Abstract
Posttranslational modifications such as ubiquitination and phosphorylation play an important role in the regulation of cellular protein function. Homeodomain-interacting protein kinase 2 (HIPK2) is a member of the recently identified family of nuclear protein kinases that act as corepressors for homeodomain transcription factors. Here, we show that HIPK2 is regulated by a ubiquitin-like protein, SUMO-1. We demonstrate that HIPK2 localizes to nuclear speckles (dots) by means of a speckle-retention signal. This speckle-retention signal contains a domain that interacts with a mouse ubiquitin-like protein conjugating (E2) enzyme, mUBC9. In cultured cells, HIPK2 is covalently modified by SUMO-1, and the SUMO-1 modification of HIPK2 correlates with its localization to nuclear speckles (dots). Thus, our results provide firm evidence that the nuclear protein kinase HIPK2 can be covalently modified by SUMO-1, which directs its localization to nuclear speckles (dots).
The ubiquitin system is one of the main posttranslational protein-modification systems that is required for the selective degradation of many short-lived proteins in eukaryotic cells, thereby regulating important cellular processes, including cell-cycle progression, signal transduction, endocytosis, and transcription (1, 2). In this system, the ubiquitin molecule is covalently attached to target proteins by the sequential reaction of the ubiquitin-activating (E1) and ubiquitin-conjugating (E2) enzymes and the ubiquitin-protein ligase (E3). Eukaryotic cells also express a set of ubiquitin-like proteins (Ubls), such as SUMO-1 (or sentrin) and Rub-1 (3, 4). Like the ubiquitin system, these Ubls can be covalently attached to target proteins by a similar but distinct enzyme machinery. Recently, it has been shown that UBC9, a functional homologue of the E2-type ubiquitin-conjugating enzymes, forms a thioester with Ubl and conjugates it to target proteins (5–7). Only a few protein targets for Ubl modification by UBC9, which include RanGAP1 (8–13), PML (14, 15), SP100 (16), and IκBα (17), have been identified to date. In contrast to ubiquitin modification, recent experimental evidence suggests a different role for Ubl modifications, such as subcellular localization or the increased stability of target proteins (8, 9, 17–20). Because this Ubl modification system is universal in eukaryotic cells, it is presently of interest to identify its novel targets as well as its role in cellular protein function.
Homeodomain-interacting protein kinases (HIPKs) are a family of recently identified nuclear protein kinases that differentially interact with homeodomain transcription factors (21) and are well conserved in various organisms (21–23). HIPK2 can act as a transcriptional corepressor for homeoproteins and contains multiple functional domains, including an interaction domain for homeoproteins, a corepressor domain (CRD), PEST sequence, and a YH domain in the C terminus in addition to a protein kinase domain (21). Most interestingly, HIPKs localize to nuclear speckles (dots) (21) that are distinct from the speckles containing splicing factors. It is unclear how HIPK2-containing nuclear speckles (dots) form. Also, how these speckles (dots) regulate the function of HIPKs within the nucleus is totally unknown. Here, we show that a nuclear protein kinase, HIPK2, is a novel target for mUBC9 and is covalently modified by the ubiquitin-like protein SUMO-1. Furthermore, we demonstrate that SUMO-1 modification of HIPK2 correlates with its localization to nuclear speckles (dots).
Experimental Procedures
Plasmid Construction and Cell Transfection.
For construction of various green fluorescent protein (GFP)-HIPK2 expression vectors, initially HIPK2 (amino acids 25–1189) was cloned into the EcoRI site of the pEGFP-C3 expression vector (CLONTECH) to generate construct B, and construct B was used for the generation of deletion constructs with unique restriction sites. The unique restriction sites used are as follows: C, BbsI (amino acid 967); D, SmaI (amino acid 859); E, PstI (amino acid 629); F, BamHI (amino acid 327); G, SapI (amino acid 139); H, ClaI (amino acid 503); I, SmaI (amino acid 860). Constructs K and L were derived from construct H. Gaps were filled by treatment with the Klenow fragment of DNA polymerase after digestion. Deletion constructs from the N terminus (G, H, I, and J) were confirmed by nucleotide sequencing to check ORFs. For the construction of the GFP-tagged SUMO-1 expression vector, first, SUMO-1 cDNA was amplified by PCR with specific primers (SUMO-53: 5′-ATGGGATCCACCATGTCTGACCAGGAGGCAAAACCTTCA-3′, and SUMO-35: 5′-ATGGTCGACAACCGTCGAGTGACCCCCCGTTTGTTC-3′) and mouse heart cDNAs as template DNA and cloned into the BamHI/SalI sites of the pBluescript vector to generate pSUMO-1. The pSUMO-1 plasmid was confirmed by nucleotide sequencing. The BamHI/SalI-codigested SUMO-1 DNA fragment was subcloned into the BglII/SalI sites of the pEGFP-C1 expression vector (CLONTECH) to generate the GFP-SUMO-1 expression vector. For the construction of the Myc-tagged HIPK2 expression vector, five copies of Myc tags were amplified from the CS3+MT vector by PCR with specific primers (MT-53: 5′-AGCTGCTAGCTGATTTAGGTGACACTATAGAATACA-3′, and MT-35: 5′-AGCTAAAGCTTGGTGAGGTCGCCCAAGCTCTCCAT-3′) and subcloned into the NheI/HindIII sites of the GFP-HIPK2 plasmid to replace the GFP region, resulting in the Myc-HIPK2 expression vector. To generate the GFP-HIPK2(K1182R) mutant form, PCR products amplified with specific primers (K2-860-53R: 5′-ATGGAATTCCGGGAGCGACAGCGGCAGACGATT-3′, and K2-1189-35X: 5′-ATGTCTAGATATGTAAGGGTATTGGTTGAC-3′ for 860-1189 construct; K2-860-53R and K2-1159-35X: 5′-ATGTCTAGAGGCAAACTGGGCTGGATACTG-3′ for 860-1159 construct; K2-860-53R and K2-K1182R-35X: 5′-ATGTCTAGATATGTAAGGGTATTGGTTGACCCGGGCAGGACTCAGTGGGTATC-3′ for the K1182R construct) were subcloned into the EcoRI/XbaI sites of pEGFP-C2. Constructs were confirmed by nucleotide sequencing. CV-1 cells were transfected with various GFP-HIPK2 constructs by the calcium phosphate precipitation method, as described previously (24), and visualized with GFP fluorescence 48 hr after transfection.
Yeast Two-Hybrid Assay.
For bait construction (construct D5), a DNA fragment (1.0 kb from amino acids 860-1189 of HIPK2) was subcloned into the SmaI site of pGBT9 (CLONTECH). Approximately 7 × 106 transformants from a mouse embryonic day 11 match-maker cDNA library (CLONTECH) were screened in the HF7C yeast strain. For the deletion analysis of the HIPK2 interaction domain with mUBC9 in yeast, DNA fragments from GFP-HIPK2 fusion constructs were generated by restriction digestion (D1, EcoRI fragment from construct B; D7, SalI/KpnI fragment from construct B). D2 was derived from construct D1 BamHI digestion and self-ligation. For generation of the D4 construct, the EcoRI DNA fragment of yeast clones obtained from the original HIPK2 screening with Nkx-1.2 (21) was subcloned into pGBT9. D6 was generated from the construct D5 by the SalI digestion and self-ligation. For construction of D3, D8, D9, and D10, DNA fragments were amplified by PCR with specific primers (D3, D3-53: 5′-ATGGAATTCATGACCAACACCTATGAGGTT-3′, and D3-35: 5′-ATGGTCGACCTTGCAAATCTCCATGTTTTG-3′; D8, D8-53: 5′-ATGGAATTCGAGCGACAGCGGCAGACGATT-3′, and D8-35: 5′-ATGGTCGACGGCCTGGGTCTTCAGTGGGGG-3′; D9, D8-53 and D9-35: 5′-ATGGTCGACGCTGGAGGAGTCAGAGTAGGG-3′; D10, D8-53 and D10-35: 5′-ATGGTCGACCTTCTGCTCCTCTTCTTCATC-3′), codigested with EcoRI/SalI and were fused in frame with the GAL4 DNA-binding domain of pGBT9. β-Galactosidase activity was measured with the liquid culture assay.
In Vitro Pull-Down Assay.
Full-length HIPK2 DNA was subcloned into a pSPUTK vector (Invitrogen) and subjected to in vitro translation with the TNT Coupled Reticulocyte Lysate System (Promega). The glutathione S-transferase (GST)-mUBC9 fusion protein was expressed in Escherichia coli by using the pGEX-5X-1 vector (Amersham Pharmacia) and purified as described (24). Pull-down assays were performed by incubating equal amounts of GST or GST-mUBC9, immobilized onto glutathione-Sepharose beads, with in vitro-translated HIPK2 protein in binding buffer A [20 mM Hepes (pH 7.5)/50 mM KCl/2.5 mM MgCl2/10% glycerol/1 mM DTT/1% Nonidet P-40/25 mg/ml BSA/200 μg/ml PMSF]. The mixtures were placed on ice for 1 hr and washed five times with Tris-buffered saline and 1% Nonidet P-40, and bound proteins were eluted, separated by 4–20% gradient SDS/PAGE, and autoradiographed.
Western Blot Analysis and Immunoprecipitation.
Transfected CV-1 cells with expression vectors for GFP-SUMO-1 and Myc-HIPK2 were lysed directly with a lysis buffer [50 mM Tris⋅HCl (pH 6.8)/2% SDS/10% glycerol/360 mM 2-mercaptoethanol] supplemented with protease inhibitors (Roche Molecular Biochemicals). Proteins (15 μg) from total cell lysates were separated on 4–20% gradient SDS/PAGE and transferred to a nylon membrane (NEN). Western blotting was performed by incubating membranes with primary Abs (anti-Myc mouse mAb from Invitrogen, 1:5,000 dilution; anti-GFP rabbit polyclonal Ab from CLONTECH, 1:1,000 dilution) in 1× TNE buffer [10 mM Tris⋅HCl (pH 7.5)/50 mM NaCl/2.5 mM EDTA] containing 3% nonfat dried milk. Anti-HIPK2 rabbit Ab was generated by using a synthetic peptide from HIPK2 (amino acids 425–443) and purified by using a protein A IgG purification kit (Pierce). The signals were detected with chemiluminescence (Supersignal; Pierce). For immunoprecipitation, cell extracts (500 μg) were diluted (1:10) in 1× NETN [50 mM Tris⋅HCl (pH 7.5)/5 mM EDTA/300 mM NaCl/1 mM DTT/1% Nonidet P-40] containing protease inhibitors and incubated with an anti-GFP rabbit polyclonal Ab (10 μg/ml) or anti-SUMO-1 goat Ab (Santa Cruz Biotechnology) for 1 hr at 4°C. After adding protein A/G plus agarose beads (Santa Cruz Biotechnology), the mixtures were further incubated for 4 hr at 4°C with mild agitation. The beads were collected and washed five times with 1× NETN using the IMMUNOcatcher (CytoSignal, Irvine, CA) as described in the manufacturer’s protocol. The bound proteins were eluted and analyzed by Western blotting with an anti-Myc mouse mAb (Invitrogen; 1:5,000 dilution). For subcellular fractionation, CV-1 cells were cotransfected with the Myc-HIPK2 and the GFP-SUMO-1 expression vectors, collected, and resuspended in buffer A [10 mM Hepes (pH 7.9)/1.5 mM MgCl2/10 mM KCl/0.5 mM DTT) containing protease inhibitors, and subcellular fractionation was performed as described previously (25). Sample fractionation with a nonionic detergent (Nonidet P-40) was performed as described (14). Briefly, the Nonidet P-40-soluble proteins were extracted in 1× NETN buffer, and the pellets (Nonidet P-40-insoluble fractions) were resuspended and extracted in SDS sample buffer [50 mM Tris⋅HCl (pH 6.8)/2% SDS/10% glycerol/360 mM 2-mercaptoethanol) by boiling.
Immunohistochemistry.
CV-1 cells were grown on coverslips and transfected with the Myc-HIK2 and GFP-SUMO-1 expression vectors. Thirty-six hours after transfection, cells were fixed with 10% formaldehyde for 15 min at room temperature and incubated with a solution containing 1× PBS and 0.5% Triton X-100. Cells were rinsed with 1× PBS containing 1% BSA, and incubated with an anti-Myc mouse mAb (1:500 dilution) for 1 hr. After washing cells five times with 1× PBS, cells were incubated with a secondary anti-mouse IgG Ab (1:1,000 dilution) conjugated with rhodamine red (Molecular Probes). Confocal laser scanning microscopy was performed with a Zeiss LSM510 microscope, using excitation wavelengths of 543 nm (rhodamine red) and 488 nm (GFP). The acquired images were processed with Adobe Photoshop and printed with Fujix pictography 3000.
Results and Discussion
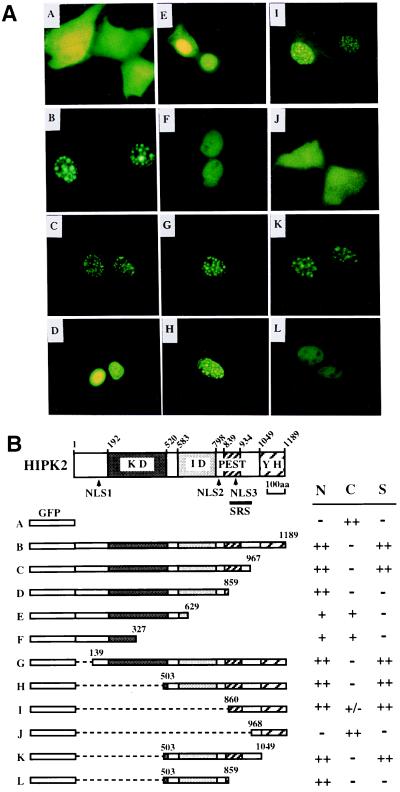
Initially, we mapped the nuclear localization signals (NLSs) and the nuclear speckle retention signal (SRS) of HIPK2. Using various GFP-HIPK2 fusion constructs, subcellular localization of GFP-HIPK2 fusion proteins was analyzed after transfection into CV-1 cells (Fig. 1). As expected, full-length HIPK2 was localized to the nuclear speckles (dots) (Fig. 1A, construct B), whereas a GFP control showed diffuse cytoplasmic localization (Fig. 1A, construct A). Serial deletion of HIPK2 from the C terminus revealed that a specific region (SRS, amino acids 860–967) is required for the localization of HIPK2 to nuclear speckles (dots) (Fig. 1A, constructs C and D). Further deletion of the SRS region prevents HIPK2 from localizing to the nuclear speckles (Fig. 1A, constructs D–F). These deletion constructs, however, still showed nuclear localization of GFP-HIPK2 fusion proteins, suggesting that HIPK2 contains a NLS(s) that is different from the SRS. The NLSs (NLS1 or NLS2) in these constructs are weak because we could also detect a signal in the cytoplasm. We confirmed these results by using additional deletion constructs from the N terminus (Fig. 1, constructs G–J). Deletion of the N terminus, including the kinase domain (constructs G–I), did not affect the nuclear speckle retention of HIPK2 until removal of the SRS region (construct J). A deletion construct that retains the SRS and the C terminus showed nuclear speckle localization but showed a weak signal in the cytoplasm, too (Fig. 1, construct I). Within the SRS, there is a weak NLS (NLS3, Fig. 1B). NLSs were confirmed by GFP-NLS constructs that were generated by fusion of specific NLS DNAs (NLS1, amino acids 97–157; NLS2, amino acids 780–840) amplified with PCR (data not shown). Thus, HIPK2 utilizes multiple NLSs (NLS1, NLS2, and NLS3) for the nuclear localization, and, at the same time, utilizes the SRS for its localization to the nuclear speckles (dots) within the nucleus (Fig. 1B). We also detected a diffuse signal in the nucleoplasm. These results suggest that HIPK2 is compartmentalized to nuclear speckles (dots) by the SRS following its nuclear localization and that speckle formation is regulated. Of note, the amino acid sequence of the SRS differs from the sequences that were shown to be required for speckle formation of the SR proteins of splicing factors (26, 27). Also, the SRS overlaps with the PEST sequence (Fig. 1B) (21, 28). Thus, it was of interest to determine whether there are cellular proteins that interact with the SRS.
Figure 1.
HIPK2 contains multiple NLSs and localizes to nuclear speckles (dots) by means of a SRS. (A) Subcellular localization of various GFP-HIPK2 fusion proteins. CV-1 cells were transfected with various GFP-HIPK2 fusion constructs, and GFP fluorescence was visualized 48 hr after transfection. Constructs A–L correspond to the construct in B used for transfection. (B) Schematics of various GFP-HIPK2 constructs (constructs A–L). Locations of NLSs and SRS are shown under the schematic of the HIPK2 protein. HIPK2 contains three NLSs (NLS1–3, arrowheads) and a SRS (amino acids 860–967) that overlaps with the PEST sequence and NLS3. The coding region that was fused to the GFP ORF is indicated by amino acid numbers in each construct. KD, protein kinase domain; ID, interaction domain with homeodomain proteins; PEST, PEST sequence; YH, a C-terminal region enriched in tyrosine and histidine residues; S, speckles; ++, strongly positive; +, positive both in the cytoplasm (C) and the nucleus (N); +/−, weakly positive; −, negative.
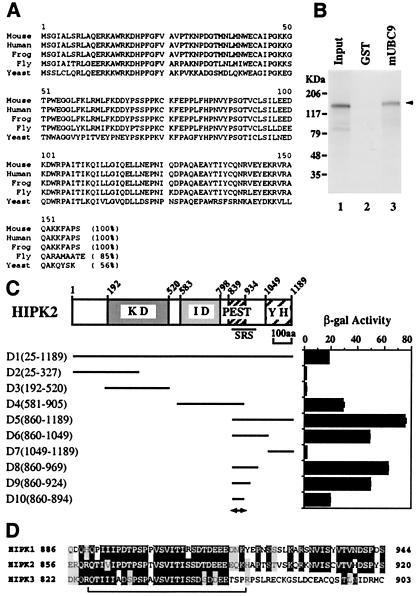
To identify potential cellular targets of HIPK2 that may facilitate the localization of HIPK2 to the nuclear speckles (dots), we have performed a yeast two-hybrid screen of mouse embryonic match-maker cDNA libraries using HIPK2 as bait. Among 28 positive clones that we have identified and sequenced, 24 clones were derived from the same gene. A GenBank search revealed that these clones are identical to the gene that was recently identified as the Ubl-conjugating (E2) enzyme (mUBC9), which is well conserved in eukaryotes (Fig. 2A). All positive clones contained full-length cDNAs coding for mUBC9, although the 5′ and 3′ ends of the cDNAs were different from each other (data not shown). mUBC9 strongly interacts with HIPK2 in yeast, and this result was confirmed by using in vitro pull-down assays, indicating that mUBC9 is a HIPK2-interacting protein (Fig. 2B). In an attempt to better understand the interaction between HIPK2 and mUBC9, we mapped the domain of HIPK2 that interacts with mUBC9 by using interaction assays in yeast. As shown in Fig. 2C, deletion analysis revealed that constructs which contain the SRS (Fig. 2C, constructs D1, D4, and D5) showed strong interactions with mUBC9. Moreover, further delineation of the SRS (constructs D6–D10) revealed that within the SRS (amino acids 860–967) there is a minimal region (amino acids 860–894) for the interaction with mUBC9 (Fig. 2C, construct D10). The amino acid sequence of this region shows strong similarity among the HIPKs (Fig. 2D) and matches the PEST sequence. HIPK1 also interacts with mUBC9 by means of this region (data not shown). These results indicate that the SRS itself contains an interaction domain with mUBC9 and suggest that the function of the mUBC9 interaction may be related to the localization of HIPK2 to nuclear speckles (dots).
Figure 2.
The SRS contains a binding site for the Ubl-conjugating (E2) enzyme mUBC9. (A) Alignment of amino acid sequences of UBC9 from different species. mUBC9 was cloned by the yeast two-hybrid screen using HIPK2 as bait, and the deduced amino acid sequence was aligned with known UBC9s from different species. Percent identity is indicated within the parentheses. (B) In vitro interaction of mUBC9 with HIPK2. In vitro pull-down assays were performed with a GST-mUBC9 fusion protein (lane 3) and in vitro-translated 35S-labeled HIPK2. Input sample (lane 1) contained 20% of the amount used for binding. GST (lane 2), pull-down assay with glutathione-Sepharose-bound GST protein as control. Bound HIPK2 is indicated by arrowhead. (C) Deletion analysis of the HIPK2 interaction domain with mUBC9. Various baits (D1–D10) were constructed, and the interactions with mUBC9 were assayed in yeast cells and are shown as β-galactosidase activity. The coding region of the HIPK2 fragment used is indicated by amino acid numbers. The mUBC9 binding domain is indicated by a double-headed arrow below D10. (D) The mUBC9 binding domain is conserved among different HIPKs. The amino acid sequence of the mUBC9 binding domain of HIPK2 (indicated by bracket) is aligned with the corresponding regions of HIPK1 and HIPK3. Black highlighting with white characters indicates identical amino acids, and light gray-highlighted amino acids represent conservative amino acid replacements. Numbers indicate amino acid residues.
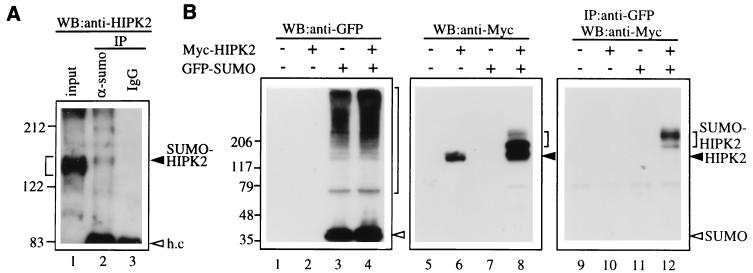
Because mUBC9 has recently been shown to act as an Ubl-conjugating (E2) enzyme and can conjugate SUMO-1 (or sentrin) to target proteins (5–7), we wondered whether HIPK2 is a target for SUMO-1 modification by mUBC9. To test whether the endogenous HIPK2 is covalently modified by endogenous SUMO-1, nuclear extracts from mouse C2C12 cells were subjected to immunoprecipitation with an anti-SUMO-1 Ab, followed by Western blot analysis with an anti-HIPK2 Ab (Fig. 3A). The input sample showed broad bands (Fig. 3A, lane1, bracketed region), including slowly migrating bands that may represent modified high molecular weight HIPK2. Only the upper bands among the broad HIPK2 bands (Fig. 3A, lane 2, arrowhead) were detected from immunoprecipitates with an anti-SUMO-1 Ab. In contrast, no band was detected in control IgG immunoprecipitates (Fig. 3A, lane 3). Thus, this result suggests that the endogenous HIPK2 can be modified by endogenous SUMO-1. To test this idea further, expression vectors for Myc-tagged HIPK2, GFP-tagged SUMO-1, and mUBC9 were generated and transfected into mammalian cells. Total cell extracts from transfected cells were analyzed by Western blotting with an anti-GFP Ab or with an anti-Myc Ab. As shown in Fig. 3B, the expressed GFP-tagged SUMO-1 was successfully conjugated to unknown target proteins (lanes 3 and 4) in CV-1 cells. Interestingly, many cellular proteins are modified by SUMO-1 (Fig. 3B, lanes 3 and 4, area indicated by large brackets). Anti-Myc Ab detected several more slowly migrating HIPK2 bands (Fig. 3B, lanes 6 and 8, area indicated by small brackets) above the 140-kDa unmodified Myc-HIPK2 band (marked with arrowhead). These slowly migrating bands are more abundant in cells cotransfected with the Myc-HIPK2 and GFP-SUMO-1 expression vectors than in cells transfected with the Myc-HIPK2 expression vector alone (Fig. 3B, lanes 6 and 8). To test whether HIPK2 is covalently modified with SUMO-1, the GFP-SUMO-1 modified proteins were immunoprecipitated with an anti-GFP Ab and were immunoblotted with an anti-Myc Ab to detect the SUMO-1-modified HIPK2 (Fig. 3B, lanes 9–12). The SUMO-1-modified HIPK2 bands were detected only in cells cotransfected with Myc-HIPK2 and GFP-SUMO-1 (Fig. 3B, lane 12, area marked by small bracket), demonstrating that the slowly migrating bands detected in lane 8 are indeed SUMO-1-modified HIPK2 bands. Taken together, these results clearly indicate that HIPK2 is covalently modified with SUMO-1 at multiple sites.
Figure 3.
Covalent modification of HIPK2 by SUMO-1. (A) Nuclear extracts from C2C12 cells were subjected to immunoprecipitation (IP) with an anti-SUMO-1 Ab (lane 2) or control IgG (lane 3) followed by Western blot analysis with an anti-HIPK2 Ab. Small bracket indicates broad bands that may include both unmodified and modified HIPK2, and the arrowhead indicates possible SUMO-1-modified HIPK2 band (SUMO-HIPK2). h.c., Ig heavy chain. (B) CV-1 cells were transfected with empty vector (lanes 1, 5, and 9) or the indicated expression vector containing a cDNA insert encoding Myc-HIPK2 (lanes 2, 6, and 10), GFP-SUMO-1 (lanes 3, 7, and 11), or both Myc-HIPK2 and GFP-SUMO-1 (lanes 4, 8, and 12). Cell lysates were analyzed by SDS/PAGE, followed by Western blotting with a mouse anti-GFP mAb (lanes 1–4) or with a mouse anti-Myc mAb (lanes 5–8). The SUMO-1-modified proteins were immunoprecipitated from the cell lysate (lanes 9–12) with a rabbit anti-GFP polyclonal Ab and analyzed by Western blotting with a mouse anti-Myc mAb. SUMO-1-modified proteins are marked by a large bracket (lanes 3 and 4), and the SUMO-1-modified HIPK2 is marked by a small bracket (lanes 8 and 12). Arrowhead indicates unmodified HIPK2 (lanes 6 and 8), and open arrowhead indicates the GFP-SUMO-1 monomer band (lanes 3 and 4). Molecular size markers are shown on the left in kDa.
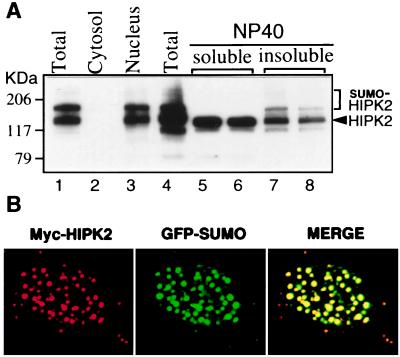
Unlike the ubiquitin-modification system, which is mainly used for protein degradations, SUMO-1 modification seems to have different functions. For example, modification of RanGAP1 is important for its targeting to the nuclear pore complex (8–13), whereas modification of IκBα increases its stability (17). In particular, it has recently been shown that covalent modification of the nuclear dot-associated protein PML correlates with nuclear body formation (14, 15, 18, 19). Because the mUBC9-binding site of HIPK2 coincides with the SRS (Fig. 2 C and D), we tested whether SUMO-1 modification of HIPK2 correlated with its localization to the nuclear speckles (dots) (Fig. 4). Initially, subcellular fractions were prepared from CV-1 cells cotransfected with the GFP-SUMO-1 and Myc-HIPK2 expression vector, and analyzed by Western blot with an anti-Myc Ab (Fig. 4A, lanes 1–3). HIPK2 bands were detected only in the nuclear fractions (Fig. 4A, lane 3), confirming that HIPK2 is a nuclear protein kinase. Furthermore, we found that, whereas the fractions soluble in the nonionic detergent Nonidet P-40 showed only unmodified HIPK2 bands that contain the soluble nuclear diffuse form of HIPK2 (Fig. 4A, lanes 5 and 6), the SUMO-1-modified HIPK2 bands were exclusively detected in the Nonidet P-40-insoluble fractions (lanes 7 and 8), suggesting that SUMO-1-modified HIPK2 is tightly associated with the nuclear matrix, along with other insoluble cellular structures.
Figure 4.
Localization of SUMO-1-modified HIPK2 to nuclear speckles (dots). (A) Fractionation of SUMO-1-modified HIPK2. Cellular extracts from CV-1 cells cotransfected with GFP-SUMO-1 and Myc-HIPK2 expression vectors were fractionated. Proteins from each fraction were separated on SDS/polyacrylamide gradient (4–12%) gels and analyzed by Western blotting using a mouse anti-Myc Ab. HIPK2 and SUMO-1-modified HIPK2 are detected in the nuclear fraction (lane 3) and in the Nonidet P-40-insoluble fractions (lanes 7 and 8). SUMO-1-modified HIPK2 and unmodified HIPK2 are marked by bracket and arrowhead, respectively. (B) Confocal microscopic image of the nucleus showing colocalization of HIPK2 and SUMO-1 to nuclear speckles (dots). CV-1 cells overexpressing Myc-HIPK2 and GFP-SUMO-1 were fixed and subjected to immunofluorescence staining with a mouse anti-Myc mAb. The red signal (HIPK2) is obtained with an anti-mouse IgG rhodamine red-conjugated secondary Ab, whereas the green signal (SUMO-1) is obtained with GFP fluorescence. Superimposing two colors (MERGE) results in a yellow signal, indicating colocalization of the two proteins.
Next, we analyzed the HIPK2 and the SUMO-1 immunofluorescence pattern in cells cotransfected with Myc-HIPK2 and GFP-SUMO-1. Labeling with an anti-Myc Ab was performed on fixed cells, and the signal was analyzed by confocal microscopy. As shown in Fig. 4B, the HIPK2 signal (red) was exclusively detected in the nuclear speckles (dots). Intensive SUMO-1 signal from GFP fluorescence (green) was detected in the nuclear speckles (dots) in addition to a diffuse signal in the nucleus. Superimposition of the HIPK2 and the SUMO-1 signals demonstrates the colocalization of HIPK2 and SUMO-1 in the nuclear speckles (dots). Because the SUMO-1 fluorescence pattern in cells transfected with GFP-SUMO-1 alone showed diffuse nuclear staining (data not shown), the speckled staining pattern in cells cotransfected with both HIPK2 and SUMO-1 most likely derives from the SUMO-1-modified HIPK2.
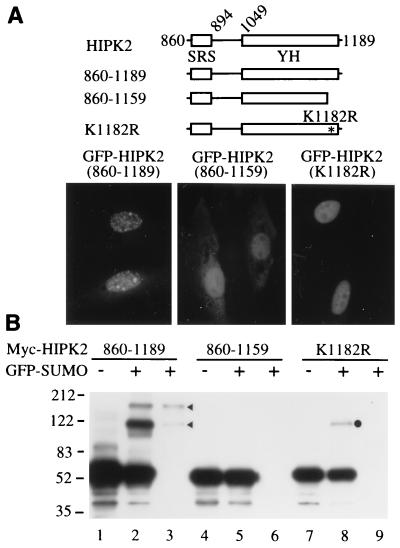
Our results suggest that mUBC9 may recognize HIPK2 via the SRS and then conjugate SUMO-1 to the target lysine residue(s) of HIPK2. Thus, if SUMO-1 modification of HIPK2 correlates with its localization to the nuclear speckles (dots), then either deletion of the SRS or disruption of the target lysine residue should be able to prevent SUMO-1 modification and consequently inhibit localization of HIPK2 to the nuclear speckles (dots). To test this idea, we mapped one possible target lysine residue for SUMO-1 modification of HIPK2 (Fig. 5). We characterized the deletion construct GFP-HIPK2(860–1189), which contains the SRS and the C terminus of HIPK2, including the YH domain (Fig. 5A). This construct successfully localized to nuclear speckles (dots) (Fig. 5A, Lower Left). Also, this construct can be covalently modified by SUMO-1, which was shown by immunoprecipitation with an anti-GFP Ab followed by Western blot analysis with an anti-Myc Ab (Fig. 5B, lane 3, arrowheads). However, we found that deletion of 30 amino acid residues from the C terminus abolished its localization to nuclear speckles (dots), although it still localized to the nucleus (Fig. 5A, Lower Middle). Furthermore, this construct cannot be modified by SUMO-1 (Fig. 5B, lanes 5 and 6). Within these 30 amino acid residues there is only one lysine residue (position 1182). Therefore, we mutated this lysine residue to an arginine residue and tested the effect of this mutation. Indeed, this mutation (K1182R) inhibited HIPK2 localization to nuclear speckles (dots) (Fig. 5A, Lower Right). We could not detect any SUMO-1-modified HIPK2 bands in immunoprecipitates generated by an anti-GFP Ab (Fig. 5B, lane 9). These results strongly suggest that inhibition of SUMO-1 modification of HIPK2 prevents localization of HIPK2 to the nuclear speckles (dots). There is one slowly migrating band (Fig. 5B, lane 8, dot), but this band was not detected in the immunoprecipitate generated by an anti-GFP Ab (lane 9). Taken together, these results indicate that SUMO-1 modification of HIPK2 correlates with its localization to the nuclear speckles (dots).
Figure 5.
Correlation of SUMO-1 modification of HIPK2 with the compartmentalization of HIPK2 to nuclear speckles (dots) within the nucleus. (A) GFP-HIPK2 constructs were transfected into CV-1 cells, and GFP fluorescence was visualized 48 hr after transfection (Lower). Schematics of GFP-HIPK2 constructs are shown (Upper). (B) Covalent modification of HIPK2 by SUMO-1. Different Myc–HIPK2 constructs were transfected into CV-1 cells in the presence (+) of GFP-SUMO-1, and SUMO-1-modified HIPK2 was detected by immunoprecipitation with an anti-GFP Ab (lanes 3, 6, and 9) followed by Western blot analysis with an anti-Myc Ab.
Based on our results described above, we conclude that SUMO-1 modification of HIPK2 plays an important role in the compartmentalization of HIPK2 within the nucleus. As far as we are aware, HIPK2 is the first example of a protein kinase that is covalently modified by the Ubl, SUMO-1. Recently, the Ubl-specific protease, Ulp1, which cleaves proteins from SUMO-1, was identified, indicating that SUMO modification of proteins has an essential role in many physiological processes (29). Of note, the SRS, which is required for the speckle retention of HIPK2 and can serve as an interacting domain with mUBC9, contains the PEST sequence (Figs. 1 and 2D). Surprisingly, we found that most proteins that showed interactions with mUBC9, such as RanGAP1 (8, 11), PML (18, 19), and E1A (30), also contain the PEST sequence. Because many unknown proteins can be modified by SUMO-1 within the cells (Fig. 3), it is tempting to speculate that proteins containing the PEST sequence may be targets of mUBC9 for Ubl modification, which is in contrast to the original PEST hypothesis (28). At present, the function of the SUMO-1-modified HIPK2-positive nuclear speckles (dots) is unknown. Perhaps they simply represent accumulations of HIPK2 following SUMO-1 modification at specific preexisting nuclear deposition sites (NUDES) (31). It was demonstrated that one of the NUDES, nuclear domain 10 (ND10), which contains proteins such as PML and Sp100, is a target for DNA viruses such as herpes simplex type 1 (32), human cytomegalovirus (33–35), and adenovirus 5 (36–38), implicating ND10 as a site for a nuclear defense mechanism (31). Although PML localized to ND10 following its modification by SUMO-1 (14, 15, 18, 19), HIPK2 did not appear to colocalize with PML (data not shown). Hence, we would like to call the SUMO-1-modified HIPK2-positive nuclear speckles (dots) the HIPK-NUDES. Given the fact that the nucleus has a dynamic, distinct substructure (39), and given the fact that HIPK2 is a protein kinase that is a component of the corepressor complex containing the Groucho corepressor and a histone deacetylase complex (C.Y.C., Y.H.K., H. J. Kwon, and Y.K., unpublished work), it will be of interest to identify the signals that regulate the formation of the HIPK-NUDES.
Acknowledgments
We thank Drs. Robert Adelstein and Marshall Nirenberg for their continuous support and encouragement during this study. This work is supported by a grant from the National Heart, Lung, and Blood Institute Intramural Research Program (to Y.K.).
Abbreviations
- Ubl
ubiquitin-like protein
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HIPK
homeodomain interacting protein kinase
- NLS
nuclear localization signal
- SRS
speckle retention signal
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Varshavsky A. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Johnson P R, Hochstrasser M. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 4.Hochstrasser M. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 5.Johnson E S, Blobel G. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 6.Desterro J M P, Thomson J, Hay R T. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz S E, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matunis M J, Coutavas E, Blobel G. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahajan R, Delphin C, Guan T L, Gerace L, Melchior F. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 10.Saitoh H, Sparrow D B, Shiomi T, Pu R T, Nishimoto T, Mohun T J, Dasso M. Curr Biol. 1997;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 11.Matunis M J, Wu J A, Blobel G. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahajan R, Gerace L, Melchior F. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee G W, Melchior F, Matunis M J, Mahajan R, Tian Q S, Anderson P. J Biol Chem. 1998;273:6503–6507. doi: 10.1074/jbc.273.11.6503. [DOI] [PubMed] [Google Scholar]
- 14.Muller S, Matunis M J, Dejean A. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamitani T, Nguyen H P, Kito K, Fukuda-Kamitani T, Yeh E T H. J Biol Chem. 1998;273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 16.Sternsdorf T, Jensen K, Will H. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desterro J M P, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 18.Kamitani T, Kito K, Nguyen H P, Wada H, Fukuda-Kamitani T, Yeh E T H. J Biol Chem. 1998;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- 19.Duprez E, Saurin A J, Desterro J M, Lallemand-Breitenbach V, Howe K, Boddy M N, Solomon E, de The H, Hay R T, Freemont P S. J Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- 20.Epps J L, Tanda S. Curr Biol. 1998;8:1277–1280. doi: 10.1016/s0960-9822(07)00538-6. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y H, Choi C Y, Lee S J, Conti M A, Kim Y. J Biol Chem. 1998;273:25875–25879. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- 22.Moilanen A M, Karvonen U, Poukka H, Janne O A, Palvimo J J. Mol Biol Cell. 1998;9:2527–2543. doi: 10.1091/mbc.9.9.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Begley D A, Berkenpas M B, Sampson K E, Abraham I. Gene. 1997;200:35–43. doi: 10.1016/s0378-1119(97)00350-8. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y M, Park T, Schulz R A, Kim Y. J Biol Chem. 1997;272:17531–17541. doi: 10.1074/jbc.272.28.17531. [DOI] [PubMed] [Google Scholar]
- 25.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Bingham P M. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 27.Hedley M L, Amrein H, Maniatis T. Proc Natl Acad Sci USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers S, Wells R, Rechsteiner M. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 29.Li S-J, Hochstrasser M. Nature (London) 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 30.Hateboer G, Hijmans E M, Nooij J B D, Schlenker S, Jentsch S, Bernards R. J Biol Chem. 1996;271:25906–25911. doi: 10.1074/jbc.271.42.25906. [DOI] [PubMed] [Google Scholar]
- 31.Maul G G. BioEssays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 32.Maul G G, Guldner N H, Spivack J G. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 33.Kelly C, van Driel R, Wilkinson G W G. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 34.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 35.Ishov A M, Sternberg R M, Maul G G. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvalho T, Seeler J S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 38.Ishov A M, Maul G G. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]