Abstract
Histone deacetylases such as human HDAC1 and yeast RPD3 are trichostatin A (TSA)-sensitive enzymes that are members of large, multiprotein complexes. These contain specialized subunits that help target the catalytic protein to histones at the appropriate DNA regulatory element, where the enzyme represses transcription. To date, no deacetylase catalytic subunits have been shown to have intrinsic activity, suggesting that noncatalytic subunits of the deacetylase complex are required for their enzymatic function. In this paper we describe a novel yeast histone deacetylase HOS3 that is relatively insensitive to the histone deacetylase inhibitor TSA, forms a homodimer when expressed ectopically both in yeast and Escherichia coli, and has intrinsic activity when produced in the bacterium. Most HOS3 protein can be found associated with a larger complex in partially purified yeast nuclear extracts, arguing that the HOS3 homodimer may be dissociated from a very large nuclear structure during purification. We also demonstrate, using a combination of mass spectrometry, tandem mass spectrometry, and proteolytic digestion, that recombinant HOS3 has a distinct specificity in vitro for histone H4 sites K5 and K8, H3 sites K14 and K23, H2A site K7, and H2B site K11. We propose that while factors that interact with HOS3 may sequester the catalytic subunit at specific cellular sites, they are not required for HOS3 histone deacetylase activity.
Keywords: acetylation, mass spectrometry
We previously have identified a yeast (Saccharomyces cerevisiae) histone deacetylase HDA1 (1) that was found to have similarity to four other histone deacetylases, RPD3, HOS1, HOS2, and HOS3, inferred from the yeast genome database (2). Of these enzymes, RPD3 is homologous to a mammalian histone deacetylase subunit, HDAC1 (3), that is recruited in a multiprotein complex (4) to various regulatory DNA sequences, such as Mad-Max binding sites, to repress adjacent genes (5, 6). This involves proteins such as mSIN3 and p48, which interact with repressor proteins and histones, respectively (5–7). Similar mechanisms are involved in repression by the unliganded thyroid hormone receptor (8), retinoblastoma protein (9–11), notch protein (12), and other proteins (13–15). In certain HDAC complexes (e.g., Mi2), ATPases are also implicated in histone deacetylase function (16–18). Surprisingly, although HDAC1 binds small-molecule deacetylase inhibitors such as trichostatin A (19), apicidin (20), and trapoxin (3), it has not yet been shown that HDAC-like proteins have intrinsic deacetylase activity. This suggests that other proteins in the HDAC complex are required for activity of the catalytic subunit.
In yeast, the RPD3 deacetylase is also found in a large, multiprotein complex even after extensive enzyme purification (2, 21, 22). This trichostatin A (TSA)-sensitive enzyme complex is targeted by SIN3 to UME6-regulated genes in a manner similar to that involving Mad-Max (23). Other yeast histone deacetylases that include HDA1, HOS1, and HOS2 are also found in large complexes after similar purification. In this paper we describe a novel histone deacetylase, HOS3, that is relatively insensitive to TSA. Although HOS3 has a predicted molecular mass of 79 kDa (2), it has been purified as a homodimer after its ectopic expression in either yeast or Escherichia coli. The homodimeric enzyme in E. coli has intrinsic catalytic activity in vitro for specific sites in each of the four core histones. It is likely that HOS3 has been purified away from a larger yeast complex; however, whereas the other proteins in the complex may sequester HOS3, they are not required for its activity as a deacetylase of acetylated substrates in vitro.
Experimental Procedures
Cloning, Disruption, and Overexpression of HOS3 in Yeast.
The full-length HOS3 gene was cloned into pBluescript II SK (Stratagene) plasmid KpnI and NheI sites by inserting a 2.5-kb fragment generated by PCR using the primers GATCGGTACCAAGGGCTCTGGAAGTAAACAG and GATCGCTAGCTGTCAGGCTGATAAAGCTTATAG. This construct then was digested with XhoI and XbaI and KanMX3 (24) was inserted to generate the HOS3 knock-out plasmid, pskH3kan. This was used to disrupt HOS3 in the parental YDS2 strain (25) to generate the yeast strain SRYH3 in which HOS3 is disrupted. Using the same primer sets, the HOS3 containing fragment also was cloned into pYES2 (Invitrogen) to generate plasmid pYHOS3 in which HOS3 is regulated by the GAL1 promoter. This plasmid was transformed into YDS2 to generate the strain SRYH3gal in which HOS3 overexpression occurs in galactose but not in glucose-containing media.
Analysis of Histone Acetylation by Western Blot.
ECL (Amersham) Western blots and use of antibodies against specific H4 sites of acetylation were similar to that described (2). The same Western blots also were reacted with 35S-labeled anti-rabbit Ig (Amersham) according to the manufacturer’s recommendations and, subsequently, were quantitated by using a Molecular Dynamics PhosphorImager and imagequant software.
Purification of HOS3 from Yeast.
Purification of HOS3 from yeast nuclear extracts was done as described previously (1), except that the high salt extraction was performed at 450 mM NaCl. The resulting extracts from YDS2 and SRYH3gal were purified on DEAE-Sepharose-ff with a 50- to 250-mM NH4Cl step, dialyzed, and rechromatographed on SP-Sepharose-ff retaining the 275- to 450-mM NaCl step gradient material. This was purified further on Mono S HR 10/10 along a linear gradient between 320 and 430 mM NaCl in 42 ml. The peak HOS3 activity was concentrated and chromatographed on a Superdex-200 (1.0 cm × 46 cm) column [all with buffers as described previously (1)]. The protein was detected both by Western blotting and deacetylase activity assays. Production of HOS3-specific polyclonal antibody (α-HOS3.640) was performed by producing a GST-fusion protein of the divergent C terminus of HOS3 between amino acids 594 and 697. This was achieved by utilizing the GST-fusion vector pGEX2T cleaved with EcoRI and BamHI (Pharmacia) and cloning a PCR product generated with the following oligonucleotides: CGTGGATCCGAAGAATTGAACAAAACTTTC and GATGAATTCCCCATCTTCCACCACTTCTTG. The GST-HOS3 fusion protein was expressed and purified on glutathione-Sepharose (Pharmacia) according to the manufacturer’s instructions, and the protein was used for antibody production in rabbits according to standard protocols (28). Enzyme assays were performed by using [3H]acetyl-lysine-labeled Hela histones as described previously (1).
Cloning, Expression, and Purification of HOS3 in E. coli.
The full-length HOS3 protein gene was amplified by PCR, using oligonucleotide primers that added BamHI (CGTGGATCCAAGCATTCAGATCCATTGG) and SmaI (GTTCCCGGGTCACCCTCTTCCACCAC) sites, and then cloned into the Stratagene E. coli expression vector pCALn. These primers allowed for the cloning of the encoded HOS3 protein from amino acid 3 to the natural stop codon at amino acid 696. The resulting plasmid was transformed and expressed in the E. coli strain BL21(DE3)pLysS. Cells with the HOS3 enzyme or vector alone were grown at 37°C in 2 liters of TYE (1% tryptone/0.5% yeast extract/170 mM NaCl) liquid medium plus 100 mg/liter ampicillin to an A600 ≈ 0.4 and then induced with 1 mM isopropyl β-d-thiogalactoside for 2 hr at 30°C. Cells were harvested and resuspended in lysis buffer (50 mM Tris⋅HCl, pH 8/250 mM NaCl/2.5 mM DTT/1.0 mM magnesium acetate/1.0 mM imidazole/2.0 mM CaCl2/0.25% Triton X-100/20 μM leupeptin/2.0 mM PMSF) at 4°C. Cells were lysed by sonication and then centrifuged in a Beckman 45Ti rotor at 40,000 rpm for 45 min. The fusion protein was affinity-purified on calmodulin affinity resin (Stratagene) according to the manufacturer’s instructions except that the wash and elution buffers contained 400 mM NaCl. This material was concentrated over a 24-hr period by vacuum dialysis (using a membrane with a 30-kDa molecular mass cutoff) in buffer DBHS (50 mM Tris, pH 8/500 mM NaCl/10 μM ZnCl2/2.5 mM DTT). The concentrated enzyme was purified further on a Superdex-200 column (1.0 cm × 46 cm) in the above dialysis buffer.
Analysis of Recombinant HOS3 Molecular Weight by Sedimentation Equilibrium Ultracentrifugation.
Sedimentation equilibrium ultracentrifugation was performed at 4°C by using a Beckman Optima XL-A analytical ultracentrifuge and software using absorption optics at 290 nm and double-sector cells. The sample was dialyzed against buffer DBHS (as above), and the dialysis buffer was used as the blank reference. Sedimentation equilibrium profiles were measured at 8,000 rpm at a concentration of 1.2 and .12 mg/ml. The XL-A data were fitted with a nonlinear least-squares exponential fit for a single, ideal species by using the Beckman origin 3.01 (Microcal Software, Northampton, MA) software. Analysis using an interacting monomer–dimer model indicates the association was high with no apparent monomer present at the lowest concentration (0.12 mg/ml) analyzed. A partial specific volume of 0.723 ml/g was used and was calculated from the amino acid composition and corrected to 4°C (29).
Synthesis of Acetylated Peptides.
The following peptides were synthesized by the Biopolymer Synthesis Center at Caltech: H2A (amino acids 1–16) SGGK-AcGGK-AcAGSAAK-AcASC, H2B (amino acids 1–25) SAK-AcAEK-AcK-AcPASK-AcAPAEK-AcK-AcPAAK-AcK-AcTSC, H3 (amino acids 1–30) ARTK-AcQTARK-AcSTGGK-AcAPRK-AcQLASK-AcAARK-AcSAC, and H4 (amino acids 1–19) SGRGK-AcGGK-AcGLGK-AcGGAK-Ac RHRC (where K-Ac is an acetylated lysine).
Deacetylation of Acetylated Histone Peptides and Mass Spectroscopy.
Analysis of deacetylated histone peptides was performed by using acetyl-lysine peptides synthesized as described above. Enzyme assays were performed with 25 nM of each peptide and assay conditions described previously (1), but reactions were stopped on dry ice-ethanol. The recombinant HOS3 enzyme was added at 0.05, 0.24, or 1.2 μg per reaction, and assays were incubated for 0, 5, 15, 30, 60, 120, and 240 min. Samples were desalted by using a C4, 300Å nanocartridge (Western Analytical Products, Temecula, CA) and screened by using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (PerSeptive Biosystems, Framingham, MA) to determine the extent of deacetylation at various time points. After MALDI-TOF-MS analysis, the specificity of deacetylation for H2A and H4 was determined directly by capillary HPLC tandem mass spectrometry (cHPLC-MS/MS) using a home-built microspray ionization source fitted on a Finnigan-MAT (San Jose, CA) LCQ quadrupole ion trap. For cHPLC-MS/MS analysis, 1–2 μl of sample was loaded by using a pressure vessel onto a reverse-phase capillary column (100 μM i.d. fused silica capillary) with 10 μM Poros R2 packing (PerSeptive Biosystems). The column was connected in-line with an Applied Biosystems 130 HPLC gradient system equipped with a flow splitter that provided ≈500-nl/min eluate that was fed directly into the LCQ. Precursor ions were isolated and fragmented by using resonance excitation with a relative collision energy of 30% and an isolation width of 3 Da. Product ion or MS/MS spectra were interpreted manually to determine the presence and exact location of deacetylated acetyl-lysine residues. Enzymatic digestion of H2B with trypsin (Promega) and H3 with endoproteinase Lys-C (Roche Molecular Biochemicals, Indianapolis) were performed before characterization by cHPLC-MS/MS. Both trypsin and endoproteinase Lys-C require an unmodified lysine in the P1 position. Acetyl-lysyl peptide bonds are not hydrolyzed by either enzyme. In all cases, the structures of internal peptides generated from proteolysis were confirmed by cHPLC-MS/MS allowing for site determination. The relative rate of catalysis of individual acetyl-lysines in the individual peptides was determined based on the relative abundance (±5%) of each species.
Results
HOS3 Disruption Results in Increased Histone H4 Acetylation in Yeast Cell Extracts.
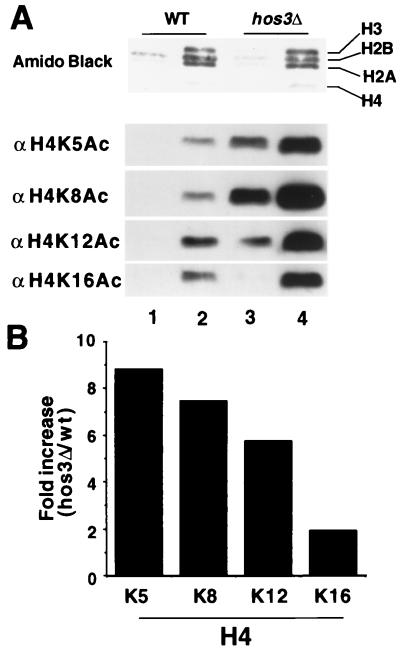
HDA1 originally was shown to have strong sequence similarity not only to RPD3, HOS1, HOS2, and HOS3 in yeast but also to an acetylpolyamine amidohydrolase (APH) of the bacterium Mycoplana bulata (2). Because this bacterium is unlikely to contain histones, there existed the possibility that HOS3 may not target chromatin. Therefore, we first asked whether HOS3 disruption increases histone acetylation in yeast. The HOS3 coding region was disrupted by insertion of the KanMX construct as described in Experimental Procedures. We found that HOS3 disruption in a haploid strain (SRYH3) did not prevent viability and had little effect on growth. For example, in rich (YPD) medium, SRYH3 had a doubling time of 109 min as compared with 108 min for the YDS2 wild-type strain. In defined medium, SRYH3 doubled in 168 min as compared with 149 min for wild type. To examine the effect of HOS3 disruption on steady-state histone acetylation, histones were purified from cell extracts of wild-type (YDS2) and hos3Δ (SRYH3) cells. Histone acetylation was analyzed by Western blotting (Fig. 1A) by using polyclonal antibodies specific for individual sites of acetylation. Antibodies used include those against histone H4 sites K5-Ac, K8-Ac, K12-Ac, and K16-Ac. In Fig. 1A, lanes 1 and 3 contain 1 μg and lanes 2 and 4 contain 3 μg of core histone protein, which were visualized by staining with amido black. The Western blots demonstrate that HOS3 disruption results in increased acetylation of all sites in H4 (Fig. 1A, compare lanes 1 and 3 and lanes 2 and 4). When the increased acetylation levels were quantitated by phosphorimaging we found that the largest fold increases in acetylation are at sites K5, K8, and K12 (Fig. 1B). These data suggest strongly that HOS3 is a histone deacetylase that affects histone H4 in vivo.
Figure 1.
HOS3 disruption results in histone H4 hyperacetylation. (A) Relative acetylation levels of histone H4 isolated from YDS2 (WT) (lanes 1 and 2) and SRYH3 (hos3Δ) yeast (lanes 3 and 4) were determined by electrophoresing 1 μg (lanes 1 and 3) or 3 μg (lanes 2 and 4) of histones on SDS-polyacrylamide gel. After transfer to PVDF membranes (Millipore) (Experimental Procedures), blots were probed with antibodies specific for H4 acetyl-lysine 5, 8, 12, or 16 (antibodies to be described elsewhere). Protein load was visualized by staining the membrane with amido black. Antibody binding then was detected by using ECL (Amersham) and anti-rabbit Ig HRP-linked antibody (Amersham) as described in Experimental Procedures. (B) Quantitation of increased acetylation levels of histone H4 sites observed for histones isolated from hos3Δ versus wild-type yeast. Western blots in A were reacted with 35S-labeled anti-rabbit Ig, detected by PhosphorImager, and analyzed by using imagequant software as in Experimental Procedures. Quantitation is given as fold increase in acetylation observed for the histones isolated from the SRYH3 strain (hos3Δ) vs. YDS2 (wt).
HOS3-Specific Histone Deacetylase Activity.
To determine whether there is an HOS3-specific histone deacetylase activity in yeast, we prepared an anti-HOS3 antibody to its divergent C terminus from residues 594–696 to minimize cross-reaction with other histone deacetylases (described in Experimental Procedures). This was used to immunoprecipitate HOS3 protein and deacetylase activity from yeast nuclear extracts. As shown in Table 1, anti-HOS3 immunoprecipitates approximately 17% of the level of deacetylase activity as does anti-HDA1 (1) from nuclear extracts of wild-type (YDS2) yeast. Anti-HOS3 does not immunoprecipitate significant deacetylase activity from nuclear extracts of hos3Δ yeast (SRYH3). This decreased, baseline level of activity is similar to that obtained when beads lacking antibody or an antibody [anti-RAP1 (30)] unrelated to histone deacetylases are used. Moreover, deletion of HOS3 had no obvious effect on the immunoprecipitation of deacetylase HDA1 when using anti-HDA1 antibody. Therefore, yeast nuclear extracts contain an HOS3-specific histone deacetylase activity.
Table 1.
Antibodies directed against HOS3 are able to specifically immunoprecipitate deacetylase activity from nuclear extracts
| Yeast strain | Anti-HOS3 | Anti-HDA1 | Anti-RAP1 | Beads alone |
|---|---|---|---|---|
| Wild type | 1,140 | 5,888 | 191 | 108 |
| hos3Δ | 131 | 6,052 | 147 | 171 |
Activity is given as total cpm release from 50,000 cpm total [3H]acetyl Hela histones. Extracts were prepared, immunoprecipitated, and assayed as described previously (1).
HOS3 (79 kDa) Can Be Fractionated as an Apparent Dimeric Complex That Is the Active Form of the Enzyme.
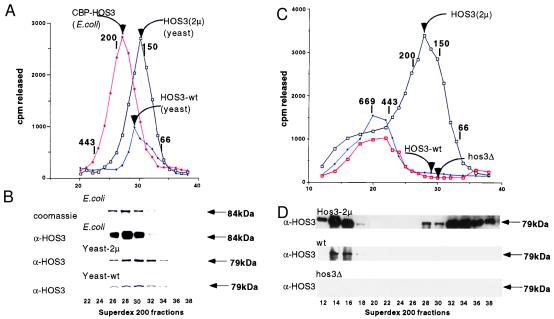
To determine the size of the complex containing the HOS3 activity, we fractionated a nuclear extract from wild-type yeast by using DEAE-Sepharose, SP-Sepharose, Mono S, and Superdex-200 gel filtration. This revealed an activity whose peak is at approximately 172 kDa (Fig. 2A). When these fractions were electrophoresed on SDS-polyacrylamide gel and reacted against anti-HOS3 antibody by Western blot we detected the HOS3 peak protein level at a similar molecular size (approximately 160 kDa) (Fig. 2B). Because the encoded molecular mass of HOS3 is 79 kDa, this suggests that HOS3 may form a homodimer.
Figure 2.
HOS3 is present in a dimer-sized complex in both yeast and E. coli. (A) E. coli-expressed HOS3 activity containing the CBP-HOS3 chromatographs on Superdex-200 at an apparent molecular mass of 190 kDa. CBP-HOS3 was purified from E. coli by affinity chromatography on calmodulin-Sepharose as described in Experimental Procedures. Twenty-five microliters of each 0.5-ml fraction were used in a 1-hr incubation with 25,000 cpm total of [3H]acetyl-Hela histones. Activity ([3H]acetic acid released) is shown as a function of fraction number. HOS3 activity from yeast strains containing HOS3(2μ) or wild-type HOS3 was purified from nuclear extracts on DEAE-Sepharose, S-Sepharose, Mono S, and Superdex 200 as described in Experimental Procedures. Protein molecular mass standards are indicated above the chromatograph as 669 kDa (thyroglobulin), 443 kDa (apoferritin), 200 kDa (β-amylase), 150 kDa (alcohol dehydrogenase), and 66 kDa (bovine serum albumin). (B) Electrophoretic (SDS/PAGE) and immunological analysis of column fractions in A. Coomassie staining of E. coli CBP-HOS3 reveals the presence of an 84-kDa protein that comigrates with deacetylase activity. This protein reacts with anti-HOS3 antibody as detected by Western blotting (ECL, Amersham). Similar electrophoresis and Western analysis of the column fractions (A) of HOS3 deacetylase from HOS3(2μ) and HOS3 (wt) yeast indicates a 79-kDa protein that comigrates with the peak yeast activities. (C) Deacetylase activities from wild-type (YDS2), hos3Δ (SRYH3), or HOS3(2μ) (SRYH3gal) strains were partially purified on DEAE-Sepharose columns and then directly chromatographed by Superdex 200 gel filtration (1.0 × 46 cm column, 0.5-ml fractions). (D) Western blotting of Superdex 200 fractions from HOS3(2μ) (SRYH3gal), HOS3 wild type (YDS2), and hos3Δ (SRYH3) yeast.
Because the HOS3 enzyme in wild-type cells is relatively rare, as compared with HDA1, making its analysis more difficult, we overexpressed the HOS3 protein ectopically. The HOS3 gene was cloned into the yeast GAL1 promoter of vector pYES2 (described in Experimental Procedures). We found that a yeast strain (SRYH3gal) containing GAL1-HOS3 and grown in galactose-containing medium contains an approximately 4-fold increase in HOS3 enzyme activity (Fig. 2A). The molecular mass of this enzyme complex is approximately 160 kDa. Therefore, it is likely that HOS3 protein forms an active homodimer when overexpressed in yeast.
To ask whether the apparent dimer of HOS3 may have been dissociated from a larger complex during purification, we used an abbreviated purification protocol. The enzymatic activity from the nuclear extract of wild-type (YDS2) cells was purified on DEAE-Sepharose, concentrated, and transferred directly to a Superdex-200 gel-filtration column. Under these conditions, most HOS3 was detected, by Western blot, in a very large complex (greater than 800 kDa) (Fig. 2C). This complex fractionates at a size that is even larger than that of the cellular deacetylase activity in this size range. This activity (peak fraction 20) is not due to HOS3 because it is found even in a hos3Δ strain; however, we cannot exclude the possibility that the larger HOS3-containing complex also contains some HOS3 deacetylase activity. Interestingly, when HOS3 is overexpressed, most of the HOS3 protein, as detected by Western blot [enhanced chemiluminescence (ECL)], is found associated with the large complex (fractions 14–16) or is apparently monomeric (fractions 32–34). However, most of the increased activity, resulting from overexpression of HOS3, lies in the location of the apparent dimer (fractions 28–30). Therefore, we conclude that HOS3 may be found in a larger complex during early stages of purification. Upon overexpression of HOS3, most of the protein is monomeric or can be associated with a very large complex. However, it is only in the location of the apparent dimer that the bulk of HOS3 activity is found.
HOS3 Forms a Homodimer and Has Intrinsic Activity in E. coli.
To determine whether HOS3 can form a homodimer with intrinsic activity we cloned the HOS3 gene into a bacterial expression vector (pCALn) that contains a tag for the calmodulin-binding protein (CBP). After expression of the CBP-HOS3 recombinant protein in E. coli, the 84-kDa fusion protein was affinity-purified on calmodulin-Sepharose. Gel filtration revealed that the peak activity chromatographed at an apparent molecular mass of 190 kDa (Fig. 2A). We found that the active peak fractions contained a single, 84-kDa CBP-HOS3 species as determined by staining an SDS/PAGE gel with Coomassie blue and by Western blotting (Fig. 2B). Because there was a slight discrepancy between the apparent molecular mass of the E. coli-produced CBP-HOS3 activity vs. the yeast HOS3 activity, we wished to determine more accurately the molecular mass of the CBP-HOS3 complex purified from E. coli. The peak Superdex fraction (28) containing CBP-HOS3 was analyzed by sedimentation equilibrium ultracentrifugation. Using this method we determined for a single, ideal species that the molecular mass of the CBP-HOS3 complex is 173 kDa ± 3.1 kDa (data not shown). This is very close to the predicted molecular mass of CBP-HOS3 homodimer (168 kDa). No other E. coli proteins were observed by SDS/PAGE after purification. Moreover, thrombin and tryptic protease digests of CBP-HOS3 were consistent with a single species (data not shown). Therefore, we conclude that HOS3 forms a homodimer in E. coli and has intrinsic deacetylase activity in this form.
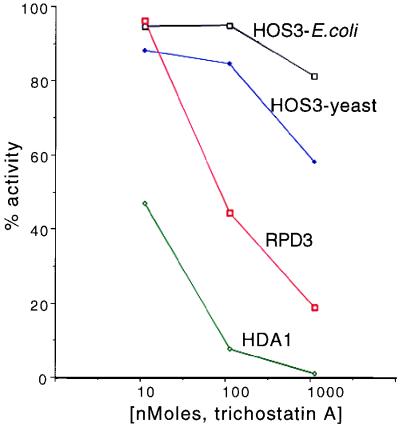
HOS3 Is Relatively Insensitive to the Histone Deacetylase Inhibitor TSA as Compared with HDA1 and RPD3.
TSA is a potent, noncompetitive inhibitor of both mammalian and yeast histone deacetylases (1, 19). To determine whether HOS3 is sensitive to TSA, we tested immunoprecipitated HDA1, RPD3, and HOS3 activities from yeast for their sensitivity to this drug by using antibodies and procedures described earlier (1, 2). HOS3 from yeast was found to be at least two orders of magnitude less sensitive to TSA than HDA1 (Fig. 3). Interestingly, CPB-HOS3 purified from E. coli appears even less sensitive to the drug. Whether there are small quantities of contaminating deacetylases in the yeast HOS3 precipitate that contribute to this difference is unknown at this time. Finally, we found that the sensitivity of RPD3 deacetylase activity to TSA is intermediate between HDA1 and HOS3. We conclude that HOS3 is relatively insensitive to TSA.
Figure 3.
HOS3 is much less sensitive to the deacetylase inhibitor TSA than the related deacetylases HDA1 or RPD3. Activities containing HOS3, HDA1, and RPD3 were obtained by immunoprecipitation from yeast nuclear extracts as described previously (1). HOS3 from E. coli was obtained from the peak fraction in Fig. 2A (fraction 28). [3H]acetyl-Hela histones (50,000 cpm) were incubated with each of the deacetylases for 30 min and assayed as described previously (1). Inhibition of deacetylase activity at various concentrations of inhibitor was calculated as a ratio of enzyme reactions containing TSA vs. no TSA. Maximal conversion of the [3H]acetyl-Hela histones to the release of free acetate was maintained below 2% to reduce the effect of substrate limitation on the apparent inhibition by TSA.
Recombinant HOS3 Shows a Distinct Site Preference for Acetylated Yeast Histone N-Terminal Peptides in Vitro.
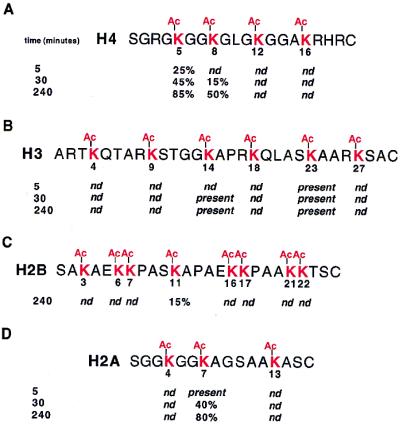
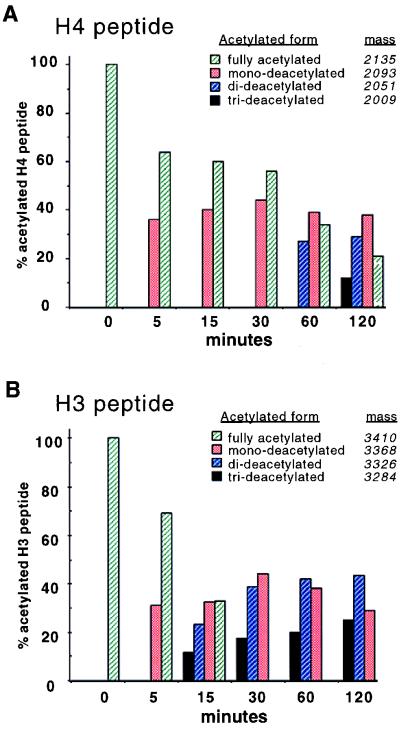
To determine which sites in each histone can be deacetylated by HOS3 in vitro we presented the E. coli-produced CBP-HOS3 enzyme with histone N-terminal peptide sequences containing acetyl groups at all lysines that are potentially acetylated in vivo. This was based on an accumulation of data (reviewed in ref. 31). After incubation with CBP-HOS3, the deacetylated peptides were analyzed by cHPLC-MS/MS (Fig. 4) and MALDI-TOF-MS (Fig. 5) to determine the precise sites most sensitive to the deacetylase. Incubating the peptides with increasing amounts of CBP-HOS3 over a time course followed by direct mass spectral analysis or a combination of proteolysis–mass spectrometry allowed us to generate a profile for deacetylation of individual sites. MALDI-TOF-MS was used to determine the number of sites. MALDI-TOF-MS analysis of the H4 peptide suggested that two major sites of deacetylation occur. The first was detected at 5 min (m/z 2093, mono-deacetylated, Fig. 5A), with the second site appearing at the 60-min incubation (m/z 2051, di-deacetylated). The later time points show a small amount of a possible third deacetylation site (m/z 2009, tri-deacetylated). The cHPLC-MS data indicate that sites K5 and K8 were deacetylated, in that order (Fig. 4A), the minor form suggesting a possible third deacetylation site generated only after longer incubation was not identified. These data demonstrate that the enzyme has a distinct preference for sites K5 and K8, with less preference for K12 or K16. Moreover, our data suggest that K5 must be deacetylated before K8 deacetylation can occur.
Figure 4.
Treatment of acetylated histone peptides with recombinant HOS3 (CBP-HOS3) results in site-specific deacetylation patterns. Fully acetylated peptides for each histone N-terminal tail were synthesized as described in Experimental Procedures. These were treated with recombinant CBP-HOS3 (peak fraction 28, Fig. 2A) for 5, 30, and 240 min with 1.2 μg of enzyme and analyzed by cHPLC-MS/MS as in Experimental Procedures. When applicable, the approximate amount of the detected ion compared with the total peptide is indicated for each site. When the value fell below the threshold of detection (≈5%), nd (not detected) is indicated. If the ion for the deacetylated species could be detected but a value could not be estimated accurately, present is indicated.
Figure 5.
MALDI-TOF-MS analysis of H4 and H3 peptides. Deacetylation of fully acetylated H4 (A) and H3 (B) peptides reveals that at least one additional site of deacetylation is present in each peptide as compared with cHPLC-MS/MS analysis. The MALDI-TOF-MS data was calibrated externally using standard peptides. Mass of the deacetylated peptides is as indicated. The presence of mono-, di-, and tri-deacetylated species is indicated for each peptide. Percentage of peptide in each acetylated state is calculated as a ratio of each acetylated form compared with the total input peptide for each reaction.
For the histone H3 peptide, two major deacetylation sites were identified. The first was detected at the 5-min incubation (m/z 3368, mono-deacetylated, Fig. 5B), and the second site was detected at 15 min (m/z 3326, di-deacetylated). This profile of deacetylation remained until the 120-min time point, in which a small amount of a possible third deacetylation site (m/z 3284, tri-deacetylated) was detected. To define more precisely the affected sites, the peptide was digested with endoproteinase Lys-C, a protease that specifically cleaves amide bonds on the carboxyl side of unmodified lysine residues, followed by analysis using cHPLC-MS/MS. Results showed that cleavage occurred only at K14 and K23 with detection of three Lys-C-generated peptides: m/z 1574 [ARTK(Ac)QTARK(Ac)STGGK], m/z 1040 [APRK(Ac)QLASK], and m/z 2639 [ARTK(ac)QTARK(Ac)STGGK(Ac)PRQK(Ac)LASK]. The existence of the overlapping peptide m/z 2639 suggests that K23 was the first site of deacetylation (Fig. 4B). We conclude that CBP-HOS3 has a distinct specificity for histone H3 K14 and K23 in vitro and is able to deacetylate one or more of the remaining sites less efficiently.
In contrast to H3 and H4, we found that, for H2A and H2B, only single sites in each were relatively efficient substrates for the enzyme. In H2A, only site K7 was deacetylated efficiently, with up to 80% deacetylation at 240 min (Fig. 4C). For H2B, only site K11 was deacetylated, but less efficiently (15% at 240 min, Fig. 4D), and all other five sites remained resistant. It is possible that the three close pairs of acetylated lysines (K6 and K7; K16 and K17; and K21 and K22) present an unnatural substrate that would not be encountered in vivo. We conclude that all four histones can be deacetylated by CBP-HOS3 in a site-specific manner.
Discussion
The data above support strongly the identification of yeast HOS3 as a new histone deacetylase. HOS3 has 38.9% similarity to RPD3 over 271 aa (HOS3 amino acids 118–389, using the bestfit program; Genetics Computer Group, Madison, WI), its disruption increases histone H4 acetylation in cell extracts, and the enzyme deacetylates core histones in vitro in a highly specific manner. However, HOS3 is a highly unique deacetylase. Under similar conditions of purification that give rise to very large and stable, multisubunit RPD3- and HDA1-containing complexes, HOS3 was purified as a homodimer. HOS3 activity is relatively insensitive to the deacetylase inhibitor TSA, demonstrating a difference in structure between the HOS3 catalytic subunit and that of HDA1 and RPD3. It also has intrinsic activity when expressed in E. coli. This may be a feature unique to HOS3. In the case of HDA1, when any of the subunits (HDA1, HDA2, or HDA3) of the HDA1 complex are disrupted, HDA1 activity is reduced completely in vitro and in vivo (J. Wu, A.A.C., and M.G., unpublished data). Therefore, HDA2 and HDA3 are essential for HDA1 activity. Similar consideration may hold for the other deacetylases. We have attempted to express the remaining histone deacetylases (RPD3, HDA1, HOS1, and HOS2) as CBP-tagged proteins in a manner identical to that described for HOS3. However, only CBP-HOS3 showed intrinsic histone deacetylase activity in E. coli (data not shown).
Interestingly, CBP-HOS3 has high specificity in vitro for restricted sites in H4 (K5, K8), H3 (K14, K23), H2A (K7), and, to a much lesser extent, H2B (K11) in acetylated histone peptides. Both the Western blot data (Fig. 1) and the MALDI-TOF-MS analysis (Fig. 5) argue for a third site that is deacetylated somewhat less efficiently in histone H4 by HOS3. This site is likely to be histone H4 K12 (Fig. 1). We do not know which other sites are deacetylated less efficiently in H3. It should be noted that one or more acetyltransferases must acetylate the sites that are subsequently deacetylated by HOS3 in vivo. ESA1 may be a candidate for such an acetyltransferase. Unlike GCN5, which has specificity for histone H3 in vitro (32) and in vivo (J. Wu and M.G., unpublished data), ESA1, like HOS3, affects H4, H3, and H2A preferentially in vitro. In particular, it acetylates H4 site K5 and H3 site K14 most efficiently (33). Whether ESA1 (or a related acetyltransferase other than GCN5) acetylates sites deacetylated by HOS3 and whether either enzyme has a different specificity in vivo remain to be determined.
HOS3 is found in a very large complex in partially purified yeast nuclear extracts. This is true whether we examine wild-type cells or cells overexpressing HOS3. In wild-type cells, most, if not all, of the HOS3 is in the larger complex. When HOS3 is overexpressed, only a small fraction (7%) is found in the size range of the homodimer. However, because the bulk of activity is in this size range it is likely that the homodimeric complex is the active form of the enzyme. Moreover, both the E. coli- and yeast-produced HOS3 enzyme are stable in very high salt concentration (1 M NaCl), suggesting that the dimer once formed is extremely stable. Interestingly, treatment of the active HOS3 complex with 100 mM 2-mercaptoethanol destroys both the complex and its activity (data not shown), arguing for disulfide bridges (there are 8 cysteines in HOS3) that stabilize the HOS3 enzyme. Because the active HDA1 complex is stable under these same 2-mercaptoethanol conditions (1), it is possible that its stability does not involve disulfide bridges in this manner but requires the noncatalytic HDA2 and HDA3 subunits. Similarly, noncatalytic subunits may be required for the stability of the other histone deacetylases (including RPD3), which are purified as large stable complexes.
Because HOS3 does not require other proteins for its in vitro activity and stability, the role of the large complex that HOS3 is found in remains a mystery. We cannot exclude the possibility that the additional proteins found in the larger complex are required for deacetylation of nucleosomal substrates in vivo, although our data would argue these factors are noncatalytic. These potentially could be required to recognize, gain access to, or target the nucleosomal bound substrate. We also cannot exclude the possibility that the large complex sequesters HOS3 from its targets in the genome until physiological conditions dictate that it be delivered in an active form to those targets. Interestingly, the amino acid sequence of HOS3 contains a nuclear DNA-binding signature motif identical to that of the myb oncoprotein family, as identified by the motif program (Genetics Computer Group). The motif of WTTEEEFNV is in a single copy starting at amino acid 319. This is similar to that of yeast REB1 (26), a DNA-binding protein that binds rDNA and the promoters of many RNA polymerase II-transcribed genes (27). Therefore, we cannot exclude the possibility that HOS3 binds DNA directly. In fact, we have used band-shift analysis and methylation interference to show that HOS3 does bind DNA upstream of the MET10 gene in vitro (data not shown). However, we have not been able to demonstrate that HOS3 regulates MET10 expression or is found at MET10 in vivo. Whether the MET10 site is recognized because it is a true binding site or because it is similar to an in vivo binding site is unknown at this time. These possibilities will be distinguished only when the in vivo sites of action of HOS3 are found. This will allow us to determine not only the specificity of HOS3 in vivo but its mode of interaction at the genes which it regulates.
Acknowledgments
We thank Suzanna Horvath for protein synthesis and Achim Wach and Peter Philippsen for KanMX constructs. We are also grateful to Martin Phillips for aiding us in the sedimentation analysis and Thomas Sutherland for assistance with fermentation. Additionally, we thank the members of the Grunstein lab for input and critical reading of the manuscript. This work was supported by a Public Health Service grant (GM23674) from the National Institutes of Health to M.G.
Abbreviations
- TSA
trichostatin A
- MALDI-TOF-MS
matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- CBP
calmodulin-binding protein
- ECL
enhanced chemiluminescence
- cHPLC-MS/MS
capillary HPLC tandem mass spectrometry
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Carmen A A, Rundlett S E, Grunstein M. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 2.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 5.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 6.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 7.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 8.Wong J, Patterton D, Imhof A, Guschin D, Shi Y, Wolffe A. EMBO J. 1998;17:520–534. doi: 10.1093/emboj/17.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 10.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 11.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 12.Kao H, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C, Evans R, Kadesch T. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 14.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 15.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 17.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Nature (London) 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 18.Wade P A, Jones P L, Vermaak D, Wolffe A P. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 20.Darkin-Rattray S J, Gurnett A M, Myers R W, Dulski P M, Crumley T M, Allocco J J, Cannova C, Meinke P T, Colletti S L, Bednarek M A, et al. Proc Natl Acad Sci USA. 1996;93:13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasten M M, Dorland S, Stillman D J. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suka N, Carmen A A, Rundlett S, Grunstein M. Cold Spring Harbor Symp Quant Biol. 1998;63:391–399. doi: 10.1101/sqb.1998.63.391. [DOI] [PubMed] [Google Scholar]
- 23.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 24.Wach A, Brachat A, Rebischung C, Steiner S, Pokorni K, te Heesen S, Philippsen P. Methods Microbiol. 1998;26:68–81. [Google Scholar]
- 25.Laman H, Balderes D, Shore D. Mol Cell Biol. 1995;15:3608–3617. doi: 10.1128/mcb.15.7.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrow B E, Ju Q, Warner J R. Mol Cell Biol. 1993;13:1173–1182. doi: 10.1128/mcb.13.2.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klempnauer K H, Sippel A E. EMBO J. 1987;6:2719–2725. doi: 10.1002/j.1460-2075.1987.tb02565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 29.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horteon J S, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 30.Hecht A, Strahl-Bolsinger S, Grunstein M. Nature (London) 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 31.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 32.Grant P, Duggan L, Cote J, Roberts S, Brownell J, Candau R, Ohba R, Owen-Hughes T, Allis C, Winston F, et al. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 33.Smith E, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R, Lucchesi J, Allis C. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]