare the proteomes of organelles isolated from the livers of juvenile and aged rats.
MATERIALS AND METHODS
Rat livers were obtained from Zivic Laboratories, Inc. (Pittsburgh, PA). Livers collected from two age groups (8–10 wk and greater than 1 y) of male Sprague-Dawley rats were snap frozen in liquid nitrogen and stored at −80°C until use. HEPES, magnesium chloride hexa-hydrate, tris(2-carboxyethyl) phosphine hydrochloride, iodoacetamide, Tween-20, 2-mercaptoethanol, and Tris-HCl (pH 8.0) were obtained from Sigma-Aldrich (St. Louis, MO), sodium chloride from Mallinckrodt (Phillipsburg, NJ), sucrose from Invitrogen (Carlsbad, CA), and methanol from VWR (West Chester, PA). PBS liquid concentrate (10X) was obtained from EMD Chemicals (Gibbstown, NJ). Criterion Precast Gels, Tris/glycine/SDS 10X buffer concentrate, Tris/glycine 10X buffer concentrate, and BioSafe Coomassie were obtained from Bio-Rad (Hercules, CA).
Homogenization
One hundred grams of frozen livers were thawed in 1X PBS buffer at 4°C and homogenized using a 1-L Waring blender in 500 mL homogenization buffer (20 mM HEPES, 5 mM MgCl2, 500 mM sucrose, pH 7.2) with protease inhibitor tablets (Roche Molecular Biochemicals, Indianapolis, IN), using the following cycle: 15 sec low-speed setting, 15 sec high-speed setting, 15 sec low-speed setting. After blending, the homogenate was filtered through two layers of cheesecloth on a double-mesh strainer. Nuclei were pelleted at 1076 g for 10 min at 4°C in a Sorvall centrifuge with an SS-34 rotor (Newtown, CT). The resulting supernatant was kept on ice, and pellets were resuspended in homogenization buffer, blended, and centrifuged at 1076 g a second time for maximal recovery of organelles. All supernatants were combined and diluted 1:1 with dilution buffer (20 mM HEPES, 5 mM MgCl2, pH 7.2) for continuous-flow ultracentrifugation.
Continuous-flow Ultracentrifugation
An Alfa Wassermann PKII centrifuge (West Caldwell, NJ) with an 800-mL rotor core was used for density gradient ultracentrifugation. The rotor was initially filled with flow buffer (20 mM HEPES, 5 mM MgCl2, 250 mM sucrose, pH 7.2). After acceleration to 20k rpm with flow to clear air from all channels, 400 mL of gradient buffer (60% w/v sucrose, 20 mM HEPES, 5 mM MgCl2, pH 7.2) was pumped with a peristaltic pump into the bottom of the rotor with the rotor at rest. Ramped acceleration to 3500 rpm established a linear 12% to 55% sucrose gradient. Following gradient formation and further acceleration to 20k rpm, freshly homogenized, diluted tissue sample was loaded into the bottom of the rotor. The flow-through was collected and reloaded at 35k rpm to maximize the entry of sample components into the gradient. The components were banded at 35k rpm for 2 h. Following controlled deceleration to minimize mixing, 25-mL fractions were collected with the rotor at rest. One-milliliter aliquots of each fraction were stored at 4°C for sample analysis. The remainder of each fraction was stored in two aliquots at −80°C until needed.
Density Gradient Calculation
The refractive index of each fraction was measured to verify the linearity of the sucrose gradient. Refractive indices were measured on a Milton Roy refractometer (Ivyland, PA). Sucrose percentages and densities were calculated using data from Griffith.14
Protein Concentration Measurement
Protein concentration of each fraction was measured by the Bradford method15 using Protein Assay Dye Reagent Concentrate (Bio-Rad) and bovine γ-globulin (Bio-Rad) as a standard.
SDS-PAGE
Samples from each fraction were adjusted to 2 mg/mL total protein solutions and then diluted 1:1 with Laemmli Sample Buffer (Bio-Rad) containing 0.7 M 2-mercaptoethanol. Bio-Rad Criterion 26-well 4–20% Tris-HCl gels and Tris/glycine/SDS buffer were used for electrophoresis. Fifteen-microgram samples were loaded into each well and electrophoresis was performed at 20 mA/gel until the dye front reached the end of the gels. One set of gels was fixed for 45 min in 50% methanol/10% CH3COOH, stained for 1 h in BioSafe Coomassie, and destained overnight in dH2O. An identical set of gels was used for Western blotting.
Western Blots
The Bio-Rad Criterion Blotter system and 40% Tris/glycine, 40% Tris/glycine/SDS, 15% methanol transfer buffer were used to transfer proteins to Bio-Rad Immun-Blot PVDF membranes at 175 mA/membrane for 90 min. Membranes were blocked overnight with 5% Carnation nonfat dry milk in 25 mM Tris-HCl (pH 8.0), 125 mM NaCl, 0.1% Tween-20. Golgi apparatus–enriched fractions were detected using mouse anti-rat p115 from Transduction Laboratories (Lexington, KY). The secondary antibody was peroxidase-conjugated AffiniPure rabbit anti-mouse IgG (H+L) from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). ER-enriched fractions were detected using rabbit anti-rat Grp-78 from Stressgen Biotechnologies (Victoria, BC, Canada). The secondary antibody was peroxidase-conjugated AffiniPure goat anti-rabbit IgG (H+L), also obtained from Jackson ImmunoResearch Laboratories. Peroxidase activity was detected by a chemiluminescent reaction using Bio-Rad Immun-Star HRP substrate. Kodak BioMax Light autoradiography film was used for imaging of membranes. Films were analyzed using Kodak1D 3.6 software (Rochester, NY).
Two-Dimensional Gel Electrophoresis
Samples from Golgi apparatus–enriched fractions were prepared by diluting solutions of 250 μg of protein with Bio-Rad Rehydration-Sample Buffer to a final volume of 190 μL. Samples from ER-enriched fractions were first concentrated at 10,000 g at 4°C for 8 h using Millipore Ultrafree-0.5 centrifugal filter units. Two hundred fifty micrograms of protein were then diluted in rehydration-sample buffer to a final volume of 190 μL. Samples containing 200–250 μg protein were placed in rehydration trays, and Bio-Rad linear-gradient IPG strips pH 5–8 were placed on top of the samples and allowed to rehydrate overnight. Isoelectric focusing was performed on the Bio-Rad Protean IEF unit using the manufacturer’s suggested program, resulting in about 30,000 V-h of focusing time. Samples immobilized on IPG strips were reduced and alkylated using Bio-Rad equilibration buffers I and II, respectively. After reduction and alkylation, Bio-Rad Criterion 8–16% IPG+1 Tris-HCl gels and Tris/glycine/SDS buffer were used for SDS-PAGE at 20 mA/gel. Gels were fixed overnight in 50% methanol/10% CH3COOH and stained as previously described. All samples were run in triplicate. The stained gels were scanned using a Umax PowerLook 1100 scanner with UTA-1100 transparency adapter (Dallas, TX). The digitized images were analyzed and compared using Nonlinear Phoretix 2D Expression software (Durham, NC).
Proteolytic Digestion
Selected spots were excised from gels. Spots were destained in 0.2 M ammonium bicarbonate (pH 8.0), 50% acetonitrile with shaking at 37°C. Gel spots were then dried, reduced with 20 mM tris(2-carboxyethyl) phosphine hydrochloride, 25 mM ammonium bicarbonate (pH 8.0) and alkylated with 40 mM iodoacetamide, 25 mM ammonium bicarbonate (pH 8.0). Gel spots were then washed and digested with 20 μL trypsin (0.8 μg/spot) overnight at 37°C. After 16 h, digests were stopped and peptides were extracted from the gel matrix with 20 μL 40 mM ammonium bicarbonate (pH 8.0) for 45 min at 37°C. Four microliters of acetic acid was added to each sample. Samples were mixed and stored at 4°C until analyzed by mass spectrometry.
Nano-LC-ESI-MS/MS
Peptide samples were analyzed on a Thermo Electron LCQ Deca XP Max with NSI source. Samples were eluted using a 30-min linear gradient of 5% solvent B (0.1% formic acid in acetonitrile) to 65% solvent B with a flow rate of 200 nL/min. The mass spectrometer was operated in a data-dependent MS/MS mode using a normalized collision energy of 35%. The capillary temperature of the ion source was set at 180°C. The mass spectra data were searched against the NCBI nonredundant protein database using SEQUEST.
RESULTS AND DISCUSSION
Continuous-flow ultracentrifugation has been tested as a method for the isolation and enrichment of organelles prior to analysis of their constituent proteins by two-dimensional gel electrophoresis and mass spectrometry. The preparative method of centrifugation is one that allows the user to compare and contrast the proteomes of individual organelles. Previously, this method has been used to characterize random proteins from rat liver mitochondria.16 Here, we carry characterization a step further in analyzing proteins from the endoplasmic reticulum and Golgi apparatus. Our attention is focused on proteins unique to the juvenile or aged rats or proteins having greater than twofold difference in expression level. Organelles were detected in the ultracentrifuge fractions by Western blot analysis of known proteins in each organelle. Narrow-range focusing was used in two-dimensional gel electrophoresis to broaden the spot array for analysis.
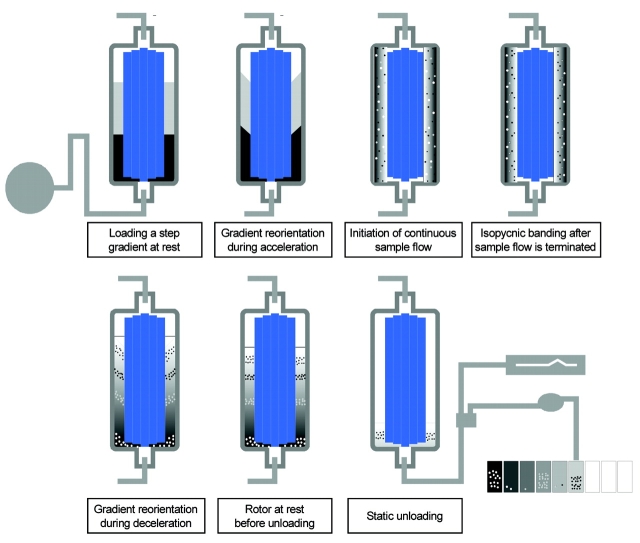
In continuous-flow ultracentrifugation (Figure 1), a step gradient is loaded with the rotor at rest. Controlled acceleration reorients the gradient vertically without disturbing the gradient. When the rotor reaches speed, a linear gradient forms by diffusion. The sample is then loaded with the rotor at speed. Due to the continuous-flow design, there is no limit to the volume of sample that can be loaded into the rotor. After loading sample, the particles are allowed to band at their buoyant density point in the gradient, resulting in separation and enrichment. When they reach their isopycnic point in the gradient, the particles no longer move, due to the equilibrium between sedimentation and floatation. With controlled deceleration, horizontal reorientation of the gradient occurs without disturbing the position of the particles. Fractions are then collected by gravity flow with the rotor at rest. This isopycnic centrifugation, as well as the continuous-flow aspects of the techniques, effectively concentrates and enriches the organelles, which allows detection of larger numbers of proteins.
FIGURE 1.
Process of gradient formation and sample separation in the vertical rotor of the PKII continuous-flow centrifuge.
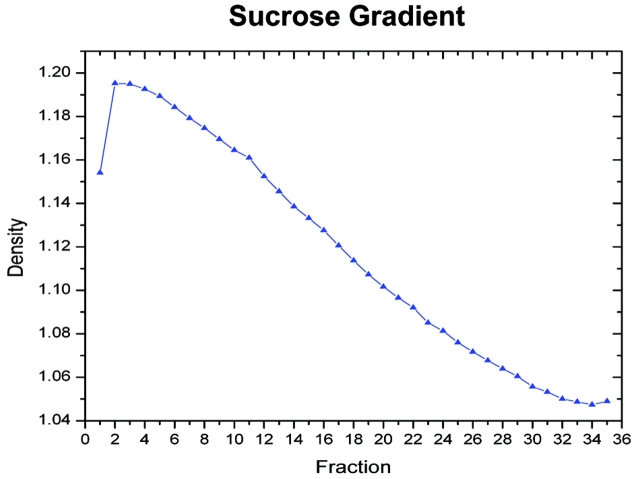
Due to the small inner to outer radial distance, a linear gradient is rapidly formed as the rotor reaches full speed. Also, the small Δr results in a much shorter run time to reach isopycnic equilibrium. A typical sucrose gradient is shown in Figure 2. The lower density in the early fraction is due to the lower sucrose concentration in the wash buffer used to load the final sample.
FIGURE 2.
Typical density profile of fractions from sucrose density continuous-flow ultracentrifugation of rat liver homogenate. The density was calculated from refractive indices.
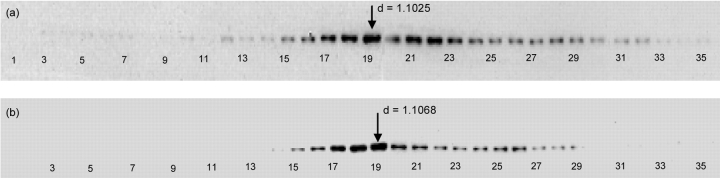
Western blot analysis was then used to detect endoplasmic reticulum and Golgi apparatus in the fractions from continuous-flow ultracentrifugation preparations of juvenile and aged rat livers. Equal amounts of protein were loaded onto the SDS-PAGE gels before running and transfer to PVDF membranes as. Figure 3 shows the chemiluminescent profiles of juvenile and aged rat liver fractions using mouse anti-rat p115 antibodies for the detection of the Golgi apparatus. Juvenile rats showed a slightly broader density range for the Golgi apparatus, but a major peak was observed at fraction 19. Fraction 19 from both juvenile and aged rat was used for further analysis using two-dimensional gel electrophoresis.
FIGURE 3.
Western blot analysis of equal protein amounts collected following continuous-flow ultracentrifugation of rat liver homogenates from (a) 8- to 10-wk-old male and (b) greater than 1-y-old male Sprague-Dawley rats. One fraction showing the highest intensity gel band in the juvenile rat liver homogenate (Fraction 19) was selected for further analysis. Fraction 19 in the aged rat liver homogenate, which also shows the highest intensity band, was selected for comparison. “d” refers to sucrose density of the fraction.
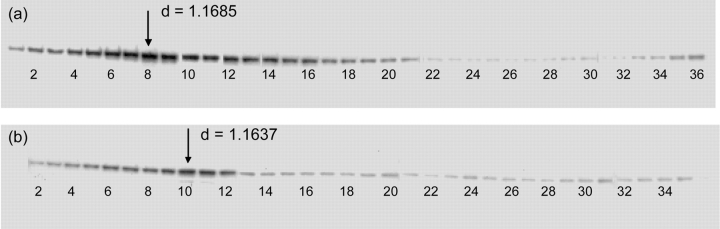
Figure 4 shows the parallel experiments using anti-rat Grp-78 (BiP) for the detection of endoplasmic reticulum. Once again, slightly different distributions of the organelle were indicated. Fractions 8 from juvenile and 10 from aged were used for two-dimensional gel electrophoresis.
FIGURE 4.
Western blot analysis of equal protein amounts collected following continuous-flow ultracentrifugation of rat liver homogenates from (a) 8- to 10-wk-old male and (b) greater than 1-y-old male Sprague-Dawley rats. The fraction showing the highest intensity gel band in the juvenile rat liver homogenate (Fraction 8) was selected for further analysis. Fraction 10 in the aged rat liver homogenate, which also showed the highest intensity gel band, was selected for comparison. “d” refers to sucrose density of the fractions.
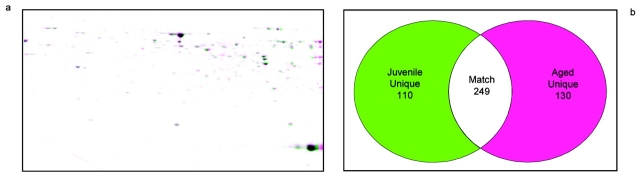
Two-dimensional gel electrophoresis (pH 5–8) was performed on fraction 19 from both the juvenile and aged rats, and the overlay of these gels is shown in Figure 5a. With the warping used from the Nonlinear Phoretix 2D Expression data analysis software, similar spots are black, and unique spots are either green (juvenile) or magenta (aged). The Venn diagram in Figure 5b shows the total numbers of unique and matching spots calculated by the software.
FIGURE 5.
(a) Comparison of 2D gels of protein mixtures arising from fraction 19 of homogenate separations from juvenile and aged rats. Isoelectric focusing on these samples was performed using the narrow pH range 5–8 strips. (b) Numerical comparison of unique and matching gel spots from (a).
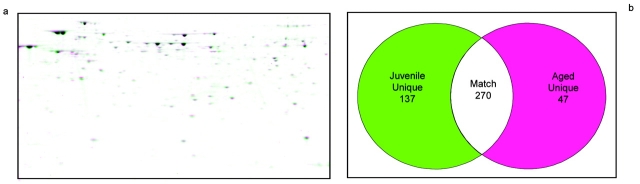
Figure 6 shows a similar analysis of fractions 8 from the juvenile rat and 10 from the aged rat. As before, a warped overlay of the two gels is shown with the calculated number of matching and unique spots.
FIGURE 6.
(a) Comparison of 2D gels of protein mixtures arising from fractions 8 and 10 of homogenate separations from juvenile and aged rats. Isoelectric focusing on these samples was performed using the narrow pH range 5–8 strips. (b) Numerical comparison of unique and matching gel spots from (a).
A sampling of spots was cut from the gels for reduction/alkylation, digestion with trypsin, and nano LC-ESI-MS/MS analysis. Spots were chosen from each gel based on the criteria that they were either unique or had a greater than twofold expression level between the juvenile and aged rats. Tables 1–3 summarize the results of proteins identified in the selected spots.
TABLE 1.
Golgi Proteins with Different Expression Levels in Fraction 19
|
Proteins identified from 2D gel spots (using IPG pH 5–8 strips) occurring in both aged (A) and juvenile (J) fraction 19 with at least a twofold difference in expression levels in two-dimensional gel electrophoresis identified by mass spectrometry. Only selected spots were analyzed.
TABLE 2.
Unique Proteins in Fraction 19
|
Proteins identified from unique two-dimensional gel electrophoresis gel spots (using IPG pH 5–8 strips) that occurred in either juvenile or aged fraction 19 using mass spectrometry. Only selected spots were analyzed.
TABLE 3.
ER Proteins with Different Expression Levels in Fractions 8 (Juvenile) and 10 (Aged)
|
Proteins identified from 2D gel spots (using IPG pH 5–8 strips) occurring in aged fraction 10 and juvenile fraction 8 with at least a twofold difference in expression levels in two-dimensional gel electrophoresis identified by mass spectrometry. Only selected spots were analyzed.
Chaperonin 60,17 α-2-microglobulin,18–20 and formiminotransferase cyclodeaminase21–23 have all been associated with the Golgi apparatus, trans-Golgi network, microtubules, or granules involved in the secretion of proteins after leaving the Golgi apparatus.
Calcium-binding protein24 and valosin-containing protein (which interacts with GRP-7825) both have been shown to be directly associated with the endoplasmic reticulum. Another protein, the proteasome subunit precursor, while not directly associated with the endoplasmic reticulum, is part of a proteolytic system able to degrade misfolded proteins.26–28 More recently, the valosin-containing protein has been associated with the proteolytic mechanism.25 Also present in the endoplasmic reticulum fractions are some known mitochondrial proteins. The presence of these may be due to a partial overlap in the fractions of these two organelles. A second possibility may be that proteins not normally found in the endoplasmic reticulum or the Golgi apparatus may be detected because they are in the process of synthesis and modification.
REFERENCES
- 1.Hunter TC, Andon NL, Koller A, et. al. The functional proteomics toolbox: methods and applications. J Chromatogr B 2002;782:165–181. [DOI] [PubMed] [Google Scholar]
- 2.Isaaq HJ. Application of separation technologies to proteomics research. Adv Prot Chem 2003;65:249–269. [DOI] [PubMed] [Google Scholar]
- 3.Isaaq HJ, Conrads TP, Janini GM, et. al. Methods for fractionation, separation an profiling of proteins. Electrophoresis 2002;23:3048–3061. [DOI] [PubMed] [Google Scholar]
- 4.Bae S-H, Harris AG, Hains PG, et. al. Strategies for the enrichment and identification of basic proteins in proteome projects. Proteomics 2003;3:569–579. [DOI] [PubMed] [Google Scholar]
- 5.Hoving H, Voshol H, van Oostrum J. Towards high performance two-dimensional gel electrophoresis using ultrazoom gels. Electrophoresis 2000;21:2617–2621. [DOI] [PubMed] [Google Scholar]
- 6.Görg A, Obermaier C, Boguth G, et. al. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 2000;21:1037–1053. [DOI] [PubMed] [Google Scholar]
- 7.Butt A, Davison MD, Smith GJ, et. al. Chromatographic separations as a prelude to two-dimensional electrophoresis inproteomics analysis. Proteomics 2001;1:42–53. [DOI] [PubMed] [Google Scholar]
- 8.Görg A, Boguth G, Köpf A, et. al. Sample prefractionation with Sephadex isoelectric focusing prior to narrow pH range two-dimensional gels. Proteomics 2002;2:1652–1657. [DOI] [PubMed] [Google Scholar]
- 9.Pasquali C, Fialka I, Huber LA. Subcellular fractionation, electromigration analysis and mapping of organelles. J Chromatogr B 1999;722:89–102. [DOI] [PubMed] [Google Scholar]
- 10.Wrzeszczynski KO, Rost B. Annotating proteins from endoplasmic reticulum and Golgi apparatus in eukaryotic proteomes. Cell Mol Life Sci 2004;61:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh BJ, Howell KE. The mammalian Golgi-complex debates. Nature Rev Mol Cell Biol 2002;3:789–795. [DOI] [PubMed] [Google Scholar]
- 12.Taylor RS, Wu CC, Hays LG, et. al. Proteomes of rat liver Golgi complex: Minor proteins are identified through sequential fractionation. Electrophoresis 2000;21:3441–3459. [DOI] [PubMed] [Google Scholar]
- 13.Bell AW, Ward MA, Blackstock WP, et. al. Proteomics characterization of abundant Golgi membrane proteins. J Biol Chem 2001;276:5152–5165. [DOI] [PubMed] [Google Scholar]
- 14.Griffith OM. Techniques of Preparative, Zonal, and Continuous Flow Centrifugation, Beckman Instruments, Fullerton, CA, 1986:9.
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–254. [DOI] [PubMed] [Google Scholar]
- 16.Drahos KL, Kiri AN, Lan W, et. al. Comparison of liver mitochondrial proteins from rats of different ages using a novel technique for fractionation and enrichment of organelles. A poster presented at the 44TH Annual Meeting for the American Society for Cell Biology, December 2004. Washington, DC.
- 17.Le Gall IM and Bendayan M. Possible association of Chaperonin 60 with secretory proteins in pancreatic acinar cells. J Histochem Cytochem 1996;44:743–749. [DOI] [PubMed] [Google Scholar]
- 18.Borregaard N, Cowland JB. Granules of the human neutrophilic polymorhonuclear leukocyte. Blood 1997;89:3503–3521. [PubMed] [Google Scholar]
- 19.Thuveson M and Fries E. Intracellular proteolytic processing of the heavy chain of rat pre-α-inhibitor. J Biol Chem 1999;274:6741–6746. [DOI] [PubMed] [Google Scholar]
- 20.Lankat-Buttgereit B, Tampé R. The transporter associated with antigen processing: function and implication in human disease. Physiol Rev 2002;82:187–204. [DOI] [PubMed] [Google Scholar]
- 21.Hennig D, Scales SJ, Moreau A, et. al. A formiminotransferase cyclodeaminase isoform is localized to the Golgi complex and can mediate interaction of trans-Golgi network-derived vesicles with microtubules. J Biol Chem 1998;273:19,602–19,611. [DOI] [PubMed] [Google Scholar]
- 22.Bashour A-M, Bloom GS. 58K, a microtubule-binding Golgi protein, is a formimiminotransferase cyclodeaminase. J Biol Chem 1998;273:19,612–19,617. [DOI] [PubMed] [Google Scholar]
- 23.Gao Y, Alvarez C, Nelson DS, et. al. Molecular cloning, characterization and dynamics of rat formiminotransferase cyclodeaminase. J Biol Chem 1998;273:33,850–33,834. [DOI] [PubMed] [Google Scholar]
- 24.Lundstrom-Ljung J, Birnbach U, Rupp K, et. al. Two resident ER-proteins, CaBP1 and CaBP2, with thioredoxin domains, are substrates for thioredoxin reductase: comparison with protein disulfide isomerase. FEBS Lett 1995;357:305–308. [DOI] [PubMed] [Google Scholar]
- 25.Zhong X, Shen Y, Ballar P, et. al. AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J Biol Chem 2004;279:45,676–45,684. [DOI] [PubMed] [Google Scholar]
- 26.Hiller MM, Finger A, Schweiger M, et. al. ER degradation of a misfolded luminal protein by the the cytosolic ubiquitin-proteasome pathway. Science 1996, 273, 1725–1728. [DOI] [PubMed] [Google Scholar]
- 27.Mayer RJ. The meteoric rise of regulated intracellular proteolysis. Nature Rev Mol Cell Biol 2000;1:145–148. [DOI] [PubMed] [Google Scholar]
- 28.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nature Rev Mol Cell Biol 2003;4:181–191. [DOI] [PubMed] [Google Scholar]