Abstract
Background
Promoter region plays an important role in determining where the transcription of a particular gene should be initiated. Computational prediction of eukaryotic Pol II promoter sequences is one of the most significant problems in sequence analysis. Existing promoter prediction methods are still far from being satisfactory.
Results
We attempt to recognize the human Pol II promoter sequences from the non-promoter sequences which are made up of exon and intron sequences. Four methods are used: two kinds of multifractal analysis performed on the numeric sequences obtained from the dinucleotide free energy, Z curve analysis and global descriptor of the promoter/non-promoter primary sequences. A total of 141 parameters are extracted from these methods and categorized into seven groups (methods). They are used to generate certain spaces and then each promoter/non-promoter sequence is represented by a point in the corresponding space. All the 120 possible combinations of the seven methods are tested. Based on Fisher's linear discriminant algorithm, with a relatively smaller number of parameters (96 and 117), we get satisfactory discriminant accuracies. Particularly, in the case of 117 parameters, the accuracies for the training and test sets reach 90.43% and 89.79%, respectively. A comparison with five other existing methods indicates that our methods have a better performance. Using the global descriptor method (36 parameters), 17 of the 18 experimentally verified promoter sequences of human chromosome 22 are correctly identified.
Conclusion
The high accuracies achieved suggest that the methods of this paper are useful for understanding the difficult problem of promoter prediction.
Background
Promoter region plays an essential role in determining where the transcription of a particular gene should be initiated. Hence, promoter recognition – the computational task of finding the promoter regions on a DNA sequence, is an important problem [1]. The accumulation of a huge amount of genome sequence data in recent years makes the annotation process more and more complicated for higher eukaryotes [2]. The RNA polymerase II (Pol II) promoter is a key region that regulates differential transcription of protein coding genes. Computational analysis of Pol II promoters may contribute to improved gene identification and to prediction of the expression context of genes [3]. There is a need for prediction techniques that can rapidly and accurately evaluate sequences for the presence of promoter sequences [1].
Existing promoter prediction methods are still far from being satisfactory [3-5]. The performance of many current eukaryote promoter prediction methods has been unreliable with poor specificity or poor sensitivity [1]. Many methods predict promoter sequences based on the regulatory sequence elements (RSEs) in them. But the RSEs are short and not fully conserved in the promoter sequences, which results in a high probability of finding similar sequence elements elsewhere in genomes, outside the promoter regions. That is why most of the promoter prediction methods end up predicting a lot of false positions [6]. Fickett and Hatzigeorgiou [3] performed an evaluation of the different promoter prediction methods on genome DNA and suggested that it would be worth attempting nonlinear recognition methods, such as neural nets or quadratic discriminant analysis. Following this direction, Gangal and Sharma [7] applied time series descriptors and machine learning methods to human Pol II promoter prediction and got a higher accuracy compared with other methods; Kanhere and Bansal [6] presented a novel prokaryotic promoter prediction method based on DNA stability showing that the changing in the stability of DNA provides a much better clue than the usual sequence motifs.
In this paper, we attempt to recognize the human Pol II promoter sequences from the non-promoter sequences which contain exon and intron sequences. It should be noted that the aim of the present paper is similar to that of Ref. [7], but the non-promoter sequences in Ref. [7] are made up of coding sequences (CDSs) and intron sequences, while we use an existing database, the Exon/Intron database, to extract non-promoter sequences. We first convert the promoter/non-promoter sequences into numeric sequences according to the 10 unified free energy parameters [8], which have been used to measure the stability of DNA [6]. Then a measure representation is introduced for the numeric sequences. Multifractal analysis of the measure is next performed, which results in the first 5 parameters. Analogous multifractal analysis [9] is also used on the numeric sequences to achieve another 4 parameters. The Z curve method, which has been used in recent years with some successes [10,11], yields 96 parameters for the promoter/non-promoter primary sequences. The protein-chain descriptor method was first proposed by Dubchak et al. [12] to predict protein folding classes. Here we propose a global descriptor for the promoter/non-promoter sequences, which yields 36 parameters for a global description of the primary sequences. Overall, a total of 141 parameters are extracted from these four different methods and categorized into seven groups (methods). Fisher's linear discriminant algorithm shows that the global descriptor method is the most effective when used separately. Complete enumerations of all the possible combinations of these seven methods (120) are tested to find possibly better results with a relatively smaller number of parameters. Numerical results show that the methods with 96 and 117 parameters can produce satisfactory results. Compared with five other existing tools, the higher sensitivity, specificity, accuracy and correlation coefficient demonstrate that the methods proposed here are useful for understanding the human Pol II promoter prediction problem. 17 of the 18 experimentally verified promoter sequences of human chromosome 22 [13] are successfully identified by the global descriptor method (with only 36 parameters).
Results
Testing
We use two different data sets downloaded from two databases. The first set is the human Pol II promoter sequences from Release 90 of the Eukaryotic Promoter Database (EPD) [14]. The EPD is an annotated non-redundant collection of eukaryotic Pol II promoters, experimentally defined by a transcription start site (TSS) [15]. The EPD is a useful database when one wants to deal with the Pol II promoter prediction problem and it is broadly tested by different prediction tools [7,16-19]. A total of 1871 entries of human Pol II promoter sequences with window size of 499 bp upstream and 100 bp downstream of TSS, which is the same as that used in Ref. [16], are obtained from EPD. The sequences containing 'N' are manually filtered out, which results in a total of 1856 sequences. The second set is the non-promoter sequences of the human genome. For this data set, we consider using the Exon/Intron Database (EID), which incorporates information on the exon/intron structure of eukaryotic genes [20] ([21], hs35p1.EID.tar.gz). Firstly, the exon/intron sequences with 'n' and length less than 600 are filtered out. Then, we randomly select 1000 intron sequences from the file hs35p1.intrEID and 500 exon sequences from the file hs35p1.exEID. A fragment of length 600 is then selected randomly from each exon/intron sequence with length larger than 600. As the intron sequences are represented by lower-case letters in the file hs35p1.intrEID, we transform them into upper-case letters to be consistent with the promoter and exon sequences.
From the four different methods described in the Methods section, we get a total of 141 parameters. We will test their contributions in the promoter/non-promoter problem. Then we will try to combine some of them to see whether better results can be achieved.
For comparison of various methods, a benchmark should be set up. We use Fisher's linear discriminant algorithm [22-24] to calculate the discriminant accuracies. We divide all promoter and non-promoter sequences into two sets randomly. A set of 90% of promoter/non-promoter sequences is regarded as a training set, and the set of remaining 10% of promoter/non-promoter sequences as a test set.
Fisher's discriminant algorithm is used to find a classifier in the parameter space for a training set. The given training set H = {x1, x2, ⋯, xn} is partitioned into n1 ≤ n training vectors in a subset H1 and n2 ≤ n training vectors in a subset H2, where n1 + n2 = n and each xi is a κ-dimensional vector, represented by one point in the κ-dimensional parameter space. Then H = H1 ∪ H2. We need to find a parameter vector w = (w1, w2, ⋯, wκ)T for the κ-dimensional space such that can be classified into two classes in the space of real numbers. If we denote
| (1) |
| (2) |
| Sw = S1 + S2, | (3) |
then the parameter vector w is estimated as [23]. As a result, Fisher's discriminant rule becomes: "assign x to H1 if and to H2 otherwise" [22].
The discriminant accuracies for resubstitution analysis are defined as
| (4) |
| (5) |
For the test analysis, the discriminant accuracies qc and qnc are defined similarly by changing "training set" to "test set" in Eqs. (4) and (5), respectively.
We first divide the data into training and test sets randomly, then we use the above algorithm to calculate the discriminant accuracies for different methods. The results are listed in Table 1.
Table 1.
The discriminant accuracies for various methods with Fisher's discriminant. The method marked "3+6+7" in the 8th row means the combination of the methods listed in the 3rd, 6th and 7th rows. The meanings of the methods marked for the 9th row is similar.
| Order | pc(%) | pnc(%) | qc(%) | qnc(%) | Method | No. of parameters |
| 1 | 73.05 | 85.63 | 74.73 | 83.33 | MFA+AMFA | 9 |
| 2 | 79.16 | 75.78 | 76.88 | 62.67 | ZC Eq.(19) | 9 |
| 3 | 78.86 | 88.00 | 79.03 | 85.33 | ZC Eq.(21), k = 12 | 12 |
| 4 | 78.62 | 89.33 | 79.57 | 89.33 | ZC Eq.(21), k = 23 | 12 |
| 5 | 80.30 | 90.74 | 80.65 | 90.00 | ZC Eqs.(20, 22) | 15 |
| 6 | 85.75 | 88.30 | 86.02 | 91.33 | GD | 36 |
| 7 | 81.92 | 91.48 | 81.72 | 89.33 | ZC Eq.(23) | 48 |
| 8 | 86.11 | 93.48 | 86.02 | 90.67 | 3+6+7 | 96 |
| 9 | 86.89 | 93.11 | 86.02 | 92.67 | 1+3+4+6+7 | 117 |
| 10 | 87.31 | 93.19 | 86.02 | 92.00 | All methods | 141 |
Firstly, seven groups of parameters are derived from the four methods: (i) 9 parameters from fractal methods (MFA and AMFA); (ii) 9 parameters from ZC representing the codon-position-dependent frequencies of mononucleotides; (iii) 12 parameters from ZC representing the frequencies of phase-specific dinucleotides (codon positions 1–2); (iv) 12 parameters from ZC representing the frequencies of phase-specific dinucleotides (codon positions 2–3); (v) 15 parameters for the phase-independent mononucleotides and dinucleotides from ZC; (vi) 36 parameters from GD; (vii) 48 parameters for the frequencies of phase-independent tri-nucleotides from ZC. From Table 1, it is seen that the results from the multifractal analyses seem to be better than that from ZC with an equal number of parameters, namely 9. We have successfully applied multifractal analyses in the clustering of large protein structures [9,25] and the distinction of coding and non-coding sequences in complete genomes [26], where the length of protein sequences and coding and non-coding sequences are larger than 300. It is well-known that the promoter sequences are highly diverse, which makes it notoriously difficult to generate patterns and rules for promoter prediction. It is expected that multifractal analyses can unfold some useful information on promoter sequences. The results from the frequencies of phase-specific dinucleotides at codon positions 2–3 in ZC indicate a better performance than that at codon positions 1–2. In addition, the accuracies from ZC with the frequencies of phase-independent mononucleotides and dinucleotides are improved but the number of parameters is increased to 15. The GD method shown in boldface in Table 1, denoted as M1, turns out to be especially useful as the accuracies are all larger that 85%. Compared with this, the results from the 48 parameters in ZC are not as good even though the number of parameters is increased.
Secondly, we want to test whether the results can be improved by increasing the number of parameters. It is not possible to test all the subsets of the 141 parameters but we can test all the combinations of the above seven methods (120 altogether). In our test, the accuracies do not simply increase as the number of parameters becomes larger, which indicates there might be some redundancy/correlation among the 141 parameters. For example, the accuracies with the 141 parameters are similar to those with only 117 parameters, suggesting the information from the mononucleotides and phase-independent dinucleotides in ZC is contained in the other methods. Therefore, all these parameters are not really needed. Nevertheless, in some circumstances the results do improve when the number of parameters is increased. Especially, among the 120 combinations, the results are relatively satisfactory in the cases of 96 and 117 parameters, which is shown in boldface in Table 1. We denote them by M2 and M3, respectively. In order to see whether multifractal analysis brings out useful information, we remove the 9 parameters of MFA and AMFA from M3 and test the results for such new combination. The pc, pnc, qc, and qnc calculated from this combination are: 86.05% 92.67%, 86.02% and 92.00% respectively. They are similar to those from M3 (86.89%, 93.11%, 86.02% and 92.67%), which demonstrates that multifractal analysis does not significantly improve the performance in M3.
In order to evaluate the correct prediction rate and reliability of a predictive method, the sensitivity (Sn), specificity (Sp), accuracy (Ac) and correlation coefficient (CC) are also used [1]:
| Sn = T P/(T P + F N), | (6) |
| Sp = T P/(T P + F P), | (7) |
| Ac = (Sn+ Sp)/2, | (8) |
| (9) |
where TP denotes the number of correctly recognized promoter sequences, FN the number of promoter sequences recognized as non-promoter sequences, FP the number of non-promoter sequences recognized as promoter sequences, TN the number of correctly recognized non-promoter sequences.
From Fisher's discriminant algorithm, we calculate the four quantities defined above. The results related to Table 1 by the "order" mark are listed in Table 2.
Table 2.
The accuracies of the prediction for promoter sequences by Fisher's discriminant algorithm. The Sn, Sp, Ac and CC are the results for the training set and S'n, S'p, A'c and CC' are the results for the test set. The rows are related to those in Table 1 according to the mark order.
| Order | Sn(%) | Sp(%) | Ac(%) | CC | (%) | (%) | (%) | CC' |
| 1 | 73.05 | 86.28 | 79.67 | 0.58 | 74.73 | 84.76 | 79.74 | 0.58 |
| 2 | 79.16 | 80.17 | 79.67 | 0.55 | 76.88 | 71.86 | 74.37 | 0.40 |
| 3 | 78.86 | 89.05 | 83.95 | 0.66 | 79.03 | 86.98 | 83.01 | 0.64 |
| 4 | 78.62 | 90.12 | 84.37 | 0.68 | 79.57 | 90.24 | 84.91 | 0.69 |
| 5 | 80.30 | 91.47 | 85.89 | 0.71 | 80.65 | 90.91 | 85.78 | 0.70 |
| 6 | 85.75 | 90.06 | 87.91 | 0.74 | 86.02 | 92.49 | 89.25 | 0.77 |
| 7 | 81.92 | 92.25 | 87.08 | 0.73 | 81.72 | 90.48 | 86.10 | 0.71 |
| 8 | 86.11 | 94.23 | 90.17 | 0.79 | 86.02 | 91.95 | 88.99 | 0.76 |
| 9 | 86.89 | 93.98 | 90.43 | 0.80 | 86.02 | 93.57 | 89.79 | 0.78 |
| 10 | 87.31 | 94.06 | 90.68 | 0.80 | 86.02 | 93.02 | 89.52 | 0.78 |
Overall, from Tables 1 and 2, when the methods are used independently, we can see that M1 is the best one. The combined methods M2 and M3 improve the results. However, the number of parameters is too high in M3. Taking this aspect into account, a preferred method would be M1 or M2.
Discussion
It is natural to ask whether the method of this paper has a better performance than the existing methods. As was done in Ref. [7], we can compare the present method with five kinds of promoter prediction tools, which are available on-line, namely Neural Network Promoter Prediction (NNPP version 2.2) [27], Soft Berry (TSSW) [28], Dragon Promoter Finder version 1.5 (DFP) [17,29], Promoter 2.0 [18,30] and Promoter Scan version 1.7 [19,31]. To be within a reasonable workload, we only compare with 10% of the promoter and non-promoter sequences used in Section 4 (186 promoter and 150 non-promoter sequences). The results are listed in Table 3. They clearly indicate that our method has a better performance than the other tools.
Table 3.
The promoter prediction accuracies for the test data set made up of 186 promoter sequences and 150 non-promoter sequences using five kinds of tools and our methods.
| Tool | Sn(%) | Sp(%) | Ac(%) | CC |
| NNPP(threshold 0.8) | 69.89 | 60.75 | 65.32 | 0.14 |
| Soft Berry(TSSW) | 67.74 | 81.29 | 74.52 | 0.48 |
| Promoter Scan version 1.7 | 67.20 | 88.65 | 77.93 | 0.57 |
| Dragon Promoter Finder version 1.5 | 30.65 | 65.52 | 48.08 | 0.12 |
| Promoter 2.0 Prediction Server | 52.15 | 91.51 | 71.83 | 0.49 |
| Our method (M3) | 86.02 | 93.57 | 89.79 | 0.78 |
However, using 90% of promoter sequences as a training set and only 10% of the promoter sequences as a test set may not provide a fair comparison against these methods. A more realistic performance would be to use 50% of the promoter sequences as a training set and the other 50% as a test set. Therefore, we use such ratio of training and test sets in Fisher's algorithm to see whether the results from our method are still satisfactory. We list the results of M1, M2 and M3 in Table 4. It shows that, with a smaller size of training set, the accuracy Ac for the test set is surprisingly better than before, suggesting that our method is robust.
Table 4.
The accuracies for M1, M2 and M3 with 50% sequences as training and the remaining 50% as test set in Fisher's discriminant algorithm.
| Order | Sn(%) | Sp(%) | Ac(%) | CC | (%) | (%) | (%) | CC' |
| M1 | 81.67 | 89.53 | 87.60 | 0.73 | 91.49 | 85.50 | 88.49 | 0.73 |
| M2 | 87.28 | 93.32 | 90.30 | 0.79 | 90.41 | 89.07 | 89.74 | 0.77 |
| M3 | 88.25 | 93.17 | 90.71 | 0.80 | 90.52 | 89.74 | 90.13 | 0.78 |
Based on support vector machine (SVM), Gangal and Sharma [7] used time series descriptors to identify promoter sequences from non-promoter sequences. They reported an accuracy of more than 85%. It will be interesting to see whether their method also works well in our test data set. But their tool Prometheus is not currently available. So it is not feasible to compare the two methods using the same data set. Nevertheless, by using 80% of data to train and the other 20% to test our method, which is the ratio used by Gangal and Sharma [7], we are able to produce a rough comparison with the results Gangal and Sharma reported (Sn = 86% and Sp = 88%). It is listed in Table 5, which shows that our results (Sn = 87.10% and Sp = 91.78%) are relatively better.
Table 5.
The accuracies for M1, M2 and M3 with 80% sequences as training and the remaining 20% as test set in Fisher's discriminant algorithm.
| Order | Sn(%) | Sp(%) | Ac(%) | CC | (%) | (%) | (%) | CC' |
| M1 | 85.78 | 89.65 | 87.71 | 0.73 | 87.10 | 88.28 | 87.69 | 0.73 |
| M2 | 86.39 | 93.71 | 90.05 | 0.79 | 87.90 | 91.09 | 89.49 | 0.77 |
| M3 | 86.86 | 93.88 | 90.37 | 0.79 | 87.10 | 91.78 | 89.44 | 0.77 |
Finally, it is important to test our method with real human DNA sequences. For example, a sliding window technique with window size of 600 bp and step size of 10 bp can be used to recognize promoter sequences in the human DNA sequences, similar to the technique adopted by Gao and Zhang [32] to recognize exons. However, because promoter sequences are not clearly marked in the human DNA sequences, we can't use this approach to test our method. Nevertheless, similar to that performed in Ref. [7,33], we use the human chromosome 22, in which 20 promoters are experimentally verified [13]. One can refer to Table 1 in Ref. [13] to get the sequences with the accession numbers. However, as AB016655 and D86746 are not clearly annotated, we do not use them in the test. We use 50% of the promoter (from EPD) and non-promoter (from EID) sequences to train M1. The coefficients in Fisher's algorithm w = (w1, w2, ⋯, w36) are determined based on the training set. The choice of a promoter/non-promoter sequence is determined by the criterion Z(x) > 0/Z(x) <0. Except for AF047576, the other 17 promoter sequences are correctly identified. This suggests that the global descriptor GD (M1), with a smaller number of parameters (36), is a practical method.
Conclusion
Promoter prediction is a difficult but important problem in gene finding, and it is critical for elucidating the regulation of gene expression [34]. We use two kinds of multifractal analysis on the free energy sequences of promoter/non-promoter, Z curve analysis, and the global descriptor for the primary sequences of promoter/non-promoter. A total of 141 parameters are extracted from these four methods. These parameters are used in both independent and combined ways to distinguish promoter sequences from non-promoter sequences.
Fisher's linear discriminant algorithm provides a quantitative assessment of the recognition methods. If we use these methods independently, the global descriptor of the promoter/non-promoter sequences is the best method based Fisher's algorithm. Combinations of various methods show that the accuracies can be improved in some cases but the improvements are not simply due to the increase of parameter numbers. With all 141 parameters together, the results are satisfactory. However, the number of parameters is too high in this condition. The number is reduced as there is some redundancy/correlation among these parameters. In the case of 117 parameters, similar results are achieved, with the discriminant accuracies pc, pnc, qc, and qnc reaching 86.89% 93.11%, 86.02% and 92.67%, respectively. The related sensitivity Sn, specificity Sp, accuracy Ac and correlation coefficient CC for the test set reach 86.02%, 93.57%, 89.79% and 0.78, respectively. A smaller number of parameters (96) also produces relatively satisfactory results. The global descriptor method with only 36 parameters successfully identifies 17 of the 18 experimentally verified promoters in human chromosome 22 [13]. Recognition of promoter sequences with such satisfactory accuracy indicates that the methods is promising for human Pol II promoter prediction.
The main aim of this work is to develop efficient algorithms that can discriminate between promoters and non-promoters in a given sequence. Another challenge being addressed is the localization of promoters rather than a simple classification considered in current methods [7]. Multifractal analysis, which is especially useful in many other fields [25,26,35,36], seems to reflect some information for promoter recognition (see first line in Table 1). But if we use method M3, multifractal analysis does not significantly improve the performance. The methods considered in this paper seem promising in enhancing the performance of biomolecular sequence analysis and promoter prediction in particular. It is a challenge to predict promoter sequences directly from the real human genome. However, it would be helpful to use first the ENCODE pilot project data set, which spans about 1% of the human genome sequence [37]. Our following work aims to contribute towards this challenging problem.
Methods
Conversion of the original data
Some studies suggested that various properties, such as stability, bendability and curvature, of the region immediately upstream of the TSS differ from that of downstream region [6,38,39]. The upstream region is less stable, more rigid and more curved than the downstream region. Kan-here and Bansal [6] predicted the prokaryotic promoter based on such difference in DNA stability. We convert the original sequences into new numeric sequences according to the free energy of dinucleotides. A sliding window with size of 2nt is used and moved one base pair forward each time. The numeric sequences can be smoothed with a larger window size. For more details on the smoothing method, one can refer to Ref. [40]. The free energy values corresponding to the 10 unique dinucleotides are taken from the unified parameters proposed in Ref. [8]. They are: AA/TT = -1.00 kcal/mol, AT/TA = -0.88 kcal/mol, TA/AT = -0.58 kcal/mol, CA/GT = -1.45 kcal/mol, CT/GA = -1.44 kcal/mol, GT/CA = -1.28 kcal/mol, GA/CT = -1.30 kcal/mol, CG/GC = -2.17 kcal/mol, GC/CG = -2.24 kcal/mol, GG/CC = -1.84 kcal/mol. The ten values are added by 2.24 kcal/mol (the negative of the smallest free energy) so that all the values are larger than or equal to zero in order to construct a measure from the time series for the multifractal method in the following analysis. For example, the free energy sequence for one of the promoter sequences with a sliding window of size 2nt is given in Figure 1.
Figure 1.
The free energy sequence of one promoter sequence. See text for a detailed description about how to get such numeric sequence.
Multifractal analysis (MFA)
Let Tt, t = 1, 2, ⋯, N, be the numeric sequence of a promoter/non-promoter with length N . First, we define
| (10) |
to be the frequency of Tt. It follows that . We define a measure μ on the interval [0, 1) by
| μ(dx) = Y(x) dx, | (11) |
where
| (12) |
We denote the interval by It. It is easy to see that μ([0, 1)) = 1 and μ(It) = Ft. We call μ(x) the measure representation [26,41] for the numeric sequence of a promoter/non-promoter.
The most common algorithms of multifractal analysis are the so called fixed-size box-counting algorithms [42]. In the one-dimensional case, for a given measure μ with support E ⊂ ℝ, we consider the partition sum
| (13) |
where the sum runs over all different nonempty boxes B of a given side ε in a grid covering of the support E, that is,
| B = [kε, (k + 1)ε). | (14) |
The mass exponent τ (q) is defined [43,44] as
| (15) |
and the generalized fractal dimensions [43,44] of the measure are defined as
| (16) |
and
| (17) |
where . The generalized fractal dimensions are numerically estimated through a linear regression of ln Zε (q)/(q - 1) against ln ε for q ≠ 1, and similarly through a linear regression of Z1, ε against ln ε for q = 1 [25,42,45]. D(1) is called the information dimension and D(2) the correlation dimension [43,44].
The concept of phase transitions in multifractal spectra was introduced in the study of logistic maps, Julia sets, and other simple systems. Evidence of a phase transition was found in the multifractal spectrum of diffusion-limited aggregation [46]. By following the thermodynamic formulation of multifractal measures, Canessa [47] derived an expression for the analogous specific heat as
| (18) |
He showed that the form of Cq resembles a classical phase transition at a critical point for financial time series.
The singularities of a measure are characterized by the Lipschitz-Hölder exponent α(q) [44], which is related to τ (q) by
| (19) |
Substitution of Eq. (15) into Eq. (19) yields
| (20) |
Again, the exponent α(q) can be estimated through a linear regression of against ln ε. The multifractal spectrum f (α) versus α can be calculated according to a relationship known as Legendre transformation [44]:
| (21) |
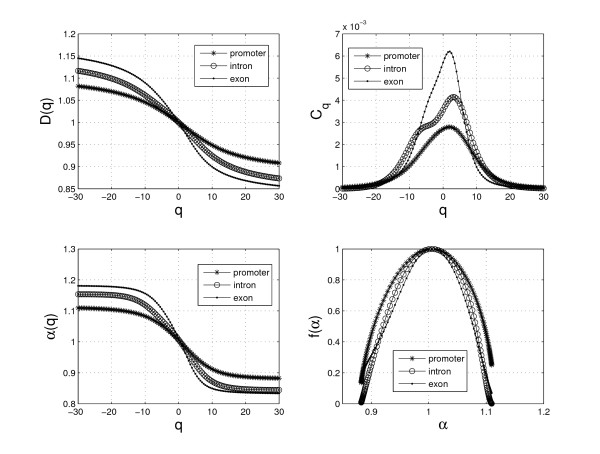
We first construct a measure for the numeric sequences according to Eq. (11), then analyze the measure with the above multifractal method. The D(q), Cq, α(q) and f (α) curves for one of the promoter, exon and intron sequences are shown in Figure 2. We select 5 parameters from MFA to distinguish between promoter and non-promoter sequences: D(2), C1, Cmax (the maximum value of Cq), Δα = αmax - αmin and Δf = f (αmax) - f (αmin).
Figure 2.
The four kinds of fractal curves for the promoter, exon and intron sequences. The figures show that there are some differences between the promoter and non-promoter (exon/intron) sequences, which suggests that it's possible to extract some values from them to distinguish the promoter sequences from the non-promoter sequences.
Analogous multifractal analysis (AMFA)
Analogous multifractal analysis is similar to multiaffinity analysis which is a useful method in many fields. It was recently proposed in [9]. We denote a time series as X(t), t = 1, 2, ⋯, N. First, the time series is integrated as
| (22) |
| (23) |
where Xave is the average over the whole time period and k ∈ [1, N]. Then two quantities Mq (L) and are defined as
| (24) |
| (25) |
where 〈〉j denotes the average over j, j = 1, 2, ⋯, N – L; L typically varies from 1 to N1 in which the linear fit is good. From the ln L vs ln Mq (L) and ln L vs ln planes, one can determine the relations:
| (26) |
| (27) |
Linear regressions of ln and ln Mq (L) against ln L will yield the exponents h' (q) and h(q) respectively.
The exponent h(q) has a nonlinear dependence on q. When q = 1, the methods are just those reported in Refs. [48,49] and these methods are used to study the length sequences from the complete genomes by Yu et al. [49]. M'(L) may be assessed to determine long-range correlation [50]. From Ref. [49], the linear fit to get the exponent h(1) is better than that to get the exponent h'(1). Our numerical results show that the exponents h(q) are more robust than the exponents h'(q), so we suggest to use the exponents h(q). We have used h(q) in clustering the structure of large proteins and it turns out to be a useful method [9].
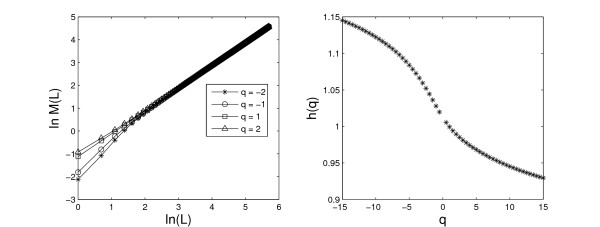
Figure 3 gives an example in applying the AMFA to the free energy sequence of a promoter sequence. It shows a good linear relationship between ln M(L) and ln(L). For different values of q, we get the exponents h(q) from linear regressions of ln M(L) against ln (L) according to Eq. (27). The exponent spectrum h(q) of the promoter sequence is shown in the right panel of Figure 3. We extract four parameters from AMFA: h(-2), h(-1), h(1) and h(2).
Figure 3.
The relationship between ln M(L) and ln(L) using the free energy sequence of one promoter (Left); the h(q) spectra for the one promoter calculated by AMFA (Right).
Z curve (ZC)
The concept of the Z curve representation of a DNA sequence was first proposed by Zhang and Zhang [51], and was used to distinguish coding and noncoding DNA sequences [52,53]. A new system based on ZC, Z CURVE 1.0, for finding protein-coding genes in bacterial and archaeal genomes has been proposed [10]. Recently, another new self-training system based on the ZC method, ZCURVE_V [11], for recognizing protein-coding genes in viral and phage genomes was reported.
In this paper, we apply the ZC method in distinguishing promoter and non-promoter sequences. For convenience, we give a brief description of the methods in Refs. [10] and [11]. The frequencies of bases A, C, G and T occurring in a promoter/non-promoter sequence with bases at positions 1, 4, 7, ⋯; 2, 5, 8, ⋯; 3, 6, 9, ⋯, are denoted by a1, c1, g1, t1; a2, c2, g2, t2; a3, c3, g3, t3, respectively. They are in fact the frequencies of bases at the first, second and third codon positions, which can be called codon-position-dependent frequencies of mononucleotides. Based on the ZC [54], ai, ci, gi, ti for each i can be used to construct three coordinates, denoted by xi, yi and zi according to the Z transform [54]:
| (28) |
where xi, yi, zi ∈ [-1, 1], i = 1, 2, 3.
We can use the above 9 parameters in the promoter/non-promoter problem. We can also consider the codon-position-independent frequencies of single bases, which results in the following three coordinates:
| (29) |
where x, y, z ∈ [-1, 1], a, c, g and t are the frequencies for the bases A, C, G and T in a promoter/non-promoter sequence, respectively.
In addition to the frequencies of codon-position-dependent mononucleotide, we also consider the frequencies of phase-specific dinucleotides. We denote the frequencies of the 16 dinucleotides AA, AC, ⋯, and TT occurring at the codon positions 1–2 and 2–3 of a promoter or non-promoter sequence by p12(AA), p12(AC), ⋯, p12(T T); p23(AA), p23(AC), ⋯, and p23(T T), respectively. Using the Z transform [54], the following 24 coordinates can be defined:
| (30) |
where pk (XY) = nk(XY)/[nk (XA) + nk (XC) + nk(XG) + nk (XT)], nk(XY) are the occurring times of dinucleotides XY, X, Y = A, C, G, T, k = 12, 23.
We can also consider the frequencies of phase-specific dinucleotides and the frequencies of phase-independent dinucleotides. For this purpose, a sliding window with size 2nt is used and moved forward one base each time to count the number of times of the occurring dinucleotides. With this method, 12 new coordinates can be defined:
| (31) |
where xX, yX, zX ∈ [-1, 1], p(XY) = n(XY)/[n(XA) + n(XC) + n(XG) + n(XT)], n(XY) are the occurring times of dinucleotides XY, X, Y = A, C, G, T.
Gao and Zhang [32] compared various algorithms for recognizing short coding sequences of human genes and they defined 48 quantities, which were the frequencies of phase-dependent tri-nucleotides. In Ref. [32], Gao and Zhang used a sliding window with size 3nt and the window was moved forward three bases each time to count the frequencies for the 64 tri-nucleotides. Now we move forward the sliding window with size 3nt one base each time. The definition for the 48 coordinates is
| (32) |
where xXY, yXY, zXY ∈ [-1, 1], p(XYZ) = n(XYZ)/[n(XYA) + n(XYC) + n(XYG) + n(XYT)], n(XY Z) are the occurring times of trinucleotides XYZ, X, Y, Z = A, C, G, T. The difference between Ref. [32] and here is in the calculation of n(XYZ); the present method can be regarded as a phase-independent method.
Global descriptor of promoter/nonpromoter sequence (GD)
Dubchak et al. [12] proposed a method for predicting protein folding classes based on a global protein chain description. The protein-chain descriptor includes overall composition, transition, and distribution of amino acid attributes. Similar methods have also been used in Refs. [55-58]. In this paper, we propose the global descriptor of promoter/non-promoter sequences.
The global description contains three parts: composition (Comp), transition (Tran) and distribution (Dist). In order to explain the method, we suppose that a sequence consists of only two kinds of letters (A and B). The composition is used to measure the frequency of occurrence of each kind of letters in the sequences. For example, for the sequence: BABBABABBABBAABABABBAAAB-BABABA, there are 14 As and 16 Bs, hence the frequencies for A and B are 100.00 × 14/(14+16) = 46.67, 100.00 × 16/(14+16) = 53.33, respectively. These two numbers represent the first part of the global description, Comp. The second part, Tran, characterizes the percent frequency with which A is followed by B or B is followed by A. For example, for the above sequence, there are 21 transitions of this type, that is, (21/29) × 100.00 = 72.14. The third part of the global description, Dist, measures the chain length within which the first, 25%, 50%, 75% and 100% of certain type of letters is located, respectively. For example, for the above sequence, the first, 25%, 50%, 75% and 100% of Bs are located within the first, 6th, 12th, 20th and 29th nucleotides, respectively. The Dist descriptor for Bs is thus: 1/30 × 100.00 = 3.33, 6/30 × 100.00 = 20.00, 12/30 × 100.00 = 40.00, 20/30 × 100.00 = 66.67 and 29/30 × 100.00 = 96.67. Likewise, the Dist descriptor for As is 6.67, 23.33, 53.33, 73.33 and 100.00. As a result, the global description for the above sequence is (Comp; Tran; Dist) = (46.67, 53.33; 72.14; 6.67, 23.33, 53.33, 73.33, 100.00, 3.33, 20.00, 40.00, 66.67, 96.67). A more detailed description of global description of sequences is given in Refs. [12,55-58].
The global description for the promoter/non-promoter sequences can be computed by a similar procedure. As the sequences consist of four types of nucleotides (A, C, G and T), there are 4 parameters for Comp, 6 parameters for Tran and 20 parameters for Dist. Overall, a total of 30 parameters are used to give a global description of a promoter/non-promoter sequence.
The Entropy Density Profile (EDP) model is a global statistical description for a DNA sequence, which employs Shannon's artificial linguistic description for a DNA sequence of finite length like an open reading frame (ORF) [59]. Zhu et al. [59] developed a new non-supervised gene prediction algorithm for bacterial and archaeal genomes based on EDP. Here we describe such method briefly. If pi(i = 1, 2, 3, 4) are the frequencies for the four types of nucleotides of a promoter/non-promoter sequence, then an EDP vector S = {si} inferred from {pi} is used to represent the sequence with an emphasis on the information content, where i is the index of the four kinds of nucleotides. The EDP si is defined as [59]
| (33) |
where is the Shannon entropy.
It was shown that is a useful statistical quantity for analysis of DNA sequences [54,60], which was called a nucleotide composition constraint of genomes [61]. As a result, we obtain 6 parameters s1, s2, s3, s4, H and P from EDP.
Overall, combining the above two description systems, we get 36 parameters for the global descriptor of a promoter/non-promoter sequence.
Authors' contributions
JYY conceived of the study, downloaded the data, analyzed the results, has been involved in programming, drafting and revising the manuscript. YZ and LQZ have been involved in the programming and discussion on the results. ZGY coordinated the study and participated in its design, analyzed the results, has been involved in drafting and revising the manuscript. VA participated in the design of the study and the results discussion, has been involved in drafting and revising the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors would like to thank Dr. Feng-Biao Guo of Tianjin University for his helpful discussions about the Z curve method and Dr. Jun Lu in Inner Mongolia University for his help with the promoter data on human chromosome 22, and the referees for their detailed comments and useful suggestions to improve the paper. Financial support was provided by the Chinese National Natural Science Foundation (grant no. 30570426), Fok Ying Tung Education Foundation (grant no. 101004), the Youth Foundation of Educational Department of Hunan Province in China (grant no. 05B007) (Z.-G. Yu), the Australian Research Council (grant no. DP0559807) (V.V. Anh), and the Scientific Research Fund of the Department of Education in Hunan Province of China (no. 06C830) (L.Q. Zhou).
Contributor Information
Jian-Yi Yang, Email: yangjianyiapple@163.com.
Yu Zhou, Email: zynova@hotmail.com.
Zu-Guo Yu, Email: yuzg1970@yahoo.com.
Vo Anh, Email: v.anh@qut.edu.au.
Li-Qian Zhou, Email: zhoulq@xtu.edu.cn.
References
- Li QZ, Lin H. The recognition and prediction of σ70 promoters in Escherichia coli K-12. J Theor Biol. 2006;242:135–141. doi: 10.1016/j.jtbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Ohler U. Promoter Prediction on a Genomic Scale-The Adh Experience. Genome Res. 2000;10:539–542. doi: 10.1101/gr.10.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett J, Hatzigeorgiou A. Eukaryotic Promoter Recognition. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- Werner T. The state of the art of mammalian promoter recognition. Breif Bioinform. 2003;4:22–30. doi: 10.1093/bib/4.1.22. [DOI] [PubMed] [Google Scholar]
- Pedersen AG, Baldi P, Chauvin Y, Brunak S. The biology of eukaryotic promoter prediction-a review. Comput Chem. 1999;23:191–207. doi: 10.1016/S0097-8485(99)00015-7. [DOI] [PubMed] [Google Scholar]
- Kanhere A, Bansal M. A novel method for prokaryotic promoter prediction based on DNA stability. BMC Bioinformatics. 2005;6:1–10. doi: 10.1186/1471-2105-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangal R, Sharma P. Human pol II promoter prediction: time series descriptors and machine learning. Nucleic Acids Res. 2005;33:1332–1336. doi: 10.1093/nar/gki271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santalucta JR. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Yu ZG, Anh V. Clustering structures of large proteins using multifractal analyses based on a 6-letter model and hydrophobicity scale of amino acids. Chaos, Solitons and Fractals. 2007 doi: 10.1016/j.chaos.2007.08.014. [DOI] [Google Scholar]
- Guo FB, Ou HY, Zhang CT. ZCURVE: a new system for recognizing proteincoding genes in bacterial and archaeal genome. Nucleic Acids Res. 2003;31:1780–1789. doi: 10.1093/nar/gkg254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FB, Zhang CT. ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. BMC Bioinformatics. 2006;7:1–11. doi: 10.1186/1471-2105-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I, Muchanikt I, Holbrook SR, Kim SH. Prediction of protein folding class using global description of amino acid sequence. Proc Natl Acad Sci. 1995;92:8700–8704. doi: 10.1073/pnas.92.19.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf M, Klingenho3 A, Frech K, Quandt K, Schneider R, Grote K, Frisch M, Gailus-Durner V, Seidel A, Brack-Werner R, Werner T. First pass annotation of promoters of human chromosome 22. Genome Res. 2001;11:333–340. doi: 10.1101/gr.154601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Website EPD http://www.epd.isb-sib.ch
- Perier R, Junier T, Bucher P. The Eukaryotic Promoter Database EPD. Nucleic Acids Res. 1998;26:353–357. doi: 10.1093/nar/26.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang V, Saeys Y, Sung WK, Mittal A. Computational modeling of oligonucleotide positional densities for human promoter prediction. Artif Intell Med. 2005;35:107–119. doi: 10.1016/j.artmed.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Bajic VB, Seah SH, Chong A, Zhang G, Koh JLY, Brusic CV. Dragon Promoter Finder: recognition of vertebrate RNA polymerase II promoters. Bioinformatics. 2002;18:198–199. doi: 10.1093/bioinformatics/18.1.198. [DOI] [PubMed] [Google Scholar]
- Knudsen S. Promoter 2.0: for the recognition of Pol II promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- Prestridge Dan S. Predicting Pol II Promoter Sequences using Transcription Factor Binding Sites. J Mol Biol. 1995;249:923–932. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Daizadeh I, Fedorov A, Gilbert W. Computational modeling of oligonucleotide positional densities for human promoter prediction. Nucleic Acids Res. 2000;28:185–190. doi: 10.1093/nar/28.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Website EID http://hsc.utoledo.edu/bioinfo/eid/index.html
- Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. Academic Press, London; 1979. [Google Scholar]
- Duda RO, Hart PE, Stork DG. Pattern Classification. 2. John Wiley & Sons, New York; 2001. [Google Scholar]
- Sneath PH, Sokal RR. Numerical Taxonomy. Freeman, San Francisco; 1973. [Google Scholar]
- Yu ZG, Anh V, Lau KS, Zhou LQ. Clustering of protein structures using hydrophobic free energy and solvent accessibility of proteins. Phys Rev E. 2006;73:031920. doi: 10.1103/PhysRevE.73.031920. Epub 2006 Mar 21. [DOI] [PubMed] [Google Scholar]
- Zhou LQ, Yu ZG, Deng JQ, Anh V, Long SC. A fractal method to distinguish coding and non-coding sequences in a complete genome based on a number sequence representation. J Theor Biol. 2005;232:559–567. doi: 10.1016/j.jtbi.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Website NNPP version 2.2 http://www.fruitfly.org/seq_tools/promoter.html
- Website TSSW http://www.softberry.com/berry.phtml?topic=tssw&group=programs&subgroup=promoter
- Website DFP version 1.5 http://www.research.i2r.a-star.edu.sg/promoter/promoter1_5/DPF.htm
- Website Promoter 2.0 http://www.cbs.dtu.dk/services/Promoter
- Website Promoter Scan version 1.7 http://www-bimas.cit.nih.gov/molbio/proscan/
- Gao F, Zhang CT. Comparison of various algorithms for recognizing short coding sequences of human genes. Bioinformatics. 2004;20:673–681. doi: 10.1093/bioinformatics/btg467. [DOI] [PubMed] [Google Scholar]
- Lu J, Luo LF. Human Pol II promoter prediction (in Chinese) Progress in Biochemistry and Biophysics. 2005;32:1185–1191. [Google Scholar]
- Zhao X, Xuan Z, Zhang M. Boosting with stumps for predicting transcription start sites. Genome Biology. 2007;8:R17. doi: 10.1186/gb-2007-8-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZG, Anh V, Wanliss JA, Watson SM. Chaos game representation of the Dst index and prediction of geomagnetic storm events. Chaos, Solitons and Fractals. 2007;31:736–746. doi: 10.1016/j.chaos.2005.12.046. [DOI] [Google Scholar]
- Tian YC, Yu ZG, Fidge C. Multifractal nature of network induced time delay in networked control systems. Phys Lett A. 2007;361:103–107. doi: 10.1016/j.physleta.2006.09.046. [DOI] [Google Scholar]
- The ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Margalit H, Shapiro B, Nussinov R, Owens J, Jernigan R. Helix stability in prokaryotic promoter regions. Biochemistry. 1998;27:5179–5188. doi: 10.1021/bi00414a035. [DOI] [PubMed] [Google Scholar]
- Vollenweider HJ, Fiandt M, Szybalski W. A relationship between DNA helix stability and recognition sites for RNA polymerase. Science. 1979;205:508–511. doi: 10.1126/science.377494. [DOI] [PubMed] [Google Scholar]
- Florquin K, Saeys Y, Degroeve S, Rouzé P, de Peer YV. Large-scale structural analysis of the core promoter in mammalian and plant genomes. Nucleic Acids Res. 2005;33:4255–4264. doi: 10.1093/nar/gki737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZG, Anh V, Lau KS. Multifractal characterisation of length sequences of coding and noncoding segments in a complete genome. Physica A. 2001;301:351–361. doi: 10.1016/S0378-4371(01)00391-0. [DOI] [Google Scholar]
- Yu ZG, Anh V, Lau KS. Measure representation and multifractal analysis of complete genomes. Phys Rev E. 2001;64:031903. doi: 10.1103/PhysRevE.64.031903. Epub 2001 Aug 24. [DOI] [PubMed] [Google Scholar]
- Mandelbrot BB. The Fractal Geometry of Nature. Academic Press, New York; 1983. [Google Scholar]
- Feder J. Fractals. Plenum, New York; 1988. [Google Scholar]
- Yu ZG, Anh V, Lau KS. Fractal analysis of measure representation of large proteins based on the detailed HP model. Physica A. 2004;337:171–184. doi: 10.1016/j.physa.2004.01.057. [DOI] [Google Scholar]
- Lee J, Stanley HE. Phase Transition in the Multifractal Spectrum of Diffusion-Limited Aggregation. Phys Rev Lett. 1988;61:2945–2948. doi: 10.1103/PhysRevLett.61.2945. [DOI] [PubMed] [Google Scholar]
- Canessa E. Multifractality in time series. J Phys A. 2000;33:3637–3651. doi: 10.1088/0305-4470/33/19/302. [DOI] [Google Scholar]
- Dunki RM, Ambuhl B. Scaling properties in temporal patterns of schizophrenia. Physica A. 1996;230:544–553. doi: 10.1016/0378-4371(96)00097-0. [DOI] [Google Scholar]
- Yu ZG, Anh V, Wang B. Correlation property of length sequences based on global structure of the complete genome. Phy Rev E. 2001;63:011903. doi: 10.1103/PhysRevE.63.011903. Epub 2000 Dec 20. [DOI] [PubMed] [Google Scholar]
- Bunde A, Havlin S, eds . Fractals in Science. Springer-verlag, Berlin; 1979. [Google Scholar]
- Zhang R, Zhang CT. Z curves, an intutive tool for visualizing and analyzing the DNA sequences. J Biomol Struct Dyn. 1994;11:767–782. doi: 10.1080/07391102.1994.10508031. [DOI] [PubMed] [Google Scholar]
- Zhang CT, Lin ZS, Yan M, Zhang R. A novel approach to distinguish between intron-containing and intronless genes based on the format of Z curves. J Theor Biol. 1998;192:467–473. doi: 10.1006/jtbi.1998.0671. [DOI] [PubMed] [Google Scholar]
- Yan M, Lin ZS, Zhang CT. A new fourier transform approach for protein. coding measure based on the format of the Z curve. Bioinformatics. 1998;14:685–690. doi: 10.1093/bioinformatics/14.8.685. [DOI] [PubMed] [Google Scholar]
- Zhang CT, Zhang R. Analysis of distribution of bases in the coding sequences by a diagrammatic technique. Nucleic Acids Res. 1991;19:6313–6317. doi: 10.1093/nar/19.22.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RJ, Dubchak I, Holbrook SR. A computational approach to identify genes for functional RNAs in genomic sequences. Nucleic Acids Res. 2001;29:3928–3938. doi: 10.1093/nar/29.19.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CZ, Han LY, Ji ZL, Chen X, Chen YZ. SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003;31:3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kochhar S, Grigorov MG. Descriptor-based protein remote homology identification. Protein Sci. 2005;14:431–444. doi: 10.1110/ps.041035505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZR, Lin HH, Han LY, Jiang L, Chen X, Chen YZ. PROFEAT: a web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res. 2006;34:W32–W37. doi: 10.1093/nar/gkl305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HQ, Hu GQ, Yang YF, Wang J, She ZS. MED: a new non-supervised gene prediction algorithm for bacterial and archaeal genomes. BMC Bioinformatics. 2007;8:1–11. doi: 10.1186/1471-2105-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CT, Wang J. Recognition of Protein Coding Genes in the Yeast Genome at Better Than 95% Accuracy Based on the Z curve. Nucleic Acids Res. 2000;28:2804–2814. doi: 10.1093/nar/28.14.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CT, Zhang R. A nucleotide composition constraint of genome sequences. Comput Biol Chem. 2004;28:149–153. doi: 10.1016/j.compbiolchem.2004.02.002. [DOI] [PubMed] [Google Scholar]