Abstract
Hematopoietic stem cell transplantation (HSCT) creates a donor-recipient cellular chimerism in the patient, which is quantitatively assayed from peripheral blood based on STR-DNA. Since chimerism values often vary across a patient’s samples, it is important to determine to what extent this variability reflects technical aspects of platform performance. This issue is systematically assessed in the current study for the first time. Using the SGM Plus multiplex PCR kit and ABI platform, the longitudinal performance of STR markers was quantitatively evaluated in two chimeric models with true values, and in patient samples (n >500 marker loci). Computation of percent chimerism for each marker, and mean (sample) percent chimerism, standard deviation, and coefficient of variance was performed by our ChimerTrack utility. In chimeric models with known values, individual markers exhibited an accuracy (observed/true) of 88–98%; replication precision was 92–100% true, with a mean error of 2%. Fragment size calling was greater than 99% accurate and precise. Patient results were comparable for markers, relaive to sample means. One source of technical variability in chimerism estimation was allelic differential amplification efficiency. The latter was influenced by signal amplitude, dye label, marker size, and allelic size interval. It can be concluded that long-term chimeric tracking is routinely feasible using this platform in conjunction with ChimerTrack software. Importantly, mean percent chimerism, for any sample, should closely approximate the true chimeric status, with a technical accuracy of 98%. Guidelines are presented for selecting an optimized marker profile.
Keywords: Chimerism, quantitation, STRs, ssmicrosatellites, stem sscell transplantion, software, hematological malignancies, genetic diseases, PCR, multiplex
Hematopoietic stem cell transplantation (HSCT) has become a successful, life-saving mode of treatment in hematological malignancies, such as leukemia. Following transplantation, one of the most useful parameters to monitor is the ratio of patient to donor cells in peripheral blood. This parameter is referred to as the patient’s chimeric status.1–12 Practically, the ratio of patient to donor DNA, extracted from blood or bone marrow cells, is estimated and expressed as percent chimerism. In cancer treatment, the ideal is to create a 100% donor chimerism, because any decrement raises the possibility that the patient may be at risk for relapse of malignancy.1
One popular approach to this type of quantitative chimerism testing is based on an analysis of microsatellite markers, or short tandem repeats (STRs).1–12 This entails PCR amplification of STR marker loci, which are short base sequences on chromosomes distributed throughout the genome. Each STR marker is actually a set (or system) of many alleles, all sharing the basic base structure of the repeat, but differing in the number of tandem repeats of this sequence. Because of this polymorphism, cells derived from either donor or recipient (pre-transplantation) can usually be distinguished. In the SGM Plus kit used in this study, there are 10 markers, each with 8–23 different-sized alleles. An individual will normally have only one or two STR alleles in a marker system, depending on whether he is homo- or heterozygous, respectively, at that marker. Similarly, a chimeric marker locus will have one to four allelic peaks (bands). In addition, for genetic and technical reasons, not all of these markers will be useable for analysis, as explained below. Those markers that are useable, or “informative,” will be referred to as the marker profile for a patient, and will function as a personalized set of chimerism markers for all samples from a specific donor-recipient pair.
Following HSCT, a dynamic relationship exists between the engrafted cells and the patient, which is often reflected in fluctuating chimeric status.3 It seems reasonable, then, to track progressive changes in chimerism values, rather than focusing on isolated values from fixed time-points following transplantation.13,14 In the early work on temporal patterns of chimerism in sequential samples,15–17 it was assumed that changes in the patient’s percent chimerism reflect actual clinical/biological variations. However, the multiplex PCR platform, usually used for STR analysis, is fraught with many sources of technical variation. Results may be influenced by biochemical, instrumental, and genetic factors.10,18–24 It therefore seems necessary to resolve a central issue in quantitative chimerism testing: To what extent do the observed fluctuations in a patient’s chimeric status reflect performance variations of the technical methodology? Specifically, this study examined three basic parameters of platform performance: accuracy of estimating percent chimerism (i.e., observed/true), precision (i.e., accuracy of results in replications), and size calling (i.e., base-pair length of STRs). Sensitivity to the minor component DNA was not examined here, since it has been evaluated previously.1–12
In undertaking such a study, it is obviously essential to have a known standard of true chimerism in order to assess the extent of variance due to the methodology. Since it is not possible to establish the true level of chimerism in a patient, two chimerism models were employed, with pre-determined standards. This provided a basis to compare the models with clinical samples in terms of the mean performance for individual markers and the marker profile overall. In another facet of this study, we documented the performance of the size identification capability of the platform for the STR-PCR products, since the genotype of an STR allele is based on this parameter. Additionally, the variable performance seen for some markers prompted us to investigate possible theoretical and technical origins of this variance. The computational burden inherent in these studies was easily managed with the aid of a new software utility, ChimerTrack.25,26 Design and functional features relevant to these studies will be briefly described.
Overall, these studies justify long-term chimerism monitoring, and provide a reasonable basis for guidelines helpful in optimized marker selection.
METHODS
Sources of Samples
Samples originated from normal paternity cases, and 48 HSCTs (1–15 exams per patient, mean = 3.2). A typical HSCT involved a matched related donor, which provided 3–5 marker loci per sample. Using donor cord blood, 7 or more loci were available. Overall, this material provided percent chimerism values for over 500 marker loci in 55 different profiles.
Extracting DNA
All evaluations were performed on EDTA-treated peripheral blood (PB), bone marrow, or T-cell fraction samples. DNA was extracted with DNA Blood Mini-kit (QIAGEN, Valencia, CA). The concentration and purity were checked by measuring absorbance at 260 nm, and the A260/A280 ratio for purity, according to the kit recommendations.
Fabricating Artificial Mixed Chimeras
PB-derived DNA is available in our laboratory from healthy unrelated male and female individuals, appearing for STR-based paternity evaluations. Based on the initial STR evaluation, pairs were chosen for this study that showed at least 6/10 marker loci with non-shared alleles, i.e., informative loci. For a complete series, DNA was mixed to achieve ratios of 1.25%, 2.5%, 5%, 10%, 30%, 50%, 70%, 90% male, with preparation accuracy estimated at 1%. The cases with male 0 and 100% served as the reference to establish the origin of each allele in the chimera.
STR Analysis
Chimeric status evaluation was based on 10 STR tetra-nucleotide markers plus amelogenin (see Table 1) amplified using the AmpFlSTR SGM Plus multiplex PCR kit (ABI, UK).
TABLE 1.
Characteristics of Markers in the AmpF/STR SGM Plus Kit Locia
| Locus | Location | Common Sequence Motif | Size Rangeb | Dye Label | Dye Color |
| D3S1358 | 3p | TCTATCTG)1-3(TCTA)n | 114–142 | 5-FAM | |
| VWA | 12p12-pter | TCTA(TCTG)3-4(TCTA)n | 157–209 | 5-FAM | |
| D16S539 | 16q24-qter | (AGAT)n | 234–174 | 5-FAM | Blue |
| D2S1338 | 2q35-37.1 | (TGCC)n(TTCC)n | 289–341 | 5-FAM | |
| Amelogenin | X: p22.1-22.3 | — | 107 | JOE | |
| Y: p11.2 | 113 | JOE | |||
| D8S117c | 8 | (TCTR)nd | 128–172 | JOE | |
| D21S11 | 21q11.2-q21 | (TCTA)n(TCTG)n(TCTA)3[TCTA)3TA (TCTA)3TCA(TCTA)2TCCA TA](TCTA)n |
187–243 | JOE | Green |
| D18S51 | 18q21.3 | (AGGAA)n | 264–345 | JOE | |
| D19S433 | 19q12-13.1 | (AAGG)(AAAG)(AAGG)(TAGG) (AAGG)n |
106–140 | NED | |
| TH01 | 11p15.5 | (AATG)n | 165–204 | NED | Yellow |
| FGA | 4q28 | (TTTC)3TTTT TTCT(CTTT)NCTCC(TTCC)2 | 215–353 | NED |
a From user’s manual.
b The size range is the actual base-pair size of sequenced alleles contained in the Ampf/STR SGM Plus Allelic Ladder. The sizes in the table include the 3 A nucleotide addition.
c In some literature references, this locus is designated as D6S502.
d R can represent either an A or G nucleotide
PCR products were labeled with one of three fluorescent dyes (Table 1), detected during electrophoresis in an ABI 3100 Genetic Analyzer, using a 36-cm capillary and the POP4 polymer. The normal signal threshold was set for 50–75 units. Run simultaneously with each sample was a GS500 ROX size standard, and positive and negative controls accompanied each plate. These raw data were analyzed offline with the macro routines of the ABI Genescan program, which produces electropherograms and tabular estimates of the quantity of DNA at each of the STR alleles, as described below. Data from Genescan were imported into ChimerTrack, our locally developed software for computation of percent chimerism and graphic and tabular display of the computed results. Further details on this utility are provided below, in Results.
Statistical Analysis
Means and standard deviations (s.d.) were computed for all groups. Evaluation of precision utilized these values to compute the percent coefficient of variance (c.v. = [s.d./mean] × 100). For situations where c.v. would not be appropriate, error was computed, defined as: Error = (absolute value of profile mean – marker value)/profile mean) × 100. For comparison of true to observed chimerism, in the artificial chimeric series, the Pearson correlation coefficient, r, was computed. Student’s t-test was used to compare means between groups.
RESULTS
Since the investigational findings and their interpretation reflect the use of ChimerTrack, the first section will describe the software. This is followed by the results of quantitative marker performance. The final section concerns factors influencing the quantitative estimations of chimerism.
Description of ChimerTrack Software
Because an understanding of the software is relevant to data interpretation, a brief description also will be provided here.
Features
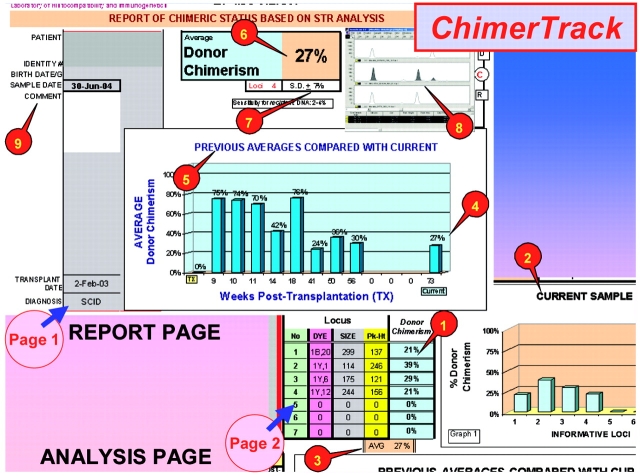
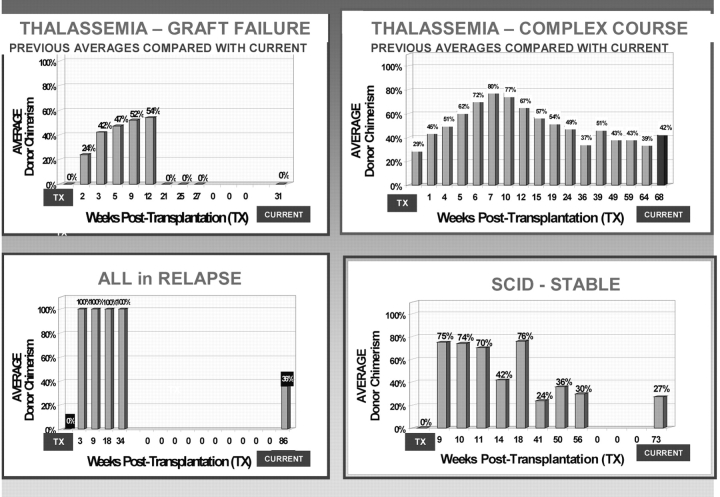
This Excel-based application computes the ratio of recipient/donor DNA, based on Genescan STR-DNA data, for alleles in chimeric loci. In order to trigger its computational algorithm to produce tabular and graphic displays (Figure 1, items #1 and #2), the Genescan DNA data are simply copied and pasted into a color-coded import block (Figure 2). For all of a sample’s informative loci, the software computes: (1) percent chimerism for each individual marker locus; (2) mean percent chimerism for sample; (3) sample s.d.; and (4) sample c.v. (Figure 1, items #6 and #7). Of particular importance for long-term tracking is that the report contains a graphic record of all previous sample results in comparison to the outcome of the current examination (Figure 1, #5). Figure 3 illustrates four graphic displays of the long-term chimerism status, as they appear on the report page in four different patients. Each record shows a clinically significant and different trend.
FIGURE 1.
Two pages from the ChimerTrack utility. The first is the report page for the current sample, which is issued to the clinicians. The second page is for posting the results of analysis, and includes data reserved for the laboratory. Nine functions are highlighted by numbered callouts: (1) Tabulated results, showing percent donor chimerism for each locus examined in current specimen; (2) bar graph for visual display of data in the table; (3) average (sample mean); (4) current mean is transferred to bar graph on the report page; (5) bar graph showing comparison of current sample mean with previous sample means, indicating the chimeric trend for the transplant; (6) current mean is automatically transferred to the report; (7) standard deviation for the profile in the current sample (right cell), and the number of informative loci (left); (8) electropherogram image, typical of the chimerism, is pasted here; (9) fields for patient and sample identification.
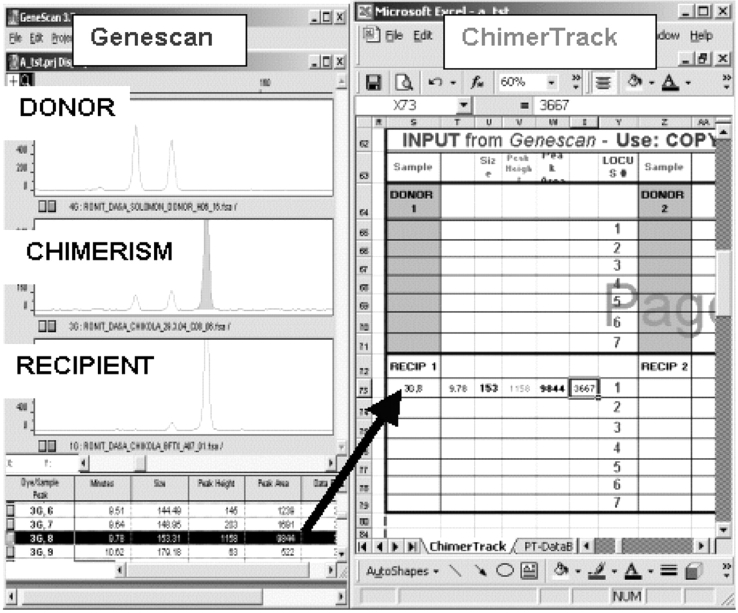
FIGURE 2.
Transfer of data from Genescan (left) to ChimerTrack. Both programs are opened side-by-side. The relevant data for a single peak is highlighted in the table below the Genescan electropherogram. Performing a copy/paste maneuver transfers the data to page 6 of the worksheet (arrow, right)
FIGURE 3.
ChimerTrack trend graphs appearing on the report page for four separate patients followed over a series of samples. All, acute lymphocytic leukemia; SCID, severe combined immune deficiency.
Computational Algorithm
A brief introduction to the algorithm is important here because it provides a basis for the criteria used to select informative loci. Additionally, it is important background for appreciating theoretical sources of error, discussed later.
The algorithm is used to calculate the ratio between donor- and recipient-derived DNA in a chimeric locus, expressed as percent chimerism. The calculation utilizes the Genescan program’s estimates for either the area or height of each electrophoretic peak (band) in a sample, which are relative measures of the quantity of DNA in each peak. The specific formulation of the ratio, according to the recommendation of a number of previous workers,3,6–8,11,12,27 is as follows:
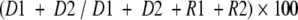 |
where D1 and D2 are donor-derived alleles in the chimeric sample, and R1 and R2 are derived from the pre-transplant recipient.
Selecting Informative Loci
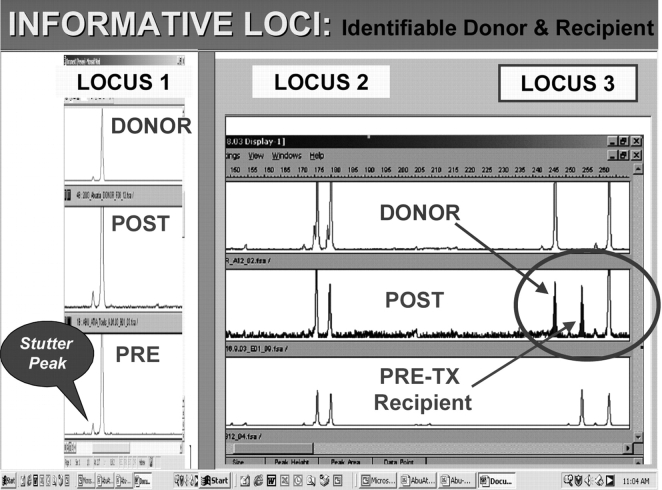
The initial stage in any form of quantitative analysis of chimerism will be the identification of informative marker loci. In such loci, recipient and donor alleles should be individually distinguishable—i.e., alleles at a locus are not completely shared between recipient and donor (Figure 4). Allelic sharing is influenced by the genetic frequencies of donor and recipient alleles in the population18,28 and the genetic relationship between these two individuals. Acceptable configurations for quantitation using ChimerTrack are summarized in Table 2. Loci with tri-allelic shared peaks (Table 2, cases 6 and 7) are not used because the estimations based on them are more variable, and less accurate, than values based on other allelic configurations (cases 1–5). This is probably primarily due to the inconsistent and non-proportional amplification of the various components in this single chimeric allele.
FIGURE 4.
Identifying informative loci. Three loci are shown from a Genescan electropherogram. Locus 3 is informative, despite a shared D-R bi-allelic peak, because it contains two other alleles that can be individually related to either the donor or the recipient. A stutter peak is shown for Locus 1. TX = transplantation.
TABLE 2.
Allelic Combinations Useable with ChimerTrack at informative Loci
| Case | Peak 1 | Peak 2 | Peak 3 | Peak 4 | Use |
| 1 | D1 | D2 | R1 | R2 | Yes |
| 2 | D1 | R1 | D2+R2 | 0 | Yes |
| 3 | D1+D2 | R1 | R2 | 0 | Yes |
| 4 | D1 | R1+R2 | D2 | 0 | Yes |
| 5 | D1+D2 | R1+R2 | 0 | 0 | Yes |
| 6 | D1 | D2+R1+R2 | 0 | 0 | No |
| 7 | D1+D2+R1 | R2 | 0 | 0 | No |
Another issue relates to stutter peaks (Figure 4, Locus 1), which are PCR-generated artifacts. When using the ABI tetranucleotide STR platform, they appear one 4-bp repeat unit smaller than an authentic allele, i.e., located to its left in the electropherogram.4,10,11,18,24 The stutter peak contains 5–15% of the DNA content ascribed to the larger peak.29 Its significance is that it may simulate a low-level mixed chimera in an allelic configuration where the donor- and recipient-derived alleles are 4 bp apart. Additionally, stutter-like peaks may occur after the main peak as well (echo peaks). Such loci generally should not be used, particularly where a definite low-level chimerism of the same magnitude is seen in another locus. However, in our experience, if the chimerism is >30%, the computational values from stutter loci do not appear less accurate, or more variable, than loci free of this configurational problem.
Based on the foregoing, in practice, a typical patient “marker profile” will usually consist of alleles from only 3–7 of the 10 marker sets (systems) in the SGM Plus kit shown in Table 1.
Quantitative Marker Performance
The following account describes platform performance in terms of three parameters: accuracy of chimerism estimate compared to true, precision (i.e., repetitive performance on the same markers and sample), and fragment size calling. Artificial and simulation chimeric models and patient material are compared in terms of how individual markers in the profile approximate the sample mean. Results are summarized in Table 3. In the final Results section, factors influencing chimerism quantitation are considered.
TABLE 3.
Summary of Marker and Profile Performance Using AmpF/STR SGM Plus Kit
| Single Patient | Simulation 50% | Artificial Chim 50% | Artificial Chim 1–100% | |
| Accuracy (marker) | 92–98% sample mean | 92–100% true | 88–98% true | true vs. observed: r = 0.99 |
| Precision (marker) | error = 2–9% | C.V. = 6% | C.V. = 4% | — |
| Precision (profile) | C.V. = 7% | C.V. = 4% | — | — |
Accuracy
The parameter is defined as the difference between true and observed values for percent donor chimerism. In artificial chimerisms comprising 10 mixtures from 0 to 100% chimerism the observed and true (fabricated) values were highly correlated (r = 0.99). Comparable results have been reported from other laboratories.15,20,22,30
Further, as a basis for comparison with clinical samples, consider the specific results for a 50% artificial chimerism with six markers in its profile. It showed a profile mean of 54% chimerism, with a range for individual markers of 51–57%, s.d.of 2%. In terms of accuracy, for this sample, individual markers had values of 102–114% of the sample mean. Some of this variance is likely to reflect slight inaccuracies attributable to pipetting small volumes of DNA. In comparison, individual markers (n = 28) from several clinical samples had values of 92–102% of the profile mean value. For another typical case, the complete results are illustrated in Table 4. This sample had a profile mean of 92%, s.d 2%, and the five individual markers had values ranging from 89 to 96% chimerism, i.e., 97 to 104% of the profile mean value. Consequently, the levels of accuracy and variance appear comparable in all material. From Table 4 it also can be seen that no significant difference results from using either peak height or area in the estimation of percent chimerism; both of these parameters are available from the Genescan data. The practical consequence of this last finding, is that when it is occasionally impossible for technical reasons to utilize peak area (our normal preference) peak height may be substituted as the input data for ChimerTrack.
TABLE 4.
Percent Chimerism in Five Loci from a Single Sample Computed Based on Peak Area vs. Peak Height Data from Genescan
| Locus 1 | Locus 2 | Locus 3 | Locus 4 | Locus 5 | Mean | s.d. | |
| Height | 91% | 90% | 92% | 96% | 90% | 92% | 2% |
| Area | 92% | 89% | 91% | 96% | 91% | 92% | 2% |
Reproducibility/Precision
There are two aspects to this issue. The first is the reproducibility of a “mean percent chimerism” value for a given marker profile, repeatedly evaluated in either the same or different samples. The second, is the repetitive performance of the same marker locus over time, in the same patient. We evaluated three sample types on this issue, summarized in Table 3.
Simulated chimerism.
This is a conceptual construct that is based on an analysis of STR-DNA from a single, normal non-chimeric individual. In such a sample, all heterozygotic loci can be considered a simulation of a 50% chimerism, according to the criteria in case 5 of Table 2. This simulation approach avoided the physical problems of actually fabricating an equivalent artificial chimerism, and therefore excluded all sources of variability except system performance. In that light, 19 replications of the same sample, with a 10-marker profile, were investigated. The mean for all sample means was 51% chimerism, mean s.d. = 2%, and mean c.v. = 4%. The data set (n = 190 marker values) should be large enough to average out intrinsic variation of individual markers. The markers themselves were separately evaluated across samples, and showed only a slightly greater degree of variability (not significant), with an overall mean c.v. of 6%. The actual range for the individual samples’ mean percent chimerism was 47–54% chimerism, or an error of 6–8% true. The mean error for the entire data set was 2% true, and may represent the optimized limit of accuracy, and hence sensitivity, for the platform
Patient material.
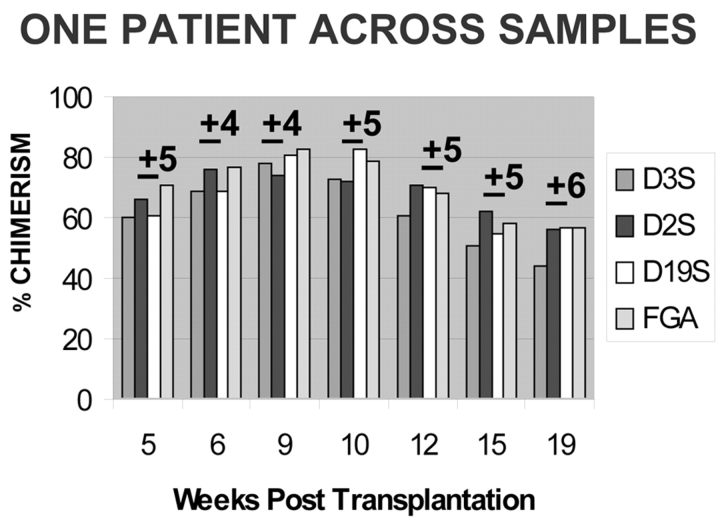
Precision was evaluated in the same marker profile over a series of samples derived from a single patient. This individual had seven sequential samples over a 98-d period, which required examining the same four markers each time, as illustrated in Figure 5. A fairly stable mid-level mixed chimerism persisted throughout that period. Although there is obviously no reference value for percent chimerism in these samples, the samples’ variance can be compared to that found for the model samples. It was found that the mean s.d. for the group of individual profile means was 5%, and the mean c.v. 7%, which is similar to performance in the models.
FIGURE 5.
Performance of four markers in the same chimeric patient across seven samples spanning a 98-d period. The patient’s clinical status was stable during this period. The sample s.d. is noted above each group of bars. Mean s.d. of group means = 5% and mean c.v. = 7%.
Fragment size calling.
In all samples, an amelogenin X marker is evaluated. The marker in >90% of samples appeared at 103 + 0.5 bp. Additionally, we inspected the ChimerTrack summary tables for three typical patients over three successive samples, using the same informative markers (Table 5) other than amelogenin. It can be seen that size calling is consistent across samples. The small variation noted is due to the integer rounding that is done by ChimerTrack for the imported Genescan data. These findings are in agreement with previous studies, which have evaluated much, but not all, of the SGM+ marker panel.24
TABLE 5.
Marker Size Calling Performance
| Locus 1 | Locus 2 | Locus 3 | |||||
| Pta | Sample | Dye | Sizeb | Dye | Size | Dye | Size |
| 1 | 1 | Blue | 299 | Yellow | 114 | Yellow | 244 |
| 1 | 2 | Blue | 298 | Yellow | 113 | Yellow | 244 |
| 1 | 3 | Blue | 298 | Yellow | 113 | Yellow | 244 |
| 2 | 1 | Blue | 119 | Blue | 253 | Green | 144 |
| 2 | 2 | Blue | 119 | Blue | 253 | Green | 144 |
| 2 | 3 | Blue | 119 | Blue | 253 | Green | 145 |
| 3 | 1 | Blue | 140 | Blue | 303 | Yellow | 129 |
| 3 | 2 | Blue | 140 | Blue | 303 | Yellow | 129 |
| 3 | 3 | Blue | 140 | Blue | 303 | Yellow | 130 |
a Three sequential samples for each of three patients (Pt).
b Allele size in base pairs (bp).
Factors influencing Quantitative Results
It is apparent from these and previous30 data that variations in marker performance are regularly occurring. Therefore, a number of factors potentially contributing to this variability were investigated.
Differential amplification efficiency of alleles.
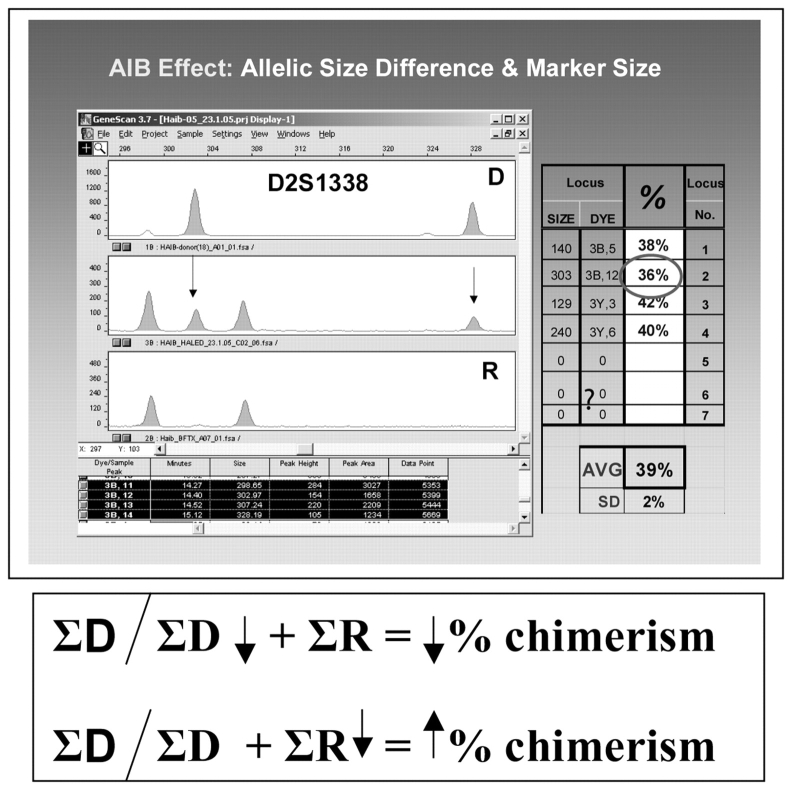
One common source of variance in our material is differential amplification efficiency of alleles, or amplification imbalances (AIB). It could be manifest either as differences between the two donor (D) alleles or two recipient (R) alleles, or between the pair of D alleles and the pair of R allelic pairs, i.e., total D vs. total R. None of these options are exclusive, and all may occur at a given locus. AIB within the D or R pairs themselves is more common,3–8,24 typically resulting in a <15% differential in 70–100% of values, depending on the marker system. A number of factors were seen associated with higher levels of AIB, including NED/yellow dye label, low signal amplitude, large allelic size, and large allelic size differential (Figure 6). However, these factors could be additive and produce uncommonly large over- or underestimations for a locus. Figure 6 illustrates probable additive effects, since marker locus exhibits low signal amplitude, allelic size differential, and large marker size among D alleles, which resulted in an AIB for the D1–D2 pair of 25%.
FIGURE 6.
Top: Chimeric sample showing differential amplification effect in the two D (donor) alleles, probably reflecting additive effects of low signal amplitude, allelic size difference, and large marker size. Since the sum of D allele peak areas will be reduced, from the ratio formula we would expect an underestimate of the true value. We do indeed see that this locus is below the profile mean. In this case the variation is sufficiently small that the results could be reported without any further changes. Bottom: Effects of reduction in quantity of R or D DNA on % chimerism.
Formula effects.
When an allele is inefficiently amplified, the resultant AIB will impact on the quantitative results. The effect will depend on whether the AIB affects just one allele in a D or R pair of alleles, or both alleles in a pair. In either case, the quantitative outcome is derived by using the specific ratio formula described above, which determines the magnitude and direction of the error in percent chimerism due to AIB (Figure 6). For instance, if the D pair is 30% reduced over true DNA quantity at these alleles, this will result in an underestimate of 10% chimerism at the locus. In contrast, a 30% reduction of one allele in the pair results in only a 4% change in percent chimerism for that marker.
Unexplained variability.
Some marker loci exhibit variability that is independent of the variables identified to date. For instance, the D3S/blue alleles in Figure 5 not only performed erractically relative to the sample, but also consistently underestimated the profile mean, despite the low frequency of AIB at that locus in normal specimens (p = 0.304, paired t-test, 1 tailed, n = 10). In contrast, the FGA marker alleles tended to overestimate the profile mean in this patient.
DISCUSSION AND CONCLUSIONS
Chimerism testing based on STRs has become an important component of the post-transplantation monitoring routine for HSCTs.1–26 Long-term monitoring enhances the predictive utility of this assay approach. Although we do not yet know whether the absolute value of chimerism is critical, relative changes are clearly important as harbingers for graft success or failure. It is currently unknown to what extent observed changes in chimerism across samples can be attributed to variation intrinsic to the technology platform. This issue is for the first time systematically assessed in the current study, which was specifically undertaken to assess the performance of the ABI multiplex STR platform, using the SGM Plus kit, particularly in regards to its suitability for long-term monitoring of hematopoietic chimerism.
Using ChimerTrack, both chimeric models and actual clinical material were evaluated. The results for each phase of the study are summarized in Table 3, and suggest several conclusions. The first is that the data are highly consistent across approaches, supporting the overall validity of the assessment. Of considerable importance is the finding that the sample mean is an extremely good estimate of the true chimerism value. One may conclude, then, that the mean percent chimerism value in clinical samples also will be a reliable estimate of the patient’s chimeric status. A practical implication of this conclusion is that reports of percent chimerism should be based on a sufficient number of optimally performing STR markers to produce a mean value with low variance. Recommendations for selecting optimized markers are described at the end of the Discussion.
Second, the levels of accuracy and precision of individual markers, and various marker profiles, are acceptable performance parameters for a laboratory assay. Sensitivity to the minor component DNA was not examined here, since it has been evaluated previously in a number of laboratories and is the range of 1–5%.1–12 These observations warrant concluding that the platform is suitable for effective tracking of clinically significant changes in chimeric status over time. On the other hand, based on the 50% simulated chimerism model, the mean estimated chimerism value was 51%, giving an error of 2% true. This value is likely to represent the optimized limit of accuracy for the platform. This value is probably a realistic estimate of the variance intrinsic to this technology platform, because the model is free of the technical variables inherent in preparing artificial chimerisms used in previous estimates.15,20,22,30
Since variability in marker performance appears intrinsic to the platform, it was of interest to attempt to identify the principal sources of this variability. Several factors were identified that impacted on quantitation by producing amplification imbalances. These factors included low signal amplitude,10,22 NED/yellow dye label,22,24,32 large marker size,31 and large allelic size interval. We believe this last factor is reported here for the first time. Nonetheless, although differential allelic amplification occurs frequently, the use of the ratio formula generally neutralizes the effect on quantitative outcome of a sample examination. An estimation of percent chimerism in a marker locus will rarely need to be rejected because of a large estimation error compared to the mean. As shown above, substantial errors of this sort typically occur only when a locus concomitantly exhibits several factors predisposing to AIB. Criteria for rejecting a value are considered below.
It is worth noting that the actual level of mixed chimerism in the sample will also influence the quantitative impact of a marker’s variability. For example, a 5% s.d. for 20% chimerism is a much greater relative error than 5% s.d. for a 90% chimerism. For this reason, we recommend that the coefficient of variability be included in the report. This statistic indicates the variance as a function of the mean, so that in this example of 5% s.d., the c.v. would be 25% and 6%, respectively.
Based on the present findings, the following guidelines have been developed for selecting profile markers for use with ChimerTrack:
Fabricate a 50%:50% artificial chimerism for each new donor-recipient pair.
Inspect the allelic configurations in all marker loci, and select loci with at least one pure donor allele and one recipient allele—i.e., loci that are informative for quantitation (Table 2).
- Select at least three high-performance and informative markers from the artificial chimera:
- —Avoid loci with alleles in stutter peak positions, or tri-allelic D-R peaks.
- —Avoid markers with overall low signal, which usually occurs among the largest NED/yellow series markers, and occasionally in D2S of the 5-FAM/blue group.
- —Assess longitudinal performance of profile markers in clinical samples, and eliminate poorly performing markers consistently >5% profile mean. Relative performance of the individual markers comprising the sample profile is automatically assessed by ChimerTrack (Figure 1, item #2).
- Delete the results for an individual marker in a profile >5% of the sample mean if it satisfies the following conditions. ChimerTrack will automatically recalculate the new mean.
- —Profile c.v. > 20% and profile mean > 40–50% chimerism.
- —Profile c.v. > 15% and profile mean < 30–40% chimerism.
- —The utility will automatically re-compute the new mean percent donor chimerism.
In the event there is only one locus informative for quantitation, as defined above, and additional support for the single value is desired, use a locus with a stutter peak configuration that is otherwise informative. However, the chimerism should be >30%. (Stutters are typically <15% of following peak).24
Although ChimerTrack is copywrited, the current version of the utility, and an illustrated tutorial, are available free from the author to hospitals and private laboratories by written request.
Acknowledgments
This work was partially supported by a grant from the Hirsh and Gania Wassermann Grant Fund for Intramural Research, Tel Aviv University.
REFERENCES
- 1.McCann SR, Lawler M. Monitoring outcome: MRD, chimaerism and relapse. In: Haematopoietic Stem Cell Transplantation, Berlin, EBMT, 2004:196–212.
- 2.Mass F, N Schaap, S Kolen, Zoetbrood A, Buno I, Dolstra H, et al. Quantification of donor and recipient hemopoietic cells by real-time PCR of single nucleotide polymorphisms. Leukemia 2003;17:621–629. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez-Aviles, F, Urbano-Ispizua A, Aymerich M, Colomer D, Rovira M, Martinez C, et al. Serial quantification of lymphoid and myeloid mixed chimerism using multiplex PCR amplification of short tandem repeat-markers predicts graft rejection and relapse respectively, after allogeneic transplantation of CD34+ selected cell from peripheral blood. Leukemia 2003;17:613–620. [DOI] [PubMed] [Google Scholar]
- 4.Schraml E, Daxberger H, Watzinger F, Lion T. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: The Vienna experience. Leukemia 2003;17:224–227. [DOI] [PubMed] [Google Scholar]
- 5.Acquaviva C, Duval M, Mirebeau D, Bertin R, Cavé H. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: The Paris-Robert Debré experience. Leukemia 2003;17:224–227. [DOI] [PubMed] [Google Scholar]
- 6.Kreyenberg H, Holle W, Mohrle S, Niethammer D, Bader P. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: The Tuebingen experience. Leukemia 2003;17:237–240. [DOI] [PubMed] [Google Scholar]
- 7.Koehl U, Beck O, Seifried E, Klingebiel T, Schwabe Seidle C. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: The Frankfurt experience. Leukemia 2003;17:232–236. [DOI] [PubMed] [Google Scholar]
- 8.Senitzer D, Gaidulis L. Short tandem repeat analysis of engraftment in allogeneic stem cell transplantation ASHI Quarterly 2001;25:49–54. [Google Scholar]
- 9.Lion T, Muller-Bérat N. Debate round table: Chimerism testing after allogeneic stem cell transplantation: Importance of timing and optimal technique for testing in different clinical-biological situations. Leukemia 2003;17:612–633. [DOI] [PubMed] [Google Scholar]
- 10.Lion T. Summary: Reports on quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection. Leukemia 2003;17:252–254 [DOI] [PubMed] [Google Scholar]
- 11.Hancock JP, Goulden NJ, Odakhill A, Steward CG. Quantitative analysis of chimerism after allogeneic stem cell transplantation using immunomagnetic selection and fluorescent microsatellite PCR. Leukemia 2003;17:247–251. [DOI] [PubMed] [Google Scholar]
- 12.Thiede C, Lion T. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection. Leukemia 2003;15:303–306. [DOI] [PubMed] [Google Scholar]
- 13.Baron F, Baker JE, Strob R, Gooley TA, Sandmaier BM, Maris MB, et al. Kinetics of engraftment inpatients with hematologic malignancies given allogeneic hematopoietic cell transplantation after non-myeloablative conditioning. Blood 2004;104:2254–2262. [DOI] [PubMed] [Google Scholar]
- 14.Lee K-H, Lee J-H, Choi S-J, Lee JH, Kim S, Seol M, et al. Monthly prospective analysis of hematopoietic chimerism after allogeneic hematopoietic cell transplantation. Bone Marrow Transplantation 2003;32:423–431. [DOI] [PubMed] [Google Scholar]
- 15.Bader P, Beck J, Frey A, Schlegel PG, Hebarth H, Handgretinger R, et al. Serial and quantitative analysis of mixed hematopoietic chimerism by PCR inpatients with acute leukemias allows the prediction of relapse after allogeneic BMT. Bone Marrow Transplantation 1998;21:487–495. [DOI] [PubMed] [Google Scholar]
- 16.Thiede C, Bornhauser M, Oelschlagel U, Brendel C, Leo R, Daxberger H, et al. Sequential monitoring of chimerism and detection of minimal residual disease after allogeneic blood stem cell transplantation BSCT using multiplex PCR amplification of short tandem repeat-markers. Leukemia 2001;15:293–302. [DOI] [PubMed] [Google Scholar]
- 17.Dubovsky J, Daxberger H, Fritsch G, Printz D, Peters C, Matthes S, et al. Kinetics of chimerism during the early post-transplant period in pediatric patients with malignant and non-malignant hematologic disorders: Implications for timely detection of engraftment, graft failure and rejection. Leukemia 1999;13:2060–2069. [PubMed] [Google Scholar]
- 18.Thiede C, Bornhauser M, Ehninger G. Evaluation of STR informativity for chimerism testing—comparative analysis of 27 STR systems in 203 matched related donor recipient pairs. Leukemia 2004;18:248–254. [DOI] [PubMed] [Google Scholar]
- 19.Thiede C. Diagnostic chimerism analysis after allogeneic stem cell transplantation: New methods and markers. Am J Pharmacogenomics 2004; 4:177–187. [DOI] [PubMed] [Google Scholar]
- 20.Thiede C, Florek M, Bornhauser M, Ritter M, Mohr B, Brendel C, et al. Rapid quantification of mixed chimerism using multiplex amplification of short tandem repeat markers and fluorescence detection. Bone Marrow Transplantation 1999;23:1055–1060. [DOI] [PubMed] [Google Scholar]
- 21.Khan F, Agarwal A, Agrawal S. Significance of chimerism hematopoietic stem cell transplantation: New variation on an old theme. Bone Marrow Transplantation 2004;34:1–12. [DOI] [PubMed] [Google Scholar]
- 22.Madeo D, Capellari A, Castaman G, Barimondi R, Rodeghiero F. Multiplex amplification and fluorimentric detection of short tandem repeats for mixed chimerism after bone marrow transplant. Leukemia & Lymphoma 2003;17:1–10. [DOI] [PubMed] [Google Scholar]
- 23.Lion T, Muller-Bérat N. Debate round table: Chimerism testing after allogeneic stem cell transplantation: Importance of timing and optimal technique for testing in different clinical-biological situations. Leukemia 2003; 17:220–254. [DOI] [PubMed] [Google Scholar]
- 24.Butler JM. Commonly used short tandem repeat markers, In: Forensic DNA Typing, San Diego, Academic Press, 2001:53–79.
- 25.Kristt D, Klein T. STR-based chimerism testing using ChimerTrack interactive-graphics software: Easing the burden. ASHI Quarterly 2004a;28:16–19. [Google Scholar]
- 26.Kristt D, J Stein, I Yaniv, T Klein. Interactive ChimerTrack software facilitates computation, visual displays and long-term tracking of chimeric status based on STRs. Leukemia 2004;18:909–911. [DOI] [PubMed] [Google Scholar]
- 27.Scharf SJ, Smith AG, Hansen JA, McFarland C, Erlich HA. Quantitative determination of bone marrow transplant engraftment using fluorescent polymerase chain reaction primers for human identify markers. Blood 1995;85:1954–1963. [PubMed] [Google Scholar]
- 28.Hantschel M, Hausmann R, Lederer T, Martus P, Betz P. Population genetics of nine short tandem repeat (STR) loci—DNA typing using the AmpFl-STR SGM plus PCR amplification kit. Forensic Sci Int 1999;112:293–395. [DOI] [PubMed] [Google Scholar]
- 29.Walsh PS, Fildes NJ, Reynolds R. Sequence analysis and characterization of stutter products at the tetra-nucleotide repeat locus vWA, Nuclei Acids Res 1996; 23:2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardiner N, Lawler M, O’Riordan JM, Duggan C, De-Arce M, McCann SR. Monitoring of lineage-specific chimaerism allows early prediction of response following donor lymphocyte infusions for relapse chronic myeloid leukaemia. Bone Marrow Transplant 1998;21:711–719. [DOI] [PubMed] [Google Scholar]
- 31.Chalandon Y, Vischer S, Helg C, Chapuis B, Roosnek E. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: The Geneva experience. Leukemia 2003;17:228–231. [DOI] [PubMed] [Google Scholar]
- 32.Lee LG, Spurgeon SL, Heiner CR, Benson SC, Rosenblum BB, Menchen SM. New energy transfer dyes for DNA sequencing. Nuclei Acids Res 1997;25:2816–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]