Abstract
Image database extensions for functional brain images were assessed by asking clinicians questions about i) diagnosis confidence level before and after using the software; ii) expected and unexpected differences between patient and control images; and iv) an overall rating of the future usefulness of this application in an everyday clinical setting. Examining the difference image of a patient compared to a normative group affects the clinicians' initial diagnosis of the patient in two-thirds of the cases. All three clinicians stated that the interface would be a useful tool when added to the clinical workup of a patient.
Keywords: PET scan, computer-assisted diagnosis, medical image database, brain, image interpretation, clinical decision support systems
Introduction
As functional neuroimaging techniques, such as positron emission tomography (PET), become increasingly important in patient diagnosis, clinicians more than ever need effective tools to quantitatively analyze and interpret these images. A quantitative analysis of a patient's PET scan involves comparing this scan to a normative database of controls, the results of which are summarized in a statistical parametric map of the t-values showing where in the brain the patient is significantly different from normal. Analyzing and interpreting these complex difference patterns associated with disorders such as Alzheimer's disease (AD) and other dementias remains in the domain of image processing experts, and is not currently available to clinicians.
PET images have been shown to be clinically useful in the management of patients with Alzheimer's disease and other forms of dementia. Silverman et al [1] assessed the sensitivity and specificity of cerebral glucose metabolic patterns in 146 AD patients undergoing evaluation for dementia. They found that PET was able to predict progressive dementia with a sensitivity of 93% and a specificity of 76%. PET is also useful in differentiating well-established cases of AD versus dementia with Lewy bodies (DLB) [2]. Even when the data are adjusted for age and MMSE scores, the PET results showed decreased cerebral metabolic rates in the DLB group compared to the AD group for visual primary and association areas of cortex. In [3], the authors examine several image classification techniques to differentiate between AD and frontotemporal dementia, and achieve diagnostic accuracies similar to and better than visual inspection alone (accuracies range from 70% to 90%). These results suggest that PET images can be used clinically to differentiate between dementias that have already caused significant decline in patients, but the changes seen in these images are often obvious and can be seen by direct visual inspection. A pressing need is for better clinical tools to aide in the diagnosis and treatment of patients whose decline is mild to moderate and where direct inspection of the PET scan will not yield useful information.
When examining patients before dementia develops, the timely evaluation of the patient's PET scan becomes even more urgent. For example, patients with Mild Cognitive Impairment (MCI) [4] present clinically with a decline in cognitive performance more pronounced than is expected for their age, and studies have shown that a certain percentage of these patients will progress to AD [5]. However, clinical criteria alone are not enough to predict which of these patients will progress [6]. In [7], PET imaging along with genetic susceptibility (patients were screened for the APOE genotype) is examined in a longitudinal, prospective study. Using genotyping alone, rates of 75% sensitivity and 56% specificity were achieved; when PET is combined with genotyping the results show either a very high sensitivity or very high specificity in detecting dementia progression. However, PET imaging alone, when patient images are compared to a normative dataset, showed the best results with sensitivity at 92% and specificity at 89%. Other studies have shown that there are detectable differences in MCI populations when compared to normal controls, particularly in the posterior cingulate cortex [8, 9, 10] and the hippocampus [11, 12, 13]. In [11], the baseline PET FDG scan predicted decline from normal aging to MCI with sensitivity of 83% and specificity 85%. These results suggest that even in the early stages of disease PET images, when compared to normal control images, give the clinician important information regarding the future prognosis of the patient. However, there is currently no accessible method for a clinician to accomplish this comparison and interpret the results in the clinical environment.
In a previous work [14], we reported our implementation of several database extensions in the form of image operators, which allow the user to perform image comparisons from within a structured query language (SQL) command. By passing in the patient image identifier, the comparison group identifiers, and several parameters and options, a complete image comparison can be run in a matter of seconds. We have simplified the execution of the comparison query by designing and implementing clinical software which allows the user to choose the inputs for the comparison, and to view the resulting t-value difference image and analysis details. In this paper, we will present the clinical software and preliminary results after having three clinicians assess the usefulness of the software.
Methods
Graphical User Interface Design
The approach to this initial interface design was purposefully generic. Because this is a novel application area, we included in the interface options and parameters which could be disabled or enabled based on the intended user group. The interface is designed to be flexible and useful to a wide range of users, from clinicians needing to interpret a single patient scan, to a radiologist or neuroscience researcher interpreting many images and re-running comparisons using different statistical and mapping techniques. The interface was also designed with remote clinician virtual labs in mind. In this context, a remote clinician without immediate access to PET image interpretation by an expert, can submit a patient scan and compare that scan to a global set of normal control scans. The remote clinician can store any number of brain scans in a “source lab” area, which could be used for online comparison projects. In this experiment, the user group is the clinician who must interpret data on a patient seen for evaluation of MCI symptoms. For this reason, several of the options not directly related to a clinicians daily work environment are disabled, and others are set to defaults.
The interface is written in JAVA and connects to the Oracle database using Oracle Application Developer Framework (Oracle ADF). The interface is designed to take advantage of the database system, and allows the user to query for the inputs to the comparison and stores the result of the comparison in the database for future reference and re-examination. The interface, shown in figure 1, has 8 sections numbered and pointed to by arrows: specifying the setup information, choosing a patient scan, choosing a comparison set of scans, setting parameters, running commands, viewing voxel peaks, viewing details on voxel peaks, and viewing patient and comparison group details. The following paragraphs describe each of these sections.
Figure 1.
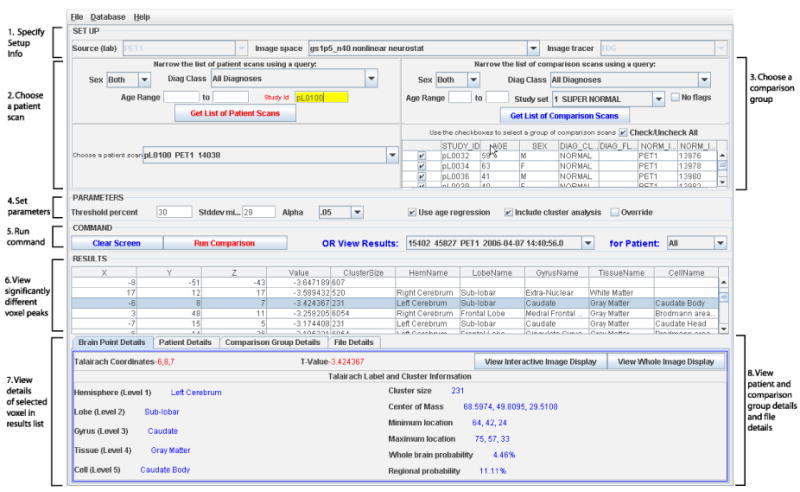
The GUI for comparing a patient brain scan to a normative reference set of scans. Clinicians use this interface to choose the patient scan, the set of comparison scans, and to view the results.
1. Specifying Setup Information
There are three options chosen before starting a comparison: the source lab, the image space, and the image tracer. The database is designed so that data can be assigned a source lab at the level of each relation. In the future, this will be important to allow for comparison of images among different labs, as well as for security purposes in case data should not be shared. For clinical testing purposes in this assessment, we have set up data in one source called ‘PET1’ and this option is disabled to the user for this work.
The image space denotes the processing technique used on the original brain image to transform it into a common coordinate system. The processing technique can include the software used for the transformation as well as any parameters or filters used. Images are stored in the database in their original format, as well as in one or more different image spaces, each achieved with a different processing technique. For the clinical user group, these processing options are not likely to be utilized, as this is more a function of an image processing expert than a clinician. Therefore, a default processing technique was used in this assessment.
The image tracer denotes the tracer used for the PET scan, which for this dataset is either [18F]flurodeoxyglucose (FDG) or [15O] water. In this assessment, we are using the interface to compare patient FDG scans, so this option is disabled to the user.
2. Choosing a patient scan
All available patient scans matching the three setup options selected in section 1 are viewable in the dropdown box in section 2 labeled ‘Choose a patient scan’. This list can be narrowed by sex, diagnosis, age, and/or study identifier using the query options, which together form an “AND” query. In most cases, the clinician knows the study identifier of the patient of interest and will type in this number to quickly select the patient.
3. Choosing a comparison group
The table at the bottom of section 3 lists scans available for comparison. To choose which scans to include in the comparison group, the user checks the checkboxes on the left hand side of the table, or, to include all the scans in the table, the ‘Check/Uncheck All’ checkbox can be used. Similar to choosing a patient scan, the user can choose the comparison group by running a query. In addition to sex, diagnosis, and age, there is a drop down list called Study Set. A Study Set is a pre-defined list of scans. This makes it easier to choose a comparison group, especially if the same group is chosen for most comparisons. By default, the Study Set titled SUPER NORMAL is chosen, so unless the clinician wishes to perform a more specific comparison, they do not have to make any changes in this section.
4. Setting parameters
In Section 4, parameters such as threshold percent, minimum contribution to standard deviation (specifies the minimum number of contributing voxels necessary for the standard deviation part of the comparison), and the statistical alpha value (alpha is the probability of rejecting the null hypothesis under null conditions, i.e. false positive) are set. By default, the threshold percent is set at 30%, the minimum contribution for the calculation of the standard deviation is set at the number of scans selected for the comparison group (the value will change automatically as new scans are added or scans are taken out of the comparison group), and alpha is set at 0.05. The checkbox ‘Use age regression’, checked by default, indicates the comparison group is first linearly age-regressed, voxel by voxel, to produce an age-regressed average image. This age-regressed average image is then used in the subtraction and calculation of the resulting t-image. The checkbox ‘Include cluster analysis’ determines whether cluster analysis will be run after the comparison is complete. Finally, the checkbox ‘Override existing comparison’ will force a comparison to run even if this same comparison has already been run in the past. If this is not checked, an error will be returned when trying to run a comparison that has already been run. This is to save disk space as well as time, since the user can simply look up any results of comparisons that have already been run.
5. Run Command
The button ‘Run Command’ runs the image comparison using the selected patient and comparison group scans and parameters. The ‘View Results’ drop down list will have the latest results selected, and the remaining sections of the interface will be filled in with the results data. The button ‘Clear Screen’ will clear the results data, and the drop down list ‘for Patient’ narrows the results drop down list to all those pertaining to a single patient.
6. View Significantly different voxel peaks
The result of an image comparison is in the form of a list of significantly different voxel peaks between the patient scan and the comparison group scans. The table displayed in section 6 of figure 1 lists the x, y and z Talairach coordinate of these voxels, along with the voxel value, the cluster size (explained later) and the Talairach label for this voxel, i.e., it's hemisphere, lobe, gyrus, tissue and cell name. The Talairach coordinate system [15] is the most commonly used method of reporting stereotactic locations. The x, y and z coordinates describe the distance in millimeters from the anterior commissure. The results list can be reordered by clicking on the column header; in combination with the CTRL key, the user can order the results by sets of columns as well. By default, the results are ordered by voxel value alone.
7. View details of the selected voxel in results list
The information in the table in section 6 of figure 1 is also displayed in section 7 for easier viewing. This display updates every time a new row is chosen in the results list. Cluster size and cluster statistics is also shown in this section, including the location of the center of mass for the cluster, the location of the minimum and maximum value in the cluster, and whole brain and regional cluster probabilities.
8. View patient and comparison group details and file details
Figure 2 shows the remaining tabs in the results pane. The top part of figure 2 shows Patient Details. Patient demographics and study protocol information can be viewed here. In addition, the button ‘View Patient Brain Scan’ can be used to view the original patient PET scan. This enables the user to verify an abnormality in the original image detected by the subtraction. Similar data can be viewed on each member of the comparison group using the Comparison Group Details tab. Finally, the File Details tab lists each file created during the comparison, along with a process identifier which can be used to track these steps individually.
Figure 2.
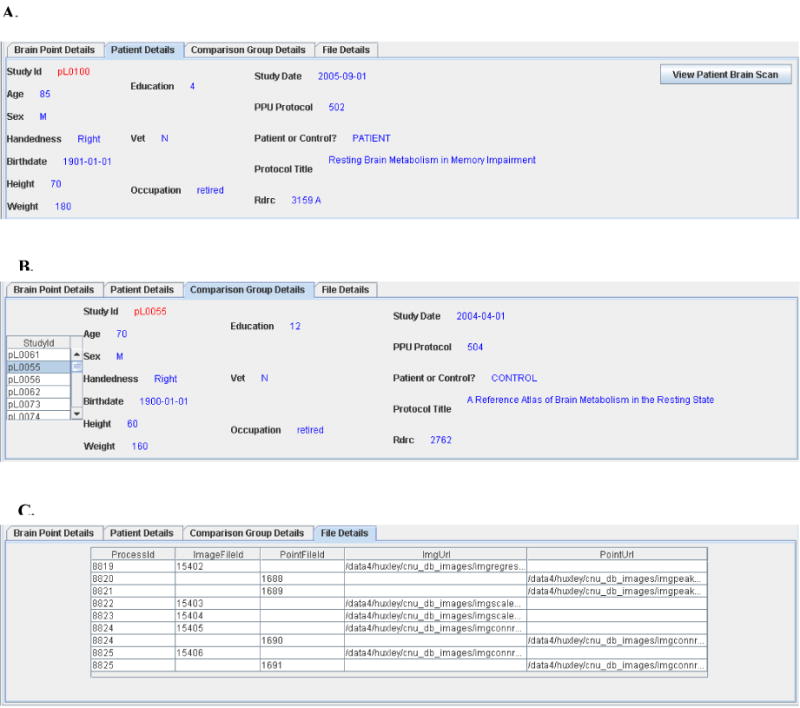
Patient and comparison group demographics and file creation details can be viewed in the lower pane of the interface. Part A displays the patient demographics. Part B displays the demographics for each member of the comparison group. Part C displays information about all physical files created during this comparison.
Viewing Results with the Interactive Image Display
Figure 3 shows how the user can view results interactively. The interactive image display allows the user to view the resulting t-image one slice at a time, from three orthogonal views (coronal, sagittal and transverse). The currently selected voxel is shown using red crosshairs on each of the three views. The color scale of the image is adjusted to negative or positive values depending on the currently selected voxel. This display is interactive because the user can choose a new result voxel from the list and the image will update the crosshairs to the new voxel location. In addition, the interaction goes both ways; the user can also click on an interesting point in one of the three views of the image and the nearest result voxel in the result list will be selected. Figure 3 shows the selected result voxel with (x,y,z) coordinates = (−25,10,36) at the red crosshairs in each of the three image views.
Figure 3.
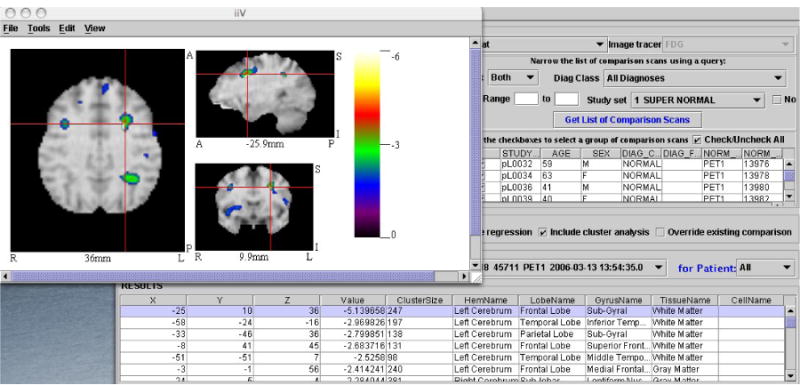
The interactive image display. The selected voxel in the result list is shown using red crosshairs on coronal, sagittal and transverse views of the voxel.
Viewing Results with the Whole Brain Image Display
In addition to the interactive image display, the user can choose to view the entire resulting t-image overlaid on an MRI image slice by slice, shown in Figure 4. The user is prompted to select whether to view the negative t-values or positive t-values. In this way, all the significantly different negative or positive areas of the patient's brain can be seen at once. Each slice has orientation labels for anterior (A), posterior (P), right (R) and left (L), as well as the millimeter labels designating the Talairach distance from the midline. The center slice label, at 0 mm, is highlighted in red.
Figure 4.
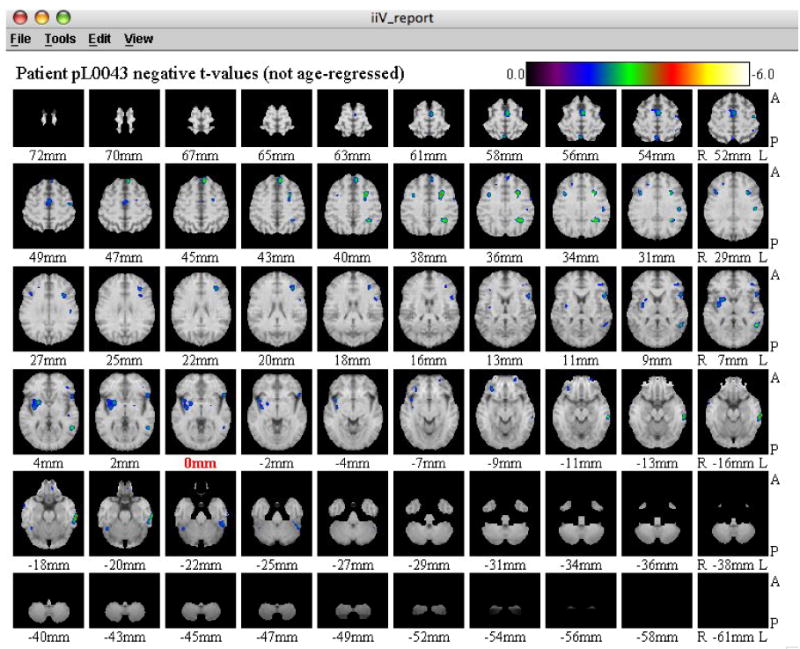
The whole brain image display. Users can view every transverse slice of the resulting t-image overlayed on a magnetic resonance image. The center slice label at 0mm is shown in red. A= anterior; P=posterior; R=Right; L=Left.
The interface can perform additional analysis tasks beyond comparing the patient scan to normal database. These tasks, which run after the image operation is complete, perform further analysis of the resulting t-value image - including analyzing the cluster size of neighboring significant voxels and analyzing cluster size statistics in relation to the whole brain and the region (defined by the hemisphere and lobe of each result voxel's Talairach location). The user can choose to run these tasks by checking the ‘Include Cluster Analysis’ checkbox, introduced in the section Setting Parameters.
The cluster size of a voxel specifies the number of connected statistically significant voxels to that voxel. Connectedness is defined by the neighboring 27 voxels in 3-D space. For example, if a voxel in the result list has cluster size 45 then there are 45 voxels, including the selected voxel, that make up a connected cluster of statistically significant voxels. The cluster size helps the clinician to determine the relevance of the currently selected voxel in the result list. A large cluster size hints at greater relevance, whereas a smaller cluster size is more likely to be due to noise in the image, depending on the image resolution.
Most brain structures of interest are of the size 5-10 mm; our image resolution is approximately 10 mm. Therefore, activations in small structures (1-2 mm) will be blurred out in our image pre-processing stages. Cluster size alone, however, is not enough to determine relevance: we need to also report cluster statistics in the form of probabilities to assess the relevance of a significant voxel.
There are two statistical values calculated from the cluster size of a voxel: the whole brain probability and the regional probability. The whole brain probability is the probability of seeing a cluster of that size or larger throughout the whole brain in a normal population. The regional probability is the probability of seeing a cluster of that size or larger in that hemisphere and lobe in a normal population. We calculated the whole brain and regional probabilities of cluster size by performing a bootstrap comparison of each normal control against the remaining normal control subjects, using a significance level of p = 0.01. The whole brain probability of a given cluster size is calculated by dividing the number of clusters greater than or equal to the given cluster size by the total number of clusters in all the normal subjects.
Similarly, the regional probability is calculated by dividing the number of clusters greater than or equal to the given cluster size in the given hemisphere and lobe by the total number of clusters in that hemisphere and lobe in all the normal subjects. The cluster probabilities enable the clinician to thoroughly assess the relevance of any significant voxel in the results list: a large cluster size with a small probability of seeing this size in the normal brain can be considered a more important result than one with a high normal brain probability.
Assessing the usefulness of the GUI in clinical practice
We asked three experienced clinicians who regularly see dementia cases in their clinic to participate in an assessment of the usefulness of the interface in clinical practice. This section presents the questionnaires used in the assessment and the results.
Each clinician filled out a pre-experiment questionnaire detailing information about their years of clinical practice, experience level in reading PET scans, experience level using computers, and experience using clinical decision support systems. The years of clinical experience ranged from 6 years to 38 years (one clinician had 6 years experience, one had 20 years and the third had 38 years). The physicians hold varying board certifications including psychiatry, neurology and internal medicine. None of the clinicians felt they had a strong expertise in reading PET scans, but they do require PET scan results about once a month in their practice. Normally, a nuclear medicine physician will send them a report after reading a PET scan and this would be incorporated into their patient work-up. Each clinician was familiar with computers enough to use applications other than email and the Internet on a daily basis, but did not routinely write their own programs or scripts. Each clinician is familiar with clinical decision support systems and uses an electronic patient record system on a daily basis.
Each clinician was asked to review excerpts from several patient charts of patients who were seen in their clinic within the last five years. The patient information was de-identified; only a numeric identifier was used to label each patient excerpt. The excerpt included the initial impression written by the clinician, initial diagnosis, and other lab results or imaging results that were available to the clinician at the time of the visit. Each patient was seen in the clinic for subjective memory complaints and/or cognitive issues.
For each patient, the clinician filled out a pre-interface questionnaire. Based only on the except given, the clinician answered the following questions:
Could there be multiple causes of the patient's complaints?
What would be your recommendations for this patient? Is further psychological testing necessary? Are further lab tests or brain scans, such as MRI, necessary?
What is your confidence level in this diagnosis? (Circle a number from 0 to 4 where 0 means “no confidence” and 4 means “very confident”)
The clinicians were encouraged to make additional notes explaining their position on the confidence level scale or to elaborate on any of the questions.
The clinicians used the interface to examine the difference between the patient's brain scan and the normative population. All defaults described in the previous section were used, including the comparison group, which was the same for each comparison. Because the interface was unfamiliar to the clinicians, they were able to ask for help in using it and could receive direction on viewing the images.
The clinician completed an after-interface questionnaire for each patient, which asked detailed questions about expected and unexpected findings and any changes in the impression of the patient. The clinician answered, among others, the following questions:
Did you see what you expected?
Did you see something unexpected?
If you previously recommended further tests, do you still think they are necessary?
What is your confidence level in the diagnosis now? (Same scale as above)
Again, they were encouraged to add additional comments where needed.
Finally, a post-experiment questionnaire was completed once by each clinician to assess opinions on the usefulness of the interface and further suggestions or comments about its future use in a clinical practice. Among the questions asked on this questionnaire were:
How useful do you think an application like this would be in an everyday clinical setting, where a physician needs to evaluate a patient's brain scan? (Answer by choosing a number from 0 to 4 where 0 means “not useful at all” and 4 means “extremely useful”)
Do you have suggestions for appropriate changes or upgrades to make this application more useful?
Results
In 10 out of 15 cases, examining the difference image of a patient compared to a normative group has an effect on the clinicians' initial diagnosis of the patient. In 7 of the 15 cases, the clinician became more confident in their initial diagnosis; in 3 cases the confidence went down; and in 5 cases there was no change. A detailed breakdown of the confidence changes is shown in Figure 5. When confidence levels went up after viewing the difference image, clinicians stated that the image confirmed or corroborated their initial impression, was suggestive of a different diagnosis (sometimes one that was not initially considered), or strengthened the probability of the initial diagnosis. In one case, the confidence went from a 2 with the comment “very uncertain about diagnosis” to a 4 (very confident) after viewing the difference image. When the confidence levels fell, clinicians stated that the image maintained their uncertainty, helped to keep them from concluding a diagnosis, or made them less certain of the diagnosis. In one such case, where the clinician saw less hypometabolism than expected, they state the image “suggests … that I should be cautious about giving a specific label for diagnosis.”
Figure 5.
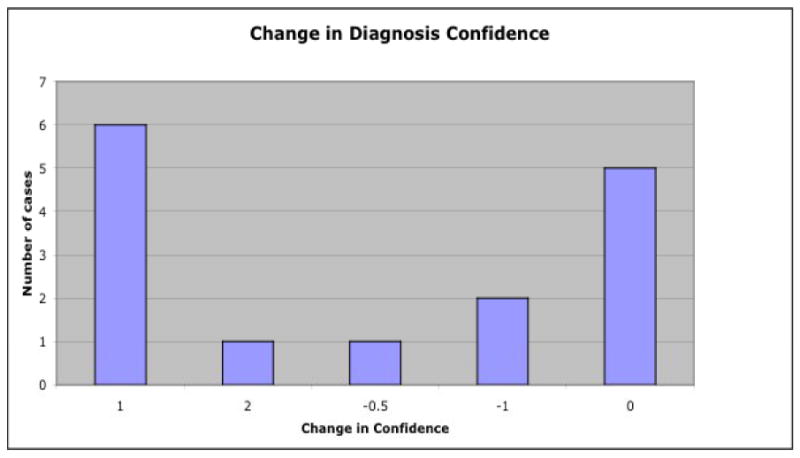
Changes in diagnosis confidence after viewing the difference image of a patient compared to normal controls. A positive change means the confidence went up after using the interface; a negative change means the confidence went down.
Of the 5 cases where confidence levels remained the same, all but one clinician still noted information was obtained from viewing the difference image. One clinician stated that their uncertainty about the diagnosis remained even after viewing the difference image. In 2 of the unchanged cases, the clinician stated that even though they were not changing their confidence level, the difference image did corroborate their initial impression. In one unchanged case the clinician stated that the difference image contradicted their initial impression and lead them to a possible new diagnosis. In only one of the unchanged cases did the clinician state that the difference image had no affect at all on their initial impression. This shows that even when the confidence level goes down or remains unchanged after viewing the image, valuable information is still provided to the clinician.
In the majority of cases, the clinician saw something unexpected in the difference image. Unexpected differences are divided into two groups: either the clinician saw hypometabolism in an unexpected area of the brain or they saw an unexpected degree of hypometabolism, either more or less than expected. In 8 cases, unexpected areas of the brain showed hypometabolism. Of these, 4 cases showed areas in addition to expected areas. In the 4 cases where only unexpected areas had hypometabolism, in 2 of these cases clinicians claimed increased confidence by providing a new possible diagnosis, and in 2 cases the clinician claimed decreased confidence and more uncertainty about the diagnosis. In one case, more profound hypometabolism was seen than expected. In this case, the clinician's initial impression was corroborated and their confidence increased, but they were surprised that the image seemed much worse than the clinical picture. Two cases showed less hypometabolism than expected; in one case the confidence level decreased, and in the other it remained unchanged.
All three clinicians stated that the interface would be a useful tool to be added to the clinical workup of a patient. On a scale from 0 to 4, with 0 meaning “Not useful at all” and 4 meaning “Extremely useful”, one clinician gave the interface a score of 2, one a score of 3, and one gave it a score of 4. However, all three clinicians stated the need for modifications, including more help in the interpretation of the difference image, better anatomical labeling, and more automated image viewing techniques.
Clinicians filled out a comments section in which they were asked to provide any suggestions for future upgrades, and any other comments about the application, their experience using it, and it's future use in a clinical setting. A common concern is the need for image interpretation. None of the three clinicians in this assessment claimed to have any experience reading PET scans. When PET scans are ordered in this clinic, a radiology specialist interprets them and a report is entered into the patient's record. This report is based solely on a visual reading of the original PET scan. The interface provides clinicians with a tool to perform a more detailed analysis of the PET scan, and the general comment among all three clinicians is that they don't feel they have the knowledge or expertise to interpret these results accurately without more background and supporting information. One way to increase the clinicians' comfort level is to provide example patterns of common psychiatric illnesses for comparison, additional tools to aide in the interpretation of the hypometabolism patterns they are seeing, and online reference information such as brain atlases and relevant literature articles. One clinician suggested that the interface be used as part of the telemedicine effort, where an online psychiatrist is available for a live, remote walk-through of the analysis of the difference image.
The interface may also have adjunct uses, such as a learning and review tool. If future versions have more informational data incorporated, such as example brain patterns for common psychiatric diseases, atlas tools for visualizing brain mapping and labels, and pointers to appropriate literature on specific diseases, clinicians can learn to use data previously unavailable to them in interpreting PET scans, as well as review their knowledge of brain anatomy, psychiatric illness, and the current relevant literature. Medical school classrooms might also benefit from a tool that provides hands-on image interpretation on real patients.
Discussion
Our intention in this work is not to compare the conventional radiologist report to the diagnostic aide, but to compare the clinician's confidence in a diagnosis before viewing the difference image analysis and after viewing the difference image analysis. Our diagnostic aide was not directly compared to the conventional radiologist report because they are not attempting the same form of analysis, and the conventional approach to reading PET scans is not part of the diagnostic process for MCI and early Alzheimer's disease. The conventional report is not reporting on differences in cognitive functioning, but rather gross anomalies that can be discerned by direct viewing of the patient scan alone. Therefore, our study is looking to complement the diagnostic process, where the conventional diagnostic process should be considered the evaluation of the patient by the clinician, including medical history, thorough neuropsychiatric testing, MRI and CPT to rule out structural problems, and a full medical exam. Our application complements this process by adding a statistical analysis of the PET scan. In this paper, we are comparing the traditional approach to diagnosis where statistical analysis of the PET scan is not done, to a new approach that includes a statistical analysis of the patient's PET scan.
There are several limitations to this assessment. Because this is a novel application area, our goal was to get feedback from a limited number of clinicians as a first step towards determining the usefulness of an application of this type; hence the number of clinicians and cases is low. Additionally, and related to the novelty of the application, there was a learning curve involved and each of the clinicians seemed to get more confident with the interface as they used it more. In light of this, it may have been helpful to have a training session beforehand where the clinicians could learn how to use the interface and the different features it provides, but time did not allow for this extra session. Another limitation is that this is a retrospective study, where the patient cases reviewed by the clinicians are historical. The clinician is not privy to the entire patient chart or other adjunct information that is normally available to them at the time of a clinical visit, such as meeting the patient and their family members. In this assessment, only a short, one paragraph excerpt from the patient chart is provided to the clinician, from which they must judge their confidence in the past diagnosis. In addition, we noted that the three clinicians participating in this assessment stated they currently interpret PET scans approximately once per month. This is not to say however, that this is a frequency representative of all clinicians, or that with the availability of better clinical tools such as the design in this paper, and the rising acceptance of PET scan evaluation into the clinical workup, that the frequency of use will not greatly increase. In future plans for a prospective study, more clinicians and patient cases will be included, better training on the interface will be provided, and the clinician will prospectively fill out the initial questionnaire when the patient first visits the clinic, and then fill out the post questionnaire after using the interface to analyze the patient PET scan.
Through this work we have brought to the clinician's desktop the ability to analyze and interpret a patient's brain scan - a previously insurmountable task. We have presented a novel software application that builds on the database extensions for image analysis work in [14] to bring online, interactive brain image analysis to the clinical environment. We have tested the use of this application with real patient cases and clinicians in an effort to assemble an initial assessment not only of the software, but also of the utility of enabling clinicians to perform brain image analysis from their own desktops. Previously, clinicians would need a computer expert to script and run several programs in order to accomplish this comparison. Now they not only can do their own image analysis, but they can re-run the comparison with new parameters or images quickly, and access associated patient information such as demographics, study protocol information, psychiatric test results, and image technical data. Any of the shortcomings noted by the clinicians in this assessment, such as lack of knowledge in reading difference images, can be easily addressed by improved clinical training in the use of brain images for diagnosis, and in appropriate upgrades to the software itself to provide supporting information and tutorials. Overall, clinicians are very receptive to the idea of having functional image analysis information at their fingertips with which to make more informed clinical decisions. With this application, we are working towards a first in clinical diagnostic aides for neuroimage analysis that will bear directly upon patient assessment, diagnosis, and treatment.
Summary
What was known before the study:
Positron emission tomography (PET) images have been shown to be clinically useful in the management of patients with Alzheimer's disease and other forms of dementia.
Early intervention and differentiation is very important, especially in cases of Mild Cognitive Impairment, where studies have shown that a substantial number of these patients will progress to AD.
Analyzing complex difference image patterns associated with disorders such as Alzheimer's disease (AD) and other dementias remains in the domain of image processing experts, and is not currently available to clinicians. These types of analyses are not regularly accomplished during the diagnosis and treatment of dementia cases.
There are currently no software packages or systems available that allow a clinician to interactively compare their own patient's PET scans to a normative control dataset, and to analyze the results as a listing of significantly different peaks, along with anatomical labeling, cluster counts, and whole brain and regional cluster statistics.
What this study has added:
In this work we have presented a novel software application that builds on previous work to bring real-time image analysis to the clinical environment.
Examining the difference image of a patient compared to a normative group has an effect on the clinicians' initial diagnosis of the patient.
According to the assessment results, clinicians are very receptive to the idea of having functional image analysis information at their fingertips with which to make more informed clinical decisions.
Acknowledgments
This work was supported by the National Institutes of Health/National Institutes of Mental Health Human Brain Project Grant R01 AG20852. We would like to thank Drs. Maurice Dysken, John R. McCarten, Howard Fink, and Michael Kuskowski for their insightful comments and participation in this study.
Biography
Kristin Munch holds a PhD in Computer Science and Computational Neuroscience from the University of Minnesota (Minneapolis, MN) and an MS in Computer Engineering from the University of Mississippi (Oxford, MS).
John Carlis is a professor in the Department of Computer Science at the University of Minnesota. His interests are in data modeling, language extensions, and scientific applications.
Jose Pardo is a professor in the Department of Psychiatry at the University of Minnesota and also the Director of the Cognitive Neuroimaging Unit at the Veterans Affairs Medical Center in Minneapolis, MN.
Joel Lee holds an MSEE from the University of Minnesota (Minneapolis, MN) and is currently a computer science researcher at the Cognitive Neuroimaging Unit at the Veterans Affairs Medical Center at the University of Minnesota.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kristin R. Munch, Computer Science and Engineering, University of Minnesota, munch@cs.umn.edu
John V. Carlis, Computer Science and Engineering, University of Minnesota
Jose V. Pardo, Department of Psychiatry, University of Minnesota Medical School and Cognitive Neuroimaging Unit, Veterans Affairs Medical Center
Joel T. Lee, Cognitive Neuroimaging Unit, Veterans Affairs Medical Center
References
- 1.Silverman DHS, Small GW, Chang CY, Lu CS, Kung de Aburto MA, Chen W, et al. Positron emission tomography in evaluation of dementia: regional brain metabolism and long-term outcome. JAMA. 2001;286:2120–2127. doi: 10.1001/jama.286.17.2120. [DOI] [PubMed] [Google Scholar]
- 2.Gilman S, Koeppe RA, Little R, An H, Junck L, Giordani B, Perstad C, Heumann M, Wernette K. Differentiation of alzheimer's disease from dementia with Lewy bodies utilizing positron emission tomography with [18F]fluorodeoxyglucose and neuropsychological testing. Exp Neurol. 2005;191:S95–S103. doi: 10.1016/j.expneurol.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Higdon R, Foster NL, Koeppe RA, DeCarli CS, Jagust WJ, Clark CM, Barbas NR, Arnold SE, Turner RS, Heidebrink JL, Minoshima S. A comparison of classification methods for differentiating fronto-temporal dementia from Alzheimer's disease using FDG-PET. Statist Med. 2004;23:315–326. doi: 10.1002/sim.1719. [DOI] [PubMed] [Google Scholar]
- 4.Petersen R. Mild Cognitive Impairment: Aging to Alzheimer's Disease. Oxford University Press; 2003. [Google Scholar]
- 5.Bruscoli M, Lovestone S. Is MCI really just early dementia? A systematic review of conversion studies. Int Psychogeriatr. 2004;16:129–140. doi: 10.1017/s1041610204000092. [DOI] [PubMed] [Google Scholar]
- 6.Friedrich MJ. Mild cognitive impairment raises Alzheimer disease risk. JAMA. 1999;282:621–622. [PubMed] [Google Scholar]
- 7.Drzezga A, Grimmer T, Riemenschneider M, Lautenschlager N, Siebner H, Alexopoulus P, Minoshima A, Schwaiger M, Kurzm A. Prediction of individual clinical outcome in MCI by means of genetic assessment and 18F-FDG PET. J Nucl Med. 2005;46(10):1625–1632. [PubMed] [Google Scholar]
- 8.Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann Neurol. 1997;42:85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]
- 9.Herholz K, Nordberg A, Salmon E, Perani D, Kessler J, Mielke R, et al. Impairment of neocortical metabolism predicts progression in Alzheimer's disease. Dement Geriatr Cogn Dis. 1999;10:494–504. doi: 10.1159/000017196. [DOI] [PubMed] [Google Scholar]
- 10.Arnaiz E, Jelic V, Almkvist O, Wahlund L-O, Winblad B, Valind S, et al. Impaired cerebral glucose metabolism and cognitive functioning predict deterioration in mild cognitive impairment. Neuroreport. 2001;12:851–5. doi: 10.1097/00001756-200103260-00045. [DOI] [PubMed] [Google Scholar]
- 11.de Leon MJ, Convit A, Wolf OT, Tarshish CY, De Santi S, Rusinek H, et al. Prediction of cognitive decline in normal elderly subjects with 2-[18F]fluro-2-deoxy-D-glucose/positron emission tomography (FDG/PET) Proc Natl Acad Sci USA. 2001;98:10966–71. doi: 10.1073/pnas.191044198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Santi S, de Leon MJ, Rusinek H, Convit A, Tarshish CY, Boppana M, et al. Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiol Aging. 2001;22:529–39. doi: 10.1016/s0197-4580(01)00230-5. [DOI] [PubMed] [Google Scholar]
- 13.Nestor PJ, Fryer TD, Smielewski P, Hodges JR. Limbic hypometabolism in Alzheimer's disease and mild cognitive impairment. Ann Neurol. 2003;54:343–51. doi: 10.1002/ana.10669. [DOI] [PubMed] [Google Scholar]
- 14.Munch KR, Carlis JV, Pardo JV, Lee JT. Novel DBMS extensions for functional brain image analysis. Proceedings 16th Intl Conf Sys Eng; 2005. pp. 301–6. [Google Scholar]
- 15.Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. Thieme Medical; New York: 1988. [Google Scholar]


