Abstract
Purpose
Monte Carlo codes can simulate the transport of radiation within matter with high accuracy and can be used to study medical applications of ionising radiations. The aim of our work was to develop a Monte Carlo code capable of generating projection images of the human body. In order to obtain clinically realistic images a detailed anthropomorphic phantom was prepared. These two simulation tools are intended to study the multiple applications of imaging in radiotherapy, from image guided treatments to portal imaging.
Methods
We adapted the general purpose code PENELOPE 2006 to simulate a radiation source, an ideal digital detector, and a realistic model of the patient anatomy. The anthropomorphic phantom was developed using computer-aided design tools, and is based on the NCAT phantom. The surface of each organ is modelled using a closed triangle mesh, and the full phantom contains 330 organs and more than 5 million triangles. A novel object-oriented geometry package, which includes an octree structure to sort the triangles, has been developed to use this complex geometry with PENELOPE.
Results
As an example of the capabilities of the new code, projection images of the human pelvis region were simulated. Radioactive seeds were included inside the phantom’s prostate. Therefore, the resulting simulated images resemble what would be obtained in a clinical procedure to assess the positioning of the seeds in a prostate brachytherapy treatment.
Conclusions
The new code can produce projection images of the human body that are comparable to those obtained by a real imaging system (within the limitations of the anatomical phantom and the detector model). The simulated images can be used to study and optimise an imaging task (i.e., maximise the object detectability, minimise the delivered dose, find the optimum beam energy, etc.). Since PENELOPE can simulate radiation from 50 eV to 1 GeV, the code can also be used to simulate radiotherapy treatments and portal imaging. Using the octree data structure, the new geometry model does not significantly increase the computing time when compared to the simulation of a much simpler quadric geometry. In conclusion, we have shown that it is feasible to use PENELOPE and a complex triangle mesh geometry to simulate real medical physics applications.
Keywords: Monte Carlo, PENELOPE, penMesh, triangle mesh, NCAT, computer-aided design
1. Introduction
Computer simulations are a valuable tool to study the medical applications of ionising radiation. There are several approaches to simulate the transport of radiation in matter and currently the most accurate codes are based on Monte Carlo (MC) algorithms. The MC simulations typically require long computing times but this fact can be overcome by executing the simulations in parallel in a multiprocessor workstation or in a computer cluster.
The improvements in computer power, notably in clock speed and physical memory, have had two important consequences in the application of MC simulation. First, some accelerated codes are becoming fast enough to be used in clinical routine. An example of this is the increasing use of MC simulation in radiotherapy treatment planning [12]. Second, more complicated physics and geometric models can be implemented in the general-purpose codes, increasing the accuracy of the simulations and allowing the study of new applications.
In this paper we describe penMesh, a MC simulation code that can simulate radiation transport in a geometry composed of objects limited by closed triangle meshes. A realistic computerised model of the human anatomy created with triangle meshes is also presented. As an example of the application of the new tools, the simulation of an x-ray imaging procedure in a prostate brachytherapy treatment is shown in the Results section.
2. Materials and Methods
2.1. The penMesh simulation code
penMesh [2,3] is a MC code that can simulate the transport of photons, electrons and positrons in a free-form geometry composed of objects limited by closed triangle meshes. This code uses the state-of-the-art physics from the general-purpose MC subroutine package PENELOPE 2006 [14,17], and the main program and tallies are based on the penEasy 2007 package [16]. The standard quadric geometry model from PENELOPE, PENGEOM, is used to transport the particles outside the triangle meshes bounding box. This facilitates the definition of some simple objects, like beam collimators or detectors. The energy range for which particles can be transported is 50 eV to 1 GeV.
penMesh and its object oriented geometric model are coded in C++. The combination of the PENELOPE code in FORTRAN 77 and the C++ code is handled by a header file that harmonises the calling conventions used by the two computer languages within the GNU compiler collection (http://gcc.gnu.org). The simulations can be easily executed in parallel taking advantage of the scripts from the clonEasy package [4].
The geometric routines used by penMesh detect the intersections between the particle trajectory and the triangle meshes. Since triangle meshes from different objects may overlap or be located inside others, it would be necessary to check the intersection with all the triangles in the geometry every time the particle moves. To avoid this time consuming task an octree structure was implemented in penMesh. An octree [8,6] is a hierarchical tree structure that recursively subdivides space in octants. This structure can be used to sort the triangles in the space in order that only those triangles located near the particle’s trajectory have to be checked for intersection. The octree requires a significant amount of computer memory but it does not introduce a significant overhead in the simulation and can increase the speed of penMesh up to 5 orders of magnitude [2]. Figure 1 shows an example of a level 9 octree structure created by penMesh to sort the triangles of the anthropomorphic phantom described in section 2.3. The displayed octree nodes contain triangles from the meshes describing the skin, the pelvis, the intestines, and a lumbar vertebra.
Fig. 1.
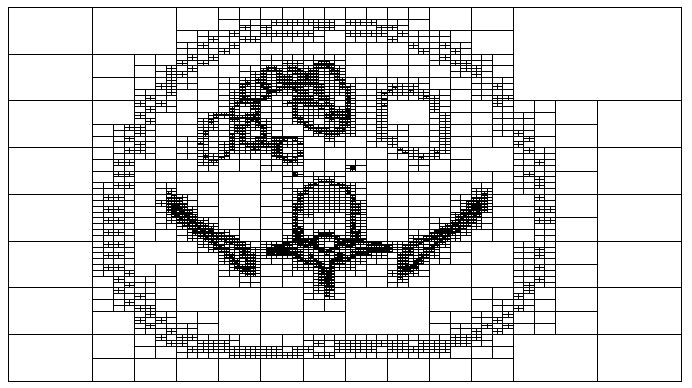
Axial slice of a level 9 octree structure sorting the triangles from the anthropomorphic phantom used in this work (see section 2.3).
2.2. Image formation model
In order to simulate a medical imaging system it is necessary to create a model of the image formation process. To create radiographic images penMesh employs a simple model in which the image is formed by scoring the energy deposited inside the detector object (typically a thin layer of CsI or any other scintillating material). The detector location, size, orientation, and the number of pixels in which it is divided, are given in the simulation input file. The detector geometry can be defined with triangle meshes or with quadric surfaces in case the detector is located outside the octree bounding box.
The implemented image model assumes that the grey level of each pixel is directly proportional to the deposited energy. The pixel value is reported in eV/cm2 per simulated particle. This allows for a straightforward comparison of images obtained using different geometries or simulation parameters. For a realistic detector geometry the detection efficiency is reproduced fairly well. However, for the sake of simplicity the detector material is usually defined to absorb all the incoming radiation. In this situation, the detector has 100% detection effciency, there is no scattering within the detector and the pixel values correspond to the energy fluence on the detector surface. penMesh includes an alternative method to generate images, which consists in a ray-tracing of the simulation geometry. In this method, which is not a MC approach, a virtual ray is cast from the focal spot to the centre of each pixel and the probability that an x ray does not interact in the full path is scored as the pixel value. The interaction probability is calculated using the cross-sections from the PENELOPE database for the mean energy of the input x-ray spectrum. The resulting image corresponds to the noise-free image that would be generated in an ideal detector disregarding any scattered radiation and beam hardening. This tally is extremely fast (requires on the order of 1 minute of computing time) and produces high contrast, noise free images because it does not include the effect of scattered radiation or the statistical uncertainty associated with a MC simulation.
2.3. Anthropomorphic phantom
The main advantage of the triangle meshes used by penMesh with respect to the quadric surfaces used by standard PENELOPE codes is that triangle meshes can represent any three-dimensional shape. This is particularly convenient for the description of organic structures [19]. Anthropomorphic phantoms have been constructed in the past using quadric surfaces, but the idealised anatomy of these phantoms is not realistic enough to be used in certain kinds of applications [19] (they cannot produce clinically realistic projection images, for example).
In order to use penMesh in medical applications we have developed a triangle mesh-based version of the NURBS-based Cardiac-Torso (NCAT) phantom [15]. The NCAT is a highly detailed male anatomical model created with Non-uniform rational B-splines (NURBS) and other computer-aided design (CAD) tools from the high-resolution CT scans of the Visible Human Project [18]. Our version of the phantom—shown in Fig. 2—is composed of 330 closed triangle meshes and comprised in total of 5 million triangles [2,10].
Fig. 2.
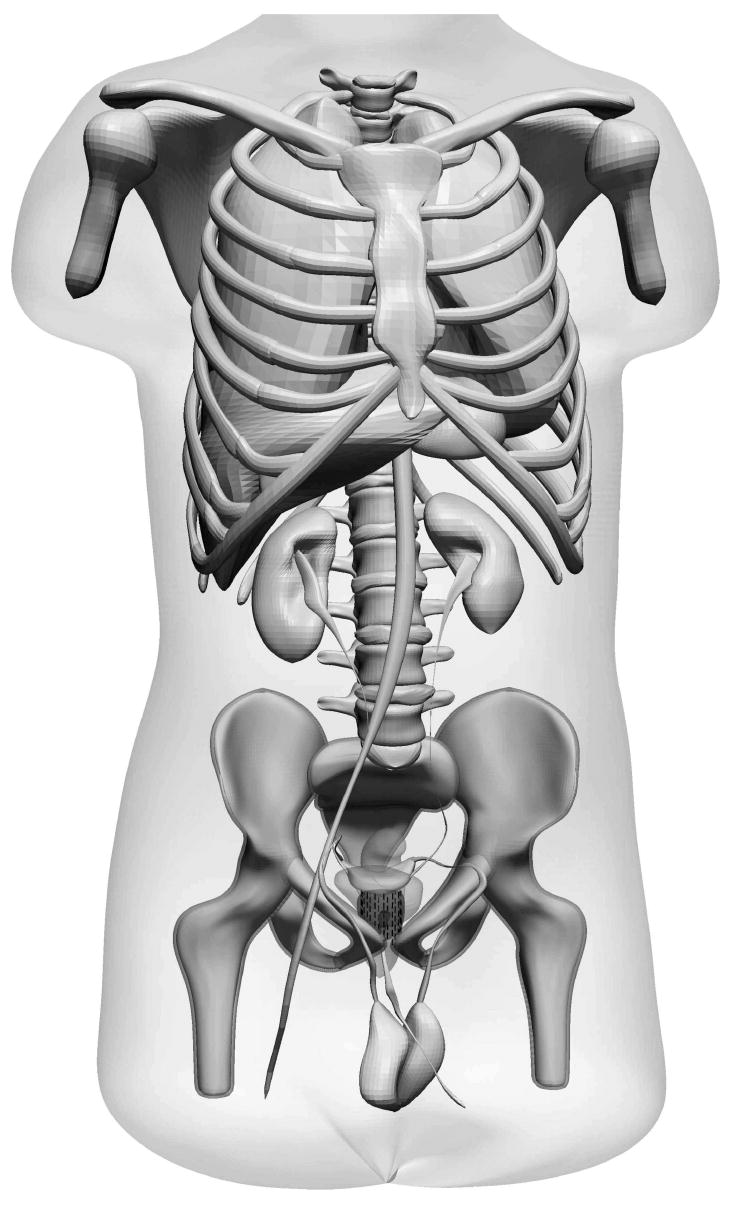
Tessellated version of the NCAT phantom [15]; 330 closed triangle meshes are used to define the shape of the organs (the intestines are not shown).
2.4. Imaging a prostate brachytherapy treatment
As an example of the use of the code and the phantom described in the previous sections, anteroposterior projection images of a male pelvis region were simulated. Radioactive seeds were included inside the phantom’s prostate to simulate the assessment of the seed positioning during a clinical prostate brachytherapy treatment [1,13].
A simplified model of a brachytherapy seed [9] was created with the open source software PARAVIEW (http://www.paraview.org) using two concentric cylinders. The internal cylinder (3 mm in length and 0.5 mm diameter) represented a Silver marker that allows the seeds to be readily seen in radiographic images, and the external cylinder (4.5 mm in length and 0.8 mm diameter) represented a Titanium encapsulation. Radioactive 125Iodine would be absorbed on the surface of the Silver rod but it was not included in our simple model. Both cylinders were tessellated into 80 triangles and stored as triangle meshes in ASCII PLY format [11], which is readable by penMesh. The meshes were cloned to produce 112 identical seeds and distributed forming a cylindrical pattern around the prostate to reproduce a brachytherapy treatment. A small random shift was applied to each seed in order to account for a small drift inside the prostate and implantation inaccuracies.
A realistic 70 kVp x-ray source [7] with a point focal spot was used for the simulations. The source-to-detector distance was 100 cm and the detector was located 20 cm behind the patient, producing a 1.3 geometrical magnification of the prostate region. A completely radio-opaque collimator was used to define 50×50 cm2 and 10×10 cm2 fields on the detector plane. The images were created with the two detector models described in section 2.2, with pixel sizes 500×500 μm2 and 200×200 μm2 for the 50×50 and 10×10 fields, respectively. The simulated detectors had a 100% detection effciency and did not include an anti-scatter grid. A dilute Barium sulphate contrast (3% weight/volume concentration) was injected inside the bladder and the urethra to facilitate the visualisation of these organs in the projection images [1].
3. Results and Discussion
The imaging procedure described in the previous section has been simulated with penMesh. The code was compiled using the GCC compilers (version 4.1.2) and executed in parallel using clonEasy [4] in a 30-node computer cluster. The computer used to report the simulation speed had an Intel® Xeon™3.0 GHz CPU and 2 GByte of RAM memory 1 . An octree with 8 levels of subdivision was used, and each execution required 260 MBytes of computer memory.
Figure 3 shows the simulated radiographic images for a 70 kVp x-ray source with a 50×50 cm2 field on a 60×60 cm2 detector with 500 μm pixels. The top image—Fig. 3 (a)—was created with the ray-tracing model. The grey scale corresponds to the probability, in %, that an x ray will arrive at the pixel without interacting. According to the obtained pixel values, more than 99.5% of the x rays that cross the phantom are absorbed or scattered before reaching the detector surface. This simulation required just 3 minutes of computing time in a single CPU. The bottom image—Fig. 3(b)—displays the full MC simulation. The image was created scoring the energy fluence on the detector plane. The grey scale has units of eV/cm2 per emitted x ray. The simulation speed was 17450 x rays per second. In total 1011 rays were simulated, what would require more than 66 days of computation in a single CPU (2.2 days in the computer cluster).
Fig. 3.
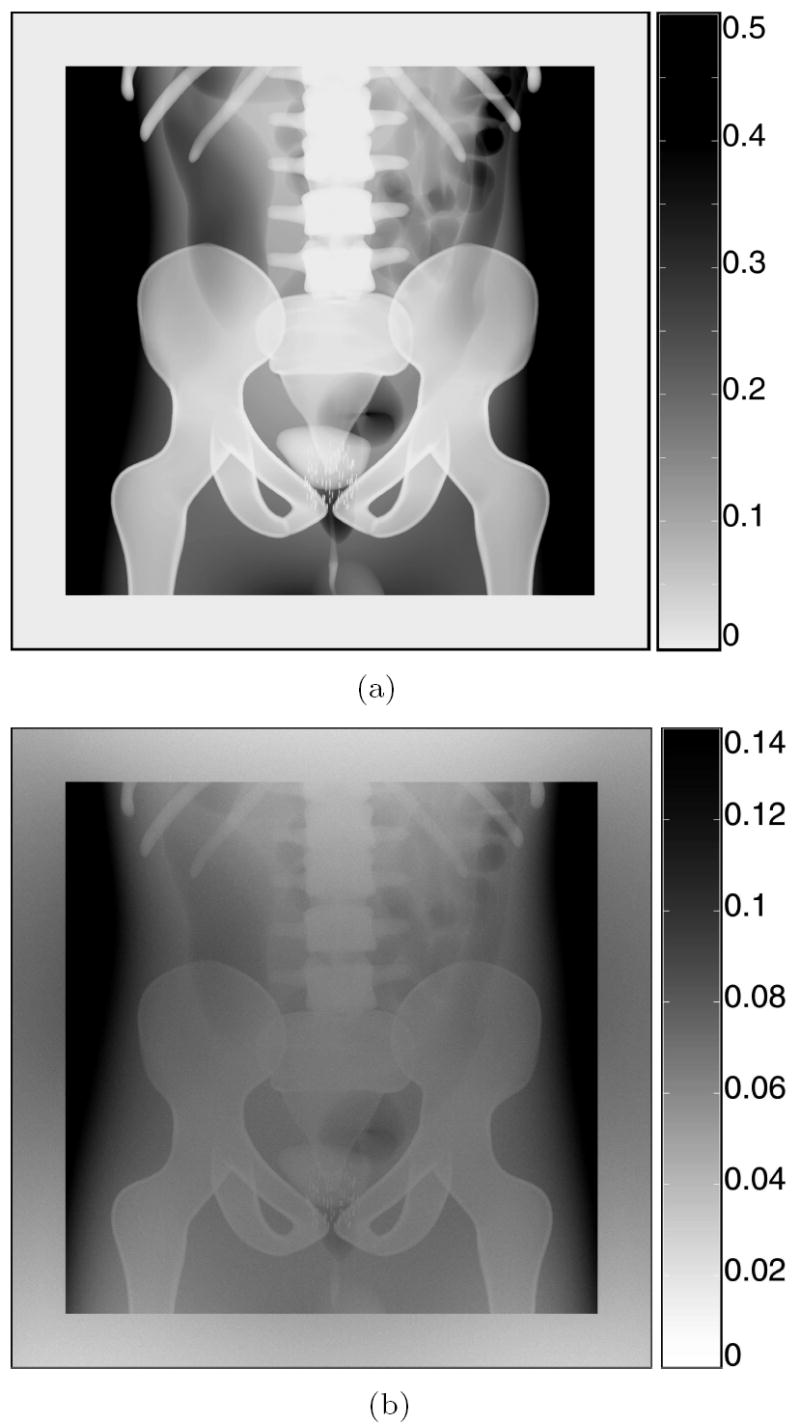
Simulated anteroposterior radiographic images produced by a 70 kVp x-ray source collimated to a 50×50 cm2 field, on a 60×60 cm2 detector with 500 μm pixels: (a) ray-traced image (grey scale: detection probability in %), (b) full penMesh simulation (grey scale: eV/cm2 per initial x ray). Pixels with values higher than the scale maximum are displayed in black.
The speed of the previous simulation using an octree with level 10, 4 and 0 (i.e., no octree) was 17725, 3105 and 4 x rays per second respectively. Therefore the octree accelerated the simulation more than 4300 times and it is definitely a key component of the algorithm. A previous attempt to use triangle meshes with PENELOPE showed a very low performance due to the lack of an acceleration data structure [5].
It has been found that the acceleration produced by the octree does not increase linearly with the number of subdivisions: the speed increases exponentially until it saturates above level 8. The saturation takes place when most of the octree nodes have a few triangles, or none, and consequently it depends on the simulation geometry. The achieved speed is comparable to the speed of an equivalent PENELOPE simulation employing a much simpler phantom described by quadric surfaces. This is caused by the fact that the most of the simulation time is spent in the physics routines and not in the geometry. In these large-field images it is possible to see the high level of detail of the phantom. The bladder and the urethra, which contain a radio-opaque contrast, can be clearly observed, as well as the intestines, which do not attenuate as much radiation as the surrounding tissue. The radioactive seeds can also be seen but not clearly because the image pixels are almost as big as the seeds.
In order to visualise the inner structure of the seeds a simulation with a higher resolution detector was performed. Figure 4 shows the simulation of a 10×10 cm2 field on a 10×10 cm2 detector with 200 μm pixels. Four images are displayed, corresponding to: (a) the ray-tracing model, (b) the full simulation, (c) the image formed only by x rays that have not interacted in the phantom (this is also the image that would be obtained with an ideal anti-scatter grid), and (d) the image created by scattered x rays and secondary radiation (including electrons). All these images have a linear grey scale with white at the value 0 and black at 0.4% for the ray-traced image (a) and 1.5 eV/cm2 per emitted x ray for the rest. In this case 1010 x rays were simulated and the speed in the reference CPU was 12175 rays per second (about 9.5 days in a single CPU).
Fig. 4.
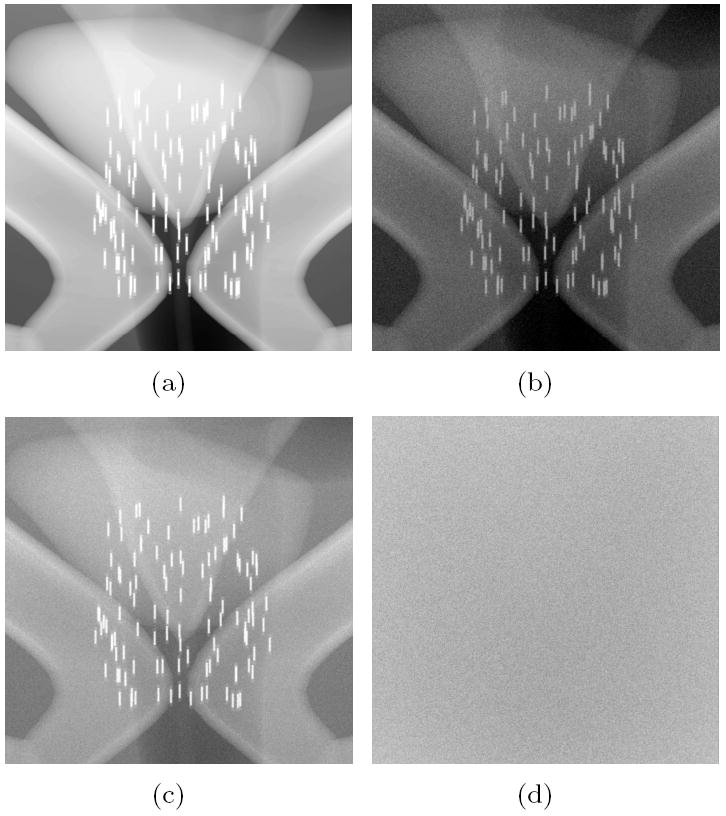
Simulated images of the prostate region for a 70 kVp source with 10×10 cm2 field, on a 10×10 cm2 detector with 200 μm pixels: (a) ray-traced image, (b) full penMesh simulation, (c) non-scattered x rays, (d) scattered x rays and secondary radiation.
Analysing the data from the previous simulation it was found that only 0.240% of the emitted x rays arrived at the detector, and 0.077% of the rays were detected after being scattered in the phantom. This gives an estimated scatter fraction of 0.32 at the detector surface.
4. Conclusions
We have described penMesh, a MC simulation code that uses the PENELOPE subroutines to transport radiation in a free-form geometry composed of triangle meshes. The performance of the new code is comparable to that of a standard PENELOPE code using a simpler geometry based on quadric surfaces. A realistic anthropomorphic phantom created with CAD tools has been adapted to penMesh. This phantom can be used to simulate medical applications that require high anatomical detail. As an example of the usage of the new tools, a simulation of an imaging procedure in a prostate brachytherapy treatment has been presented.
Acknowledgments
This project was supported in part by an appointment to the Research Participation Program at the Center for Devices and Radiological Health administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration (FDA). The authors also gratefully acknowledge partial financial support from the FDA Office of Women Health, the NIH-NIBIB, and the Spanish Ministerio de Educación y Ciencia project FIS2006-07016.
Footnotes
Submission details:
Ms. Ref. No.: RO-D-07-00520
The present revised version of the paper includes most of the corrections suggested by the journal reviewers.
The manuscript was prepared with the Elsevier LaTeX template ’elsart3p’ and includes 7 EPS figures (with JPEG compression).
This work was presented as a Proffered Paper during the 9th Biennial ESTRO Physics Meeting in Barcelona 2007.
The mention of commercial products herein is not to be construed as either an actual or implied endorsement of such products by the U.S. Department of Health and Human Services.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ash D, Flynn A, Battermann J, et al. ESTRO-EAU-EORTC recommendations on permanent seed implantation for localized prostate cancer. Radiother Oncol. 2000;57:315–321. doi: 10.1016/s0167-8140(00)00306-6. [DOI] [PubMed] [Google Scholar]
- 2.Badal A, Kyprianou I, Badano A, Sempau J, Myers KJ. Monte Carlo package for simulating radiographic images of realistic anthropomorphic phantoms described by triangle meshes. In: Hsieh J, Flynn MJ, editors. Medical Imaging 2007: Physics of Medical Imaging. 6510 of Proc SPIE. 2007. [Google Scholar]
- 3.Badal A, Kyprianou I, Badano A, Sempau J. Monte Carlo simulation with PENELOPE and a realistic anthropomorphic phantom described by triangle meshes. Radiother Oncol. 2007;84(Supplement 1):S46. [Google Scholar]
- 4.Badal A, Sempau J. A package of Linux scripts for the parallelization of Monte Carlo simulations. Comput Phys Commun. 2006;175:440–450. [Google Scholar]
- 5.Borglund N, Eriksson J, Fumero E, et al. Geometry package for Monte Carlo simulations on CAD files. Nucl Instrum Meth Phys Res A. 2004;525:417–420. [Google Scholar]
- 6.Chang A. A Survey of Geometric Data Structures for Ray Tracing, Tech Rep TR-CIS-2001-06. Polytechnic University; New York: 2001. [Google Scholar]
- 7.Cranley K, Gilmore BJ, Fogarty GWA, Desponds L. Medicine Report 78. Institute of Physics and Engineering; 1997. Catalogue of Diagnostic X-ray Spectra and Other Data. [Google Scholar]
- 8.Glassner A. Space Subdivision for Fast Ray Tracing. IEEE Computer Graphics and Applications. 1984;4:15–22. [Google Scholar]
- 9.ICRU Report 72, Dosimetry of Beta Rays and Low-Energy Photons for Brachytherapy with Sealed Sources. Journal of the ICRU. 2004;4:21–28. doi: 10.1093/jicru/ndh018. [DOI] [PubMed] [Google Scholar]
- 10.Kyprianou IS, Badal A, Badano A, et al. Monte Carlo simulated coronary angiograms of realistic anatomy and pathology models. In: Cleary KR, Miga MI, editors. Medical Imaging 2007: Visualization and Image-Guided Procedures. 6509 of Proc SPIE. 2007. [Google Scholar]
- 11.PLY format, The Stanford 3D Scanning Repository. http://www-graphics.stanford.edu/data/3Dscanrep/
- 12.Reynaert N, van der Marck SC, Schaart DR, et al. Monte Carlo treatment planning for photon and electron beams. Rad Phys Chem. 2007;76:643–686. [Google Scholar]
- 13.Salembier C, Lavagnini P, Nickers P, et al. Tumour and target volumes in permanent prostate brachytherapy: A supplement to the ESTRO-EAU-EORTC recommendations on prostate brachytherapy. Radiother Oncol. 2007;83:3–10. doi: 10.1016/j.radonc.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Salvat F, Fernández-Varea JM, Sempau J. PENELOPE–2006: A code system for Monte Carlo simulation of electron and photon transport. Nuclear Energy Agency (OECD); Issy-les-Moulineaux: 2006. available in PDF format at http://www.nea.fr. [Google Scholar]
- 15.Segars WP. Ph.D. thesis. University of North Carolina; 2001. Development of a new dynamic NURBS-based cardiac-torso (NCAT) phantom. [Google Scholar]
- 16.Sempau J. penEasy — Modular generic main program for PENELOPE. 2007 source code available at www.upc.es/inte/downloads/penEasy.htm.
- 17.Sempau J, Acosta E, Baró J, et al. An algorithm for Monte Carlo simulation of coupled electron-photon transport. Nucl Instrum Meth B. 1997;132:377–390. [Google Scholar]
- 18.Visible Human Project, National Library of Medicine. http://www.nlm.nih.gov/research/visible.
- 19.Zaidi H, Xu XG. Computational Anthropomorphic Models of the Human Anatomy: The Path to Realistic Monte Carlo Modeling in Radiological Sciences. Annu Rev Biomed Eng. 2007;9:1.1–1.30. doi: 10.1146/annurev.bioeng.9.060906.151934. [DOI] [PubMed] [Google Scholar]


