Abstract
Knockdown of the insulator factor CCCTC binding factor (CTCF), which binds XL9, an intergenic element located between HLA-DRB1 and HLA-DQA1, was found to diminish expression of these genes. The mechanism involved interactions between CTCF and class II transactivator (CIITA), the master regulator of major histocompatibility complex class II (MHC-II) gene expression, and the formation of long-distance chromatin loops between XL9 and the proximal promoter regions of these MHC-II genes. The interactions were inducible and dependent on the activity of CIITA, regulatory factor X, and CTCF. RNA fluorescence in situ hybridizations show that both genes can be expressed simultaneously from the same chromosome. Collectively, the results suggest a model whereby both HLA-DRB1 and HLA-DQA1 loci can interact simultaneously with XL9, and describe a new regulatory mechanism for these MHC-II genes involving the alteration of the general chromatin conformation of the region and their regulation by CTCF.
MHC class II (MHC-II) genes encode cell-surface glycoproteins that present antigens to CD4 T cells to initiate and control adaptive immune responses (1). The expression of MHC-II genes and accessory genes necessary for antigen processing (Ii and HLA-DM) is regulated in a developmental fashion in antigen-presenting cells and by external signaling through cytokines, among which IFN-γ is a powerful inducer of their expression (2). The transcription factors regulatory factor X (RFX), cAMP response element binding protein (CREB), and nuclear factor–Y (NF-Y) bind to proximal promoter conserved upstream sequences (termed X1, X2, and Y boxes, respectively) of all MHC-II genes (for review see references 3–5), and are required and constitutively expressed, but are not sufficient, to initiate transcription of MHC-II. Instead, expression is controlled by the class II transactivator (CIITA), a highly regulated transcriptional coactivator that interacts with the X-Y box DNA-bound factors (6, 7). At MHC-II promoters, CIITA serves as an adaptor molecule by recruiting several chromatin remodeling factors and the general transcription machinery (for review see references 3, 8). Although this paradigm is widely accepted, it is not complete.
Several lines of evidence suggest that MHC-II gene expression is controlled by a much more complex system. A transgenic mouse developed and characterized >10 yr ago in which upstream sequences of the Eα gene were deleted displayed aberrant MHC-II expression patterns, suggesting that the deleted elements may serve a role in tissue-specific expression (9, 10). Analysis of this region found a distal X-Y box sequence. A similarly located X-Y element was found upstream of the HLA-DRA gene. The distal X-Y box region of the HLA-DRA gene displayed robust acetylation, produced low levels of transcripts, and was suggested to function as a locus control region (11). Additional regions of potential control were also found upstream of the HLA-DRB1 gene (12). A bioinformatics search of the human MHC-II region for nonpromoter-associated X and X-Y boxes revealed ∼40 such sequences with varying degrees of homology (13). Two of these, termed X box–like region 7 (XL-7) and XL-8, positioned between HLA-DRA and HLA-DRB3, were active with respect to their binding of RFX and CIITA and histone modifications. These sequences are likely to be remnants of the promoters of HLA-DRB pseudogenes, but their activity suggests that they may serve to increase the accessibility of the MHC-II region chromatin in CIITA-expressing cells.
Another of these elements, XL-9, located in the intergenic DNA between HLA-DRB1 and HLA-DQA1 (Fig. 1 A), was unusual in that it did not appear to bind RFX or CIITA but displayed high levels of histone acetylation and a histone modification profile that was associated with accessible chromatin (14). Further investigation of the region showed that the acetylation extended for several kilobases in both directions from the originally identified sequence. The peak of histone acetylation was located ∼0.24 kb away from the XL-9 sequence in a region designated XL9d. XL9d was bound by the transcriptional insulator factor CCCTC binding factor (CTCF). When placed between an SV40 enhancer and a promoter, XL9d-containing DNA functioned as a potent enhancer-blocking element, preventing the expression of the reporter gene. XL9d was found in nuclear matrix preparations, a property associated with CTCF binding regions. These findings suggested that the XL-9 region (henceforth referred to as XL9) exhibits properties and activities of a transcriptional insulator; however, the role of XL9 and CTCF in the expression of the flanking MHC-II genes has not been elucidated.
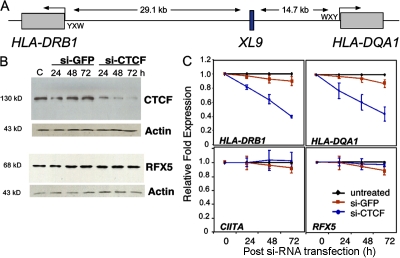
Figure 1.
CTCF knockdown reduces HLA-DRB1 and HLA-DQA1 mRNA levels. (A) An overall schematic of the HLA-DRB1 and HLA-DQA1 gene and XL9 region is shown. The conserved proximal promoter elements of the MHC-II genes, W-X-Y, where RFX and CIITA interact are indicated. (B) siRNA to CTCF but not an irrelevant siRNA knocked down the expression of CTCF protein. Raji cells were transiently transfected with SMART pool siRNAs to CTCF or GFP and assayed by Western blotting for CTCF, RFX5, and β-actin expression at the indicated time points. (C) CTCF siRNA–transfected Raji cells displayed a marked decrease in HLA-DRB1 and HLA-DQA1 mRNA levels as determined by real-time RT-PCR. The decrease in expression of these genes was specific, as a nonspecific siRNA had no effect and CIITA and RFX5 gene expression were not significantly altered. This analysis was performed three times. The mean of each experiment (with SEM) is shown with the data normalized to the levels of GAPDH. GAPDH mRNA levels were unaltered during the course of this analysis.
CTCF is a ubiquitous mammalian transcriptional insulator factor with >13,000 binding sites in human cells (15). As a transcriptional insulator, CTCF binding prevents the actions of an enhancer from acting on the promoter of a downstream gene (16). This action is responsible for the expression patterns associated with the imprinting control region in the mouse H19/Igf2 genes, where CTCF binding and gene expression are controlled by methylation of the imprinted region (17–19). CTCF binding can also prevent the encroachment of heterochromatin to an active gene, and therefore functions as a boundary element (20). CTCF binding sites are degenerate because of the fact that CTCF can use any number of its 11 zinc fingers to interact with DNA. The exact mechanisms by which CTCF functions in each of these events is not known. Although these examples of CTCF biology are extraordinary, other potential roles for CTCF are likely.
In this paper, the hypothesis that CTCF regulates HLA-DRB1 and HLA-DQA1 expression is tested and the mechanism is explored. Small interfering RNA (siRNA) to CTCF was effective at reducing HLA-DRB1 and HLA-DQA1 expression. Coimmunoprecipitation experiments showed that CTCF, CIITA, and RFX5 (a subunit of the X1 box factor RFX) were in the same complex, suggesting that the CTCF-bound region (XL9) and the flanking HLA-DRB1 and HLA-DQA1 proximal promoters may interact. To determine if this was the case, the chromatin conformation capture (3C) assay was used to determine if such interactions existed. Distinct interactions were observed, suggesting a role for XL9 in the regulation of these genes. Interactions with XL9 were dependent on active transcription of MHC-II genes and the presence of the MHC-II–specific transcription factors CIITA and RFX. Moreover, XL9 interactions were inducible by IFN-γ in nonimmune cells, supporting a role for XL9 in HLA-DRB1 and HLA-DQA1 expression. CTCF was found to be required for the interactions, as siRNA-mediated knockdown of CTCF expression resulted in the loss of the 3C product and a reduction in histone modifications associated with transcription at the promoters of the HLA-DRB1 and HLA-DQA1 genes. RNA fluorescence in situ hybridization (FISH), used to assess the expression patterns of these genes, showed that both HLA-DRB1 and HLA-DQA1 expression could be detected in some cells simultaneously. The data describe a novel mechanism for the regulation of these immune system genes, a mechanism that may be common to other MHC-II genes.
RESULTS
Loss of CTCF results in the reduction of HLA-DRB1 and HLA-DQA1 expression
CTCF was found to interact with XL9 by in vivo (chromatin immunoprecipitation [ChIP]) and in vitro (electrophoretic mobility shift assay) assays (14) (15). To test the hypothesis that CTCF plays a role in the regulation of the flanking MHC genes HLA-DRB1 and HLA-DQA1, siRNAs were used to knockdown the expression of CTCF. The efficacy of transiently transfecting SMART pools of CTCF siRNAs into Raji cells was tested in the MHC-II–expressing Burkitt's lymphoma B cell line Raji. As shown by the knockdown time course (Fig. 1 B), CTCF protein was significantly but not completely reduced at 72 h. siRNAs to GFP did not affect CTCF protein levels, and β-actin levels were not affected by either of the siRNA pools (Fig. 1 B). Similarly, neither the control nor CTCF siRNAs altered the expression of the RFX subunit factor RFX5 (Fig. 1 B). Assessment of HLA-DRB1 and HLA-DQA1 mRNA expression during the knockdown time course assays showed a marked reduction in steady-state mRNA levels of both HLA-DRB1 and HLA-DQA1 when siRNAs to CTCF but not GFP were used (Fig. 1 C). This reduction in expression had no effect on the expression of RFX and CIITA mRNA (Fig. 1 C), or the expression of GAPDH, CARM1, Cox2, cMyc, or PRMT1 mRNAs (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20071843/DC1), suggesting that a reduction in CTCF expression did not produce a general inability to express genes transcribed by RNA polymerase II. Although CTCF was shown to affect cMyc expression and, consequently, cell growth (21, 22), the lack of CTCF control of cMyc in Raji cells likely reflects the Burkitt lymphoma translocation of these cells, whereas cMyc is not under its normal regulation or chromosome environment (23). Additionally, the in vivo occupancy of RFX5 and CIITA on the proximal promoters of these genes was unchanged, indicating that the ability of MHC-II–specific transcription factors to bind their regulatory sites was not altered by CTCF knockdown (Fig. S2).
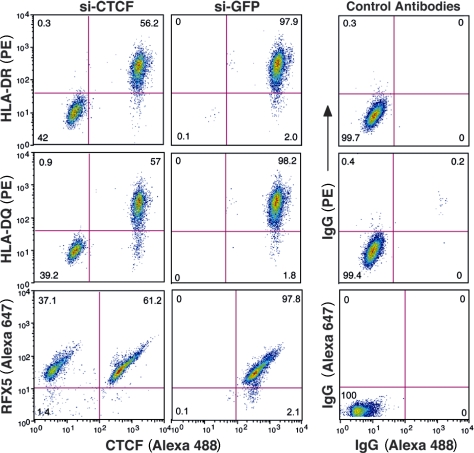
The presence of CTCF protein and HLA-DRB1 or HLA-DQA1 mRNA after siRNA treatment may have been caused by the efficiency of the transient transfection of the siRNAs into the cells. To examine only those cells that have lost CTCF, an intracellular staining flow cytometry assay was developed for CTCF and RFX5. When combined with HLA-DR and HLA-DQ surface expression staining, the data showed that cells that lost CTCF expression also lost surface HLA-DR and HLA-DQ but not RFX5 expression (Fig. 2). siRNA to GFP had no effect on HLA-DR or -DQ surface or intracellular RFX5 expression. The combined data demonstrate a role for CTCF in the expression of these MHC-II genes.
Figure 2.
CTCF knockdown results in a loss of surface HLA-DR and HLA-DQ expression. Raji cells transiently transfected for 72 h with siRNA to CTCF or to GFP as in Fig. 1 were stained for intracellular CTCF and surface MHC-II expression. Two populations of cells exist for the CTCF siRNA panels, identifying those cells that were efficiently transfected from those that were not. GFP siRNA had no effect on the levels of MHC-II or CTCF. siRNA-transfected cells were also stained for CTCF, followed by RFX5 staining. No change in RFX5 levels were observed. The fluorophores used for detection are indicated (PE and Alexa Fluor 488). Control staining patterns for intracellular and surface antibodies are shown. This figure is representative of two independent experiments.
CTCF associates with CIITA
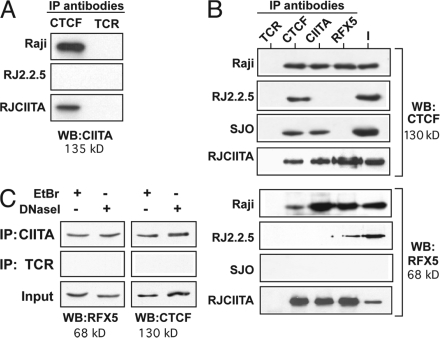
One way in which CTCF could mediate this effect was if it associated with MHC-II transcription factors or a complex containing these factors. To determine if this could be the case, coimmunoprecipitation experiments were performed to test the hypothesis that CIITA and CTCF associate. Immunoprecipitations using antibodies to CTCF, CIITA, and RFX5 with lysates from the MHC-II–expressing Raji B cells were performed (Fig. 3). In Raji cell lysates, CTCF and CIITA were found to associate, as antibodies to each could coprecipitate the other (Fig. 3, A and B). CTCF and CIITA antibodies each coprecipitated RFX5, and RFX5 antibodies coprecipitated CTCF (Fig. 3 B). Although CIITA is known to interact directly with RFX5 in Raji cells (24, 25), interactions between CTCF and RFX5 or CIITA were not previously known. Clues to the nature of these interactions were derived from CIITA and RFX5 mutant cells. RJ2.2.5 cells were derived directly from Raji cells by mutagenesis and are MHC-II deficient because of debilitating deletions in CIITA (6, 26–28). RJ-CIITA cells are RJ2.2.5 cells that were stably complemented with a CIITA-expressing vector (28) and express similar amounts of CIITA as Raji cells (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20071843/DC1). SJO cells, derived from a bare lymphocyte syndrome patient, express CIITA but lack RFX5, one of the RFX subunits (29). Lysates from the mutant cells were examined and showed that although CIITA and CTCF associate in the RFX5-deficient cell line, RFX5-CTCF coimmunoprecipitates were absent in the CIITA-deficient cell line (Fig. 3 B, top). This association is restored in the RJ-CIITA–complemented cell line (Fig. 3, A and B). The lack of CIITA and RFX5 immunoprecipitates in RJ2.2.5 and SJO, respectively, demonstrates the specificity of the antibodies, as does the failure of the nonspecific antibody to pull down the factors. Thus, RFX5 and CTCF immunoprecipitates are dependent on the presence of CIITA, suggesting that the three proteins can associate in a complex. It should be noted that these coimmunoprecipitates were observed with endogenously expressed proteins.
Figure 3.
CIITA associates with CTCF. Cellular lysates from Raji (wild-type), RJ2.2.5 (CIITA−, RFX5+), RJ-CIITA (CIITA+, RFX5+), and SJO (CIITA+, RFX5−) cells were prepared and immunoprecipitated with the indicated antibodies. The immunoprecipitates were analyzed by Western blotting for the presence of (A) CIITA, (B, top) CTCF, and (B, bottom) RFX5. CIITA was found in immunoprecipitates using CTCF, and CTCF was found in coimmunoprecipitates using CIITA and RFX5 antisera in Raji cells, indicating an association between the factors. The association was not observed in RJ2.2.5 cells with either CIITA or RFX5 antisera, suggesting that the associations between RFX5 and CTCF were dependent on the presence of CIITA. As in earlier papers (references 24, 25), RFX5 and CIITA interactions were also observed. No interactions with the nonspecific antibody to TCR were observed. 10% of the input is indicated (I). (C) Coimmunoprecipitation experiments using either CIITA or TCR antisera in the presence of ethidium bromide or treated with DNase I were performed as described in Materials and methods. The precipitations were analyzed by immunoblotting for RFX5 and CTCF as indicated. Each panel in this figure set is representative of at least two independent experiments.
The possibility that the observed complexes coprecipitated because of the binding of the components to DNA was eliminated by including ethidium bromide, an intercalating agent that disrupts protein–DNA complexes (30), into the precipitation reactions. Treatment of lysates with DNase I was also performed. Both treatments did not alter the coprecipitation results (Fig. 3 C), demonstrating that CIITA–CTCF complexes formed independent of DNA.
The proximal promoters of HLA-DRB1 and HLA-DQA1 interact directly with XL9
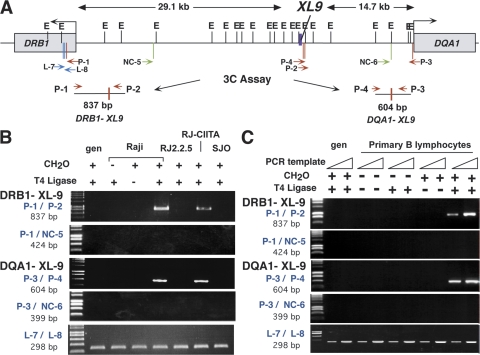
These coimmunoprecipitation results suggest that interactions between XL9 and the proximal promoters of the HLA-DRB1 and HLA-DQA1 genes may occur. To test this hypothesis the 3C assay (31–33) was developed for this system and used. In the 3C assay, cellular components are cross-linked with formaldehyde to stabilize in vivo interactions. After the isolation of the cross-linked chromatin, the DNA is restriction digested, diluted, and ligated under conditions that favor intramolecular ligation products. PCR is conducted on the purified DNA to detect whether a novel ligation junction is formed between the regions being assayed (Fig. 4). The formation of a novel, specific ligation product signifies that long-range chromatin interactions occurred in vivo between the cis-acting sequences assayed (31–36).
Figure 4.
Long-range chromatin interactions form between XL9 and the proximal promoter regions of the HLA-DRB1 and HLA-DQA1 genes. (A) A schematic of the HLA-DRB1 and HLA-DQA1 gene region depicts the locations of the EcoRI sites (top tick marks) and the primers used (bottom tick marks) in the 3C assays. The position of XL9 is indicated by a thick purple vertical bar. In the 3C assay, chromatin isolated from formaldehyde–cross-linked cells was digested overnight with a large excess of EcoRI. After inactivation of the enzyme, samples were diluted and T4 DNA ligase was added. Novel ligation junctions were detected by PCR using the primer sets indicated and 35 cycles of amplification. 3C PCR primers P-1 with P-2 and P-3 with P-4 (red) allow detection of the XL9 restriction fragment with HLA-DRB1 and HLA-DQA1, respectively. (B) Interactions between XL9 and HLA-DRB1 or HLA-DQA1 are specific and dependent on the presence of CIITA and RFX5. A 3C product was observed in Raji cells when formaldehyde (CH2O) cross-linking and T4 DNA ligase were included during the assay. Intact chromatin was necessary for the formation of the 3C product, as no product was observed with purified genomic DNA. 3C product formation required CIITA and RFX5, as no products were observed for RJ2.2.5 or SJO cells, which are deficient for CIITA and RFX5, respectively. A 3C product was observed in RJ-CIITA cells, which are RJ2.2.5 cells stably complemented with CIITA. PCR assays using nonspecific control primers NC-5 (A, green) with P-1 or NC-6 (A, green) with P-3 were used to demonstrate that random ligation of their encoding restriction fragments did not occur with HLA-DRB1 and HLA-DQA1, respectively. Loading control primers L-7 and L-8 (blue) amplify DNA contained within a single EcoRI fragment and serve as a loading control. Fivefold less DNA was added to the PCR reaction for the loading controls. (C) The 3C assay was performed on freshly isolated CD19+ human peripheral B lymphocytes. The experimental conditions were identical to those described in B, with two concentrations (50 and 100 ng) of genomic DNA used for the PCR step. gen, genomic DNA.
Thus, to determine if the HLA-DRB1 and HLA-DQA1 proximal promoter regulatory regions interacted with XL9, 3C assays were performed using Raji cells. A map of the DR/DQ intergenic region with the location of the restriction sites, PCR primers, and the predicted novel 3C products is shown in Fig. 4 A. 3C analysis showed the formation of a novel ligation product between the EcoRI restriction fragments encoding XL9 and the conserved proximal regulatory region of HLA-DRB1. This “3C product” was dependent on formaldehyde cross-linking, restriction digestion, and DNA ligation (Fig. 4 B). Similarly, a novel ligation product between the HLA-DQA1 W-X-Y– and XL9–encoding restriction fragments was detected. The 3C products were cloned, and their sequence matched the predicted novel ligation product. No 3C products were detected if purified genomic DNA was used (Fig. 4 B), suggesting that the observed products required intact chromatin and interactions between the factors bound to the respective DNA fragments. The generation of a PCR amplicon that was independent of the assay established that equal amounts of DNA were present in all reactions and conditions (Fig. 4 B, L-7/L-8). To control for nonspecific ligation products, 3C was conducted on randomly chosen restriction fragments within the intergenic regions between XL9 and each gene. No 3C products were detected (Fig. 4 B, primers P-1/NC-5 and P-3/NC-6). Several additional controls were conducted (31). PCR amplification by each primer set was tested for efficacy on a mix of genomic bacterial artificial chromosome (BAC) DNA templates from the HLA-DR/DQ region generated by restriction digestion and religation. PCR showed that the primer sets were functional and had similar efficiencies (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20071843/DC1). Additionally, no differences in the ability of EcoRI to cleave the assayed sites in the chromatin preparations were detected in Raji cells or the other cell lines examined (unpublished data).
To demonstrate that the 3C interactions were not specific to the model B lymphocyte Raji cell line and occurred in primary human B cells, the 3C assay was performed on CD19+ purified peripheral B cells of healthy volunteers. Identical results were observed (Fig. 4 C). Thus, two long-range chromatin interactions occur between XL9 and the regulatory regions of HLA-DRB1 and HLA-DQA1 genes in MHC-II–expressing B lymphocytes.
Interactions with the XL9 region are dependent on CIITA and RFX
To ascertain the relationship between MHC-II gene transcription and XL9, B lymphocyte cell lines, which are MHC-II negative because of mutations in either CIITA or RFX5, were used. No 3C PCR product associated with XL9 and the flanking MHC-II genes was observed in the CIITA-deficient RJ2.2.5 cells (Fig. 4 B), suggesting that the observed interactions occurred during MHC-II transcription and required the presence of CIITA. Moreover, when the 3C assay was conducted on RJ2.2.5 cells that had been stably complemented with CIITA (RJ-CIITA) (28), interactions between XL9 and the regulatory regions of HLA-DRB1 and HLA-DQA1 were restored, demonstrating that CIITA expression was required for interactions with XL9. 3C interactions were not observed in the MHC-II–negative, RFX5-deficient but CIITA-positive SJO cell line (Fig. 4 B), suggesting that assembly of the transcription regulatory factors at the HLA-DRB1 and HLA-DQA1 promoters was required for interactions with XL9. RFX subunit–deficient cells do not assemble the other RFX subunits (CREB, NF-Y, or CIITA) at their X-Y box sequences (37, 38).
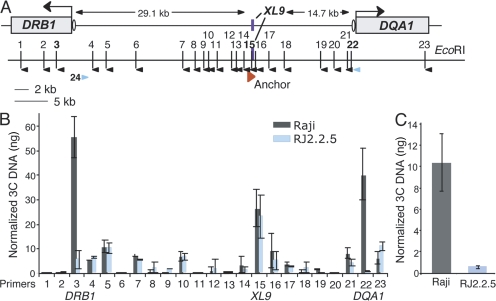
To determine if XL9 interacted with other regions between HLA-DRB1 and HLA-DQA1, real-time PCR primer sets spanning the HLA-DRB1 and HLA-DQA1 intergenic region were designed, tested for amplification, and used to detect 3C products with a series of XL9 anchor primers (Fig. 5). Test primers and XL9 anchor primers (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20071843/DC1) were chosen based on relative efficiency and the ability to produce a single amplicon on EcoRI-digested/religated BAC DNA encoding the region. Two major peaks were observed with primers 3 and 22 and their XL9 anchors, which denote the EcoRI fragments encoding the W-X-Y box regions of HLA-DRB1 and HLA-DQA1, respectively (Fig. 5 B). Importantly, the 3C products for these primer sets were the only sets that exhibited a significant reduction in RJ2.2.5 cells, denoting their relationship to MHC-II expression and confirming the qualitative data presented in Fig. 4. Background interactions with EcoRI fragments close to HLA-DRB1 and HLA-DQA1 can be observed with equal levels between Raji and RJ2.2.5 cells. Such 3C background products are seen in other systems (39). Equal amounts of a control 3C product of the XL9 EcoRI fragment ligating to itself were observed with an XL9 anchor primer and primer 15, demonstrating that equal preparations were used. Collectively these data (Figs. 4 and Figs.5) demonstrate that long-range chromatin interactions form between the 5′ proximal regulatory regions of the HLA-DRB1 and HLA-DQA1 genes and the XL9 intergenic region in a manner that is dependent on the MHC-II–specific transcription factors CIITA and RFX. Importantly, these data provide clear genetic proof that the formation and detection of 3C products are not random or an artifact of the 3C assay.
Figure 5.
XL9 interacts with HLA-DRB1 and HLA-DQA1. To ascertain if XL9 interacted with other restriction fragments within the HLA-DRB1, HLA-DQA1 region, a quantitative 3C assay was used across the region. (A) A schematic showing the subregion and each EcoRI site numbered 1–23 is shown. The orientation of primers specific to these sites is indicated by black arrowheads. XL9 primers, which served as “anchors” for each 3C PCR, are shown by a single red arrowhead. Because of the differences in optimal annealing temperature between the restriction fragment primers, multiple XL9 anchor primers were required to produce single PCR products with similar efficiencies. Ovals represent the respective W-X-Y box regions. Primers 22 and 24 (blue arrowheads) were used to detect interactions between HLA-DRB1 and HLA-DQA1, as described in C. (B) 3C assays were performed on Raji and RJ2.2.5 cells as in Fig. 4. Each 3C assay was compared with its own standard curve generated from the amplification of an HLA-DRB1– and HLA-DQA1–containing BAC that had been EcoRI digested and ligated. The results from three separate experiments were averaged and normalized against the BAC DNA used in the standard curve. The results are plotted with the standard deviation observed. Primer set 15 with its XL9 anchor serves as a loading control for the system, as this ligation product is derived from a single EcoRI fragment (XL9). Note that primers 6 and 20 represent the same EcoRI fragments as primers NC-5 and NC-6 from Fig. 4. (C) Raji and RJ2.2.5 cells were used to examine whether the HLA-DRB1 and HLA-DQA1 promoter regions interact. A 3C product between HLA-DRB1 (primer 24) and HLA-DQA1 (primer 22) promoter-containing restriction fragments was detected that was dependent on the presence of CIITA. Student's t tests were used to determine the significance of differences between Raji and RJ2.2.5 samples for each primer set. In B, only 3C samples using XL9 anchors with primers 3 and 22 showed significance (P < 0.04). The differences in C were found to be significant (P < 0.03).
IFN-γ induces interactions with XL9
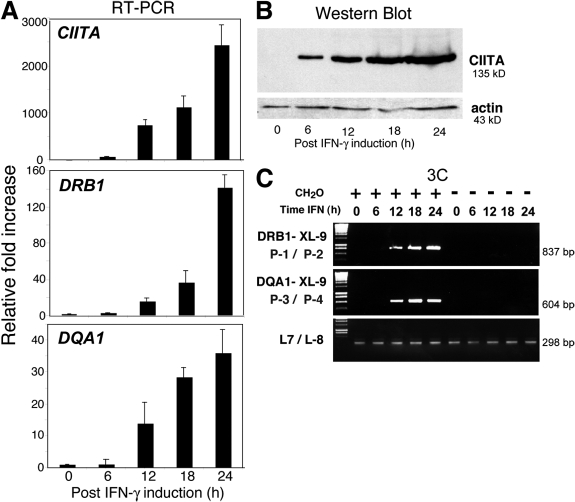
To further demonstrate that MHC-II transcription was coincident with interactions with the XL9 region, the 3C assay was used on A431 epithelial cells after IFN-γ induction of MHC-II genes. IFN-γ regulates MHC-II expression by controlling the expression of CIITA (7). Thus, if MHC-II transcription and interactions with XL9 are concurrent events, then after IFN-γ treatment, MHC-II expression and XL9 interactions should occur with similar kinetics. A431 cells were treated with IFN-γ for 0, 6, 12, 18, and 24 h before 3C analysis. The mRNA and protein levels for CIITA first appear at ∼6 h and increase during the time course (Fig. 6, A and B) (38). The expression of HLA-DRB1 and HLA-DQA1 occurs with CIITA mRNA appearance and reaches a maximal level of expression in this time course at 24 h. The levels of RFX5 and GAPDH were unchanged during the treatment (unpublished data). Interactions between XL9 and HLA-DRB1 and HLA-DQA1 were detected after the 6-h time point (Fig. 6 C), a time coincident with high levels of CIITA expression and, importantly, CIITA assembly at MHC-II promoters (38). MHC-II expression was also observed after 12 h. Interactions with XL9 were maintained during the 24-h time course of the assay. These results demonstrated that interactions between XL9 and the proximal promoters of the HLA-DRB1 and HLA-DQA1 genes were synchronized with active MHC-II expression and were absent in MHC-II–negative cell lines. This inducible system also highlighted the specificity of the assay by demonstrating that in the absence of the inducer (IFN-γ) and in the presence of all other required elements, no XL9–MHC-II promoter interactions were detected.
Figure 6.
IFN-γ induces long-range interactions between XL9 and the proximal promoter regions of HLA-DRB1 and HLA-DQA1. MHC-II–negative A431 epithelial cells were treated with IFN-γ in a time course that extended to 24 h. (A) mRNA analysis, (B) Western blots, and (C) 3C analysis were conducted at the indicated time points. (A) Real-time RT-PCR was performed with CIITA-, HLA-DRB1–, and HLA-DQA1–specific primers on RNA isolated at the indicated time points after IFN-γ treatment. The crossover threshold (Ct) real-time PCR values were normalized to the Ct values of those obtained for GAPDH mRNA and presented as the mean fold induction with standard error. (B) A Western blot shows that CIITA protein can be detected as early as 6 h after IFN-γ induction and that the expression of CIITA is maintained over the 24-h time course. (C) The 3C assays were performed on the A431 cells as in Fig. 4. Interactions between XL9 and the HLA-DQA1 and HLA-DRB1 promoter regions were observed at 12 h after the induction time point. All assays were performed at least three times from independently treated cultures.
CTCF is required for interactions between XL9 and HLA-DRB1 and HLA-DQA1 and for maximal histone modification of the promoter regions
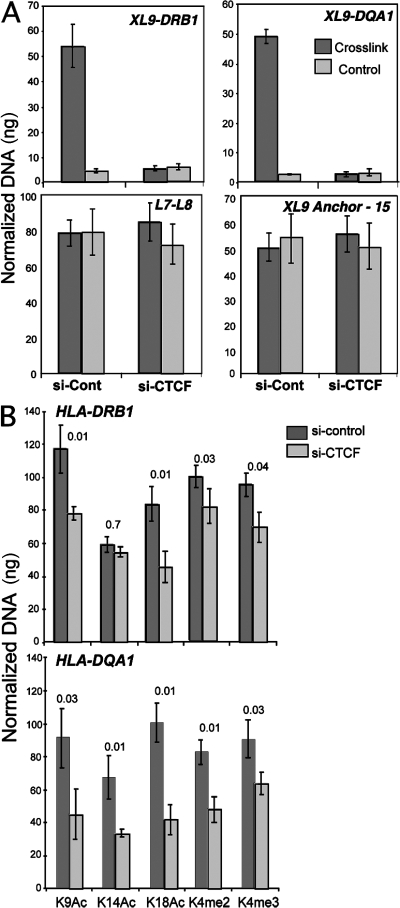
To test whether CTCF was important for the long-range interactions observed, siRNAs to knockdown the expression of CTCF were used as described in Fig. 1. When assayed after siRNA transient transfection of Raji cells, the HLA-DRB1/XL9 and HLA-DQA1/XL9 3C products were decreased substantially but not completely (unpublished data). Incomplete loss of the 3C product was likely caused by the siRNA transfection efficiency, as described in Fig. 2. To overcome this technical issue, siRNA-transfected cells were separated from nontransfected cells by using HLA-DR antibody–coated magnetic beads. The HLA-DR–negative cells were assayed for formation of the HLA-DRB1/XL9 and HLA-DQA1/XL9 3C products (Fig. 7 A). A complete loss of the 3C product was observed in the CTCF siRNA– but not control GFP siRNA–transfected cells. Control primer amplifications (L7/L8) and the intramolecular ligation product (P15/anchor) demonstrate equal loading of the samples. In total, these results suggest that CTCF is required for interactions between the proximal promoter regions of HLA-DRB1 and HLA-DQA1 with XL9.
Figure 7.
CTCF is required for interactions between XL9 and the HLA-DRB1 and HLA-DQA1 promoter regions. 72 h after CTCF siRNA transfection, Raji cells were labeled with anti–HLA-DR magnetic beads and separated from nontransfected cells by MACS column separation. The HLA-DR–negative cells were assayed by 3C and compared with siRNA GFP–transfected cells. Cells were either cross-linked or left untreated (control) before 3C. Quantitative 3C primer sets to detect XL9/HLA-DRB1 and XL9/HLA-DQA1, as indicated in the figure, were used as described in Fig. 5 and Fig. S5. Loading control primer sets from Fig. 4 (L-7/L-8) and 3C ligation control primers (15/XL9 anchor) from Fig. 5 were also used. 3C products were quantitated as in Fig. 5. Data from three independent chromatin preparations were averaged and plotted with their standard deviation. (B) ChIP assays for histone modifications at the HLA-DRB1 and HLA-DQA1 promoter regions were conducted using the indicated antibodies 72 h after cells were transfected with either control or CTCF siRNAs. The real-time PCR data were normalized to input and to a standard curve for each locus, and the mean of three independent assays is shown with standard deviation. Student's t test p-values are shown above each comparison.
To ascertain a consequence of CTCF interaction with the MHC-II promoters, CTCF and control GFP siRNA knockdown cells were assayed by ChIP for histone modifications that are associated with transcriptional activation (Fig. 7 B). For the HLA-DRB1, a reduction in histone H3K9 and K18 acetylation was observed in CTCF but not GFP siRNA–transfected cells (33 and 45%, respectively). Histone H3 K14 acetylation was not affected. A small reduction in H3 K4 dimethylation (18%) and a modest reduction in H3 K4 trimethylation (27%) were observed. At the HLA-DQA1 promoter, more substantial reductions in the histone modifications tested were observed in CTCF siRNA–treated cells. GFP siRNA–treated cells showed the same level of histone modifications as untreated cells (unpublished data). These data suggest that CTCF contributes to the modification of the chromatin structure at the promoters of the HLA-DRB1 and HLA-DQA1 genes in a manner that is favorable to transcription.
Expression of HLA-DRB1 and HLA-DQA1 alleles
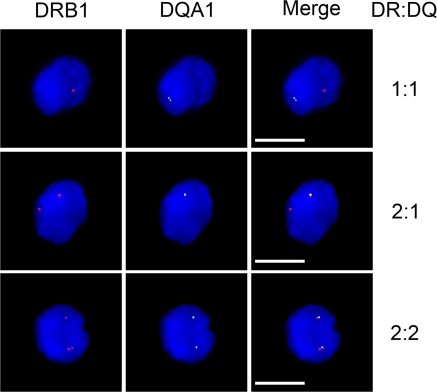
It is generally accepted that all MHC-II genes and alleles are coordinately expressed in mature B cells, including Raji cells, as determined by surface expression and mRNA levels. Such experiments view the steady state of populations or individual cells, and thus, it is not known if both alleles are being transcribed simultaneously or whether each locus is active at the same time. To determine the allele expression patterns of the HLA-DRB1 and HLA-DQA1 genes, RNA FISH was performed on Raji and RJ2.2.5 cells using probes for HLA-DRB1 and HLA-DQA1 transcripts (Fig. 8). A range of expression patterns was observed in Raji cells (Table I). No RNA FISH signals were observed in the CIITA-deficient and MHC-II–negative RJ2.2.5 cells (unpublished data). HLA-DRB1 was expressed in almost all cells (98.7%). In 77% of the cells, HLA-DRB1 was expressed from both alleles. HLA-DQA1 was expressed in 80% of the cells, but never without HLA-DRB1 expression. In contrast to HLA-DRB1, only 23% of the cells expressed HLA-DQA1 from both alleles. With 77% of the cells expressing both genes from at least one chromosome, these data argue that HLA-DRB1 and HLA-DQA1 are for the most part expressed at the same time, suggesting the possibility that both promoter regions could interact with XL9 simultaneously (Fig. 8).
Figure 8.
Transcription of HLA-DRB1 and HLA-DQA1 genes. RNA FISH for HLA-DRB1 and HLA-DQA1 mRNA in Raji cells displayed a variety of expression patterns, suggesting that the neighboring genes could be coexpressed. Three HLA-DRB1 and HLA-DQA1 coexpression patterns are shown. The complete analysis of all expression patterns is shown in Table I. PCR probes corresponding to the middle of each of these genes, including intronic sequences, were used. Cells are counterstained with DAPI to highlight the nucleus. No expression was observed in RJ2.2.5 cells (not depicted). Bars, 10 μM.
Table I.
Expression of MHC-II alleles and genes by RNA FISH
| Alleles expressed | Numbera | % |
|---|---|---|
| Neither | 3 | 1.3 |
| One DRB1 only | 22 | 9.9 |
| One DQA1 only | 0 | 0 |
| Two DRB1 only | 20 | 9 |
| Two DQA1 only | 0 | 0 |
| One of each: distinctb | 7 | 3.1 |
| One of each: coincident | 18 | 8.1 |
| Two DRB1 with one DQA1 | 101 | 45.3 |
| One DRB1 with two DQA1 | 2 | 0.9 |
| Two DRB1 with two DQA1 | 50 | 22.4 |
Based on 223 Raji cells observed from two independent experiments. No signal was observed in RJ2.2.5 cells.
Distinct and coincident refer to whether the HLA-DRB1 and HLA-DQA1 signals were separate or overlapping, respectively.
One prediction of these observations is that if the HLA-DRB1 and HLA-DQA1 promoter regions are being expressed at the same time, they may be in close proximity to each other when associated with XL9. This prediction would suggest that a 3C product could be detected between the two regions. To ascertain if this was the case, 3C was performed between the restriction fragments encoding the promoters of HLA-DRB1 and HLA-DQA1 (Fig. 5 C). When 3C interactions were compared between Raji and RJ2.2.5 cells, a 20.6-fold difference was observed. The lack of 3C products in RJ2.2.5 cells demonstrated that CIITA was necessary and that the interaction was specific. Although the overall magnitude observed for the 3C product in Raji cells in Fig. 5 C was not as robust as the 3C products formed between the promoter fragments and XL9 (Fig. 5 B), this was expected because the promoter–promoter interactions (Fig. 5 C) require two independent interaction events to occur, whereas the promoter–XL9 interactions require only a single event. Moreover, the RNA FISH data (Fig. 8 and Table I) indicate that not all chromosomes express both HLA-DRB1 and HLA-DQA1 simultaneously.
DISCUSSION
We demonstrate in this paper that XL9 forms interactions with the promoter regions of the two flanking MHC-II genes: HLA-DRB1 and HLA-DQA1. These interactions span ∼25 and 15 kb, respectively, and likely form a loop-type structure connecting the promoter regions of these genes to XL9 (Fig. 9). The interactions were only observed in cells expressing MHC-II gene products constitutively or when induced with IFN-γ. Importantly, the interactions were dependent on the presence of factors that interact with XL9 (CTCF) and the proximal regulatory region of the MHC-II genes CIITA and RFX. Interactions between CIITA and CTCF or complexes containing these factors were observed. The results therefore describe a new model for the control of MHC-II gene expression and a novel role for CTCF in regulating MHC-II gene activity.
Figure 9.
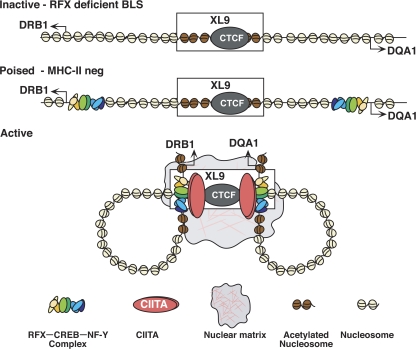
Models of interactions between HLA-DRB1 and HLA-DQA1 and the XL9 region. Three potential states are shown. The inactive state represents RFX-deficient cell types as in SJO cells from bare lymphocyte syndrome patients with no factors bound to the MHC-II proximal regulatory regions. In the inactive state, no interactions with XL9 occur and the proximal regulatory regions of MHC-II genes display background chromatin marks associated with the gene activation. A poised state is suggested for normal MHC-II–nonexpressing cells. In such cells, RFX–CREB–NF-Y but not CIITA are assembled at the MHC-II proximal regulatory regions. No interactions with XL9 occur in the poised state, but chromatin marks associated with genes that can be activated are present. In MHC-II–expressing cells, CIITA is expressed and an “active state” is proposed with the proximal promoter regions of the HLA-DRB1 and/or HLA-DQA1 genes interacting directly with XL9. The potential for both genes to interact with XL9 is depicted, but it is possible that only one gene interacts or is transcribed at a time.
The current model predicts that several chromatin organizational states may exist: inactive, poised, and active. As depicted, the active state may occur in MHC-II constitutively expressing B lymphocytes and cells induced to express MHC-II genes by IFN-γ. The active state would feature interactions between XL9 and the promoter regions of the HLA-DRB1 and HLA-DQA1 genes. The data collected using the 3C assay represent a population view and not individual haplotypes or alleles. Thus, with a recent report suggesting that active genes do not continuously transcribe their DNA (40), it was possible that only one of the two flanking genes was transcribed at a time or interacting with XL9. However, the RNA FISH data argue that all combinations of expression occur, with the predominant combination including HLA-DRB1 and HLA-DQA1 expression from the same chromosome. The observation of a CIITA-dependent 3C product between the promoter regions of HLA-DRB1 and HLA-DQA1 suggests that both promoter regions are in close proximity when expressed, and supports the model of both promoters interacting with XL9. Most nonimmune cells are CIITA negative and do not express MHC-II genes. In such cells, RFX, CREB, and NF-Y are found bound to their X-Y box sites, as demonstrated by in vivo genomic footprinting and ChIP (37, 38, 41). ChIP analyses also demonstrated that the promoter region–associated nucleosomes in MHC-II–negative cells still bear accessibility marks, such as histone H3 lysine 42me (13). Such data suggest that MHC-II genes are likely organized in a poised state in cells that lack CIITA and do not express MHC-II genes. An inactive state may therefore only exist in bare lymphocyte syndrome patient cells that are deficient in one of the RFX subunits, as cells mutant for subunits of the RFX complex display unoccupied MHC-II regulatory regions (37, 41). All nucleosome modifications associated with accessible and active chromatin were near background levels in RFX-deficient cells (13, 38).
The model also predicts that RFX and CIITA should be detected at XL9 by ChIP. Although this was not observed under the standard conditions in previous experiments (14), extending the cross-linking time resulted in moderate detection of both RFX and CIITA at XL9 (unpublished data). The values were ∼5–20-fold less than the robust binding observed at X-Y box regions. This was expected, as RFX and CIITA binding to XL9 is indirect and would require multiple protein–protein cross-linking events to detect by ChIP.
CTCF siRNA knockdown was found to diminish all assayed activities associated with XL9 and HLA-DRB1 and HLA-DQA1 gene expression; however, the activities never approached background levels. This was most likely caused by the efficiency of transfection of the siRNAs. By intracellular staining, cells were either positive or negative for CTCF after 72 h. CTCF-negative cells did not display MHC-II on their surfaces. This suggests that the decreases in CTCF function assayed were likely greater than those observed in the quantitative assays in which populations were examined (RNA expression and histone modification), and argues that CTCF is critical for expression of these genes. Although one might predict, because of its binding to ∼13,000 sites in the human genome (15) and the effects of CTCF and cellular activation on inducing apoptosis in some B cell lines (21), that CTCF knockdown would result in numerous off-target effects, this was not the observed case. CTCF siRNA–transfected cells proliferated during the time course of assay, implying that a large number of genes were expressed, that the cells did not enter a quiescent phase, and that the cells did not induce a programmed cell death pathway in response to aberrant gene expression. This difference from a previous report (21) in which CTCF regulates cMyc activity, cell-cycle progression, and programmed cell death is likely caused by the Burkitt lymphoma translocation in Raji cells that removes cMyc from its normal regulatory environment (23). Additionally, of the 10 genes assayed, only the 2 HLA class II genes showed significant decreases in expression. This may be because of the fact that the affected genes are highly regulated and those not affected are likely to be constitutively expressed. This being said, we predict that many genes will ultimately be found that are regulated by CTCF in these cells.
How could XL9 contribute to the transcription of MHC-II genes? XL9 has several activities. Previously, we showed that when placed in a reporter assay vector, XL9 can block the activity of the SV40 enhancer with a similar efficiency as the insulator from the chicken β-globin locus (14). This suggested that in this context XL9 contains insulator activity. As with some other CTCF binding regions (42–44), XL9 associates also with the nuclear matrix (14). Here, we found that in a CTCF-dependent manner XL9 participates in the spatial reorganization/orientation of the promoters of the neighboring MHC-II genes when they are expressed through association with the MHC-II–specific transcription factors. We also found that a loss in CTCF activity correlated directly with a decrease in some histone modifications, including H3 K9 and K18 acetylation, as well as H3 K42me and K43me. These marks are associated with actively transcribing genes (45, 46) and suggest that interactions with XL9, which are also dependent on the presence of CTCF, function to increase histone acetyltransferase recruitment or activity, stabilize the modifications, or prevent histone deacetylases from removing the marks at the promoters of HLA-DRB1 and HLA-DQA1. These activities are consistent with two conclusions. The first is that XL9 is critical to the activation of the neighboring MHC-II genes whereas the second is that its activity is a consequence of active transcription. Although the current dataset argue for the former, the conclusion is not certain, as experimentation involving deletion of the element in human cells is not currently feasible.
In Drosophila, insulator elements like gypsy are responsible for the formation of chromatin loops that organize DNA in such a manner that enhancers only affect the genes within their own loop and not on adjacent loops (47). In Drosophila, the foci of the loops appear to be organized with respect to the nuclear matrix (47, 48). Some of the mechanistic aspects of this paradigm may exist here as well with the control of the HLA-DR and HLA-DQ gene expression occurring in separate chromatin loops. Although the HLA-DR and HLA-DQ genes are considered to be coregulated, they can be found to be discordantly regulated in certain non-Hodgkin's lymphomas (49, 50). Aberrant regulation of the XL9 element may allow these regulatory differences to be manifested.
In addition to functioning as an insulator at the chicken β-globin and mammalian Igf2/H19 loci, CTCF also functions in organizing higher-order chromatin structure. A recent report demonstrated that CTCF binding to the imprinting center of the Igf2/H19 locus enabled direct interactions with the Wsb/NF1 locus, bridging chromosomes 7 and 11 (19). Although it is not known whether XL9 forms additional intra- or interchromosomal interactions, it is unlikely that it functions to form bridges between expressed MHC alleles, as distinct foci were observed for the different expressed alleles. Microsatellite DNA in the second intron of the HLA-DRB1*0401 gene was found to bind CTCF in an in vitro electrophoretic mobility shift assay (51), suggesting that additional CTCF sites may exist in the human MHC. Although this site was not identified in a recent ChIP-on-chip analysis for CTCF binding in human cells, several other CTCF binding sites were observed within the MHC-II region (15). It is intriguing that these CTCF sites appear between clusters of MHC-II genes and, if active, may serve to coordinate the expression of the closest MHC-II gene in a manner similar to XL9. Alternatively, it may be possible that multiple CTCF binding sites may contribute to the regulation of the MHC-II genes. In either case, the regulation of this gene system may depend on having accessible CTCF binding sites to reorganize the chromatin structure, activate, and regulate transcription of the MHC-II gene system.
MATERIALS AND METHODS
Cell culture.
Raji cells, a Burkitt's lymphoma B cell line that is wild type for MHC-II expression, were obtained from the American Type Culture Collection. Derived by mutagenesis from Raji cells, RJ2.2.5 cells (provided by R. Accolla, University of Insubria, Varese, Italy) are mutant for CIITA (6, 26). RJ-CIITA are RJ2.2.5 cells that have been stably complemented with a CIITA expression vector (28). These three cell lines were grown in RPMI supplemented with 5% fetal bovine serum, 5% bovine calf serum (HyClone, Inc.), 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.29 mg/ml l-glutamine (Invitrogen). SJO cells were derived from a patient with the bare lymphocyte syndrome and are deficient for MHC-II expression because of mutations in their RFX5 genes (29). SJO cells were grown under the described conditions except that fetal bovine serum was used at 15%. A431 is an epithelial cell line that is negative for MHC-II expression. A431 cells were grown in DME with the described supplements and 10% fetal bovine serum. For some assays, A431 cells were treated with 500 U/ml IFN-γ for the time indicated in the figures to induce CIITA and MHC-II expression, as previously described (52).
Flow cytometry.
Cells were harvested, washed with PBS, and incubated with PE-conjugated anti–human HLA-DR antibody or unconjugated anti–human HLA-DQ antibody (BD Biosciences). Anti–HLA-DQ–stained cells were incubated with PE-conjugated anti–mouse IgG secondary antibody (SouthernBiotech). Cells were washed with PBS and fixed with 2% paraformaldehyde in PBS at room temperature for 10 min, followed by permeabilization with 0.05% saponin in PBS for 30 min. Permeabilized cells were incubated in blocking buffer (1% BSA, 5% normal goat serum, and 0.05% saponin in PBS) for 30 min at room temperature. Blocked cells were incubated with anti-CTCF (Chemicon) at 4°C overnight, and Alexa Fluor 488 goat anti–rabbit IgG (Invitrogen) was used to detect CTCF. Some CTCF-stained cells were washed and blocked as described and subsequently stained for RFX5 for 1 h using an RFX5 antibody (53) that was conjugated to Alexa Fluor 647 according to the manufacturer's directions (Invitrogen). Control stained samples were processed in parallel using only the secondary antibodies indicated in the figures. IgG Alexa Fluor antibodies were purchased from Invitrogen.
siRNA treatment.
siRNA treatment was performed to knockdown CTCF expression in Raji (wild-type) cells. SMART pool siRNAs (Dharmacon) specific for CTCF were transfected into 4 × 106 cells using a Nucleofection apparatus and transfection reagents (kit V) from Amaxa Biosystems. siRNA to GFP was transfected as a negative control. After transfection, siRNA-treated and -untreated cells were harvested at the indicated times. Each of these experiments was performed at least three times with separate transfections.
3C assay.
3C assays were performed as previously described (34), with some modifications. 107 cells were resuspended in 50 ml DME supplemented with 10% fetal bovine serum. Formaldehyde was added to the cells to a final concentration of 1% and incubated at room temperature for 10 min, after which the reactions were quenched by the addition of glycine to a final concentration of 0.125 M. The cells were lysed using ice-cold lysis buffer (0.34 M sucrose, 10 mM Tris, 10 mM NaCl, 1% NP-40) containing protease inhibitors. The nuclei were collected from the lysates and washed once with restriction enzyme buffer and resuspended in restriction enzyme buffer containing 0.1% SDS. The samples were incubated on a rotator for 10 min at 37°C. Triton X-100 was added to a final concentration of 1%, and the nuclei were incubated for an additional 10 min at 37°C to sequester the SDS. The cross-linked DNA was digested overnight with 1,000 U EcoRI. EcoRI was heat inactivated by incubation at 65°C for 20 min. Samples were diluted 1:40 with ligase buffer (New England Biolabs, Inc.) and 1% Triton X-100 and then incubated for 10 min at 37°C. T4 DNA ligase was added, and the samples were incubated for 4 h at 16°C, followed by a 30-min incubation at room temperature. 10 μg/ml of proteinase K was added to the ligation reactions and incubated overnight at 65°C to reverse the cross-links and digest the proteins. The DNA was extracted with phenol and chloroform and concentrated by ethanol precipitation. 50 ng of the precipitated DNA was analyzed by PCR, with an annealing temperature of 62°C for 30 s and 45-s extension times at 68°C. 35 cycles of PCR were performed. The locations of the primer sets used in these assays are shown in Fig. 4 A, and their sequences are provided in Fig. S5. HLA-DRB1 and XL9 3C products were detected using primers P-1 and P-2; HLA-DQA and XL9 interactions were detected using primers P-4 and P-3. Primers to demonstrate that nonspecific interactions were not occurring included NC-5 and NC-6, which were used with P-1 and P-3, respectively. Loading controls were derived by PCR amplification of sequences contained within a single restriction fragment using primers L-7 and L-8. All PCR products were analyzed on 1.5% agarose gels stained with ethidium bromide. Each of the PCR products was cloned and sequenced to confirm its identity. All 3C assays were repeated at least three times from independent cultures with identical results.
Quantitative 3C assays were performed as described using an XL9 anchor primer with each of the 23 primers described in Fig. S5 and real-time instrumentation (iCycler; Bio-Rad Laboratories). To derive primer sets that would work for all 23 fragments, a total of six XL9 anchor primers (Fig. 5 A, red arrowhead) were designed and tested with the restriction fragment–specific primers (Fig. 5 A, black arrowheads). Primer sequences are provided in Fig. S5. All real-time PCR primers were tested on human BAC DNA (RP11-257p24) that was EcoRI digested and religated to generate all possible products. Only primer sets that produced a single amplicon were used. To quantitate 3C products, a standard curve was generated for each PCR primer/anchor amplicon using a known amount of digested/ligated BAC DNA. Data were derived from three independent chromatin samples, averaged, normalized to the relative efficiency of amplification and ligation of the BAC DNA, and plotted with their standard deviation. The Student's t test comparing Raji and RJ2.2.5 samples was used to assess statistical significance for each 3C primer set.
In some experiments, CTCF siRNA–transfected cells were separated from nontransfected cells based on their loss of HLA-DR surface expression. In these experiments, 107 cells were transfected with CTCF siRNAs as described in the previous section. Cells were washed twice with cold 1× PBS and resuspended in 80 μl PBE buffer (1× PBS, 0.5% BSA, and 2mM EDTA). 20 μl of MACS HLA-DR microbeads (Miltenyi Biotec) were added and incubated at 4°C for 20 min. Magnetically labeled cells were resuspended twice with PBE buffer and collected by centrifugation at 300 g for 10 min. The separation column was simultaneously prepared by washing with PBE buffer. Cells were applied to the column, and the HLA-DR negative cells representing the siRNA-transfected cells were collected and used for the 3C assay as described.
ChIP assays.
ChIP assays were performed as described previously (13, 38). Immunoprecipitations in a 50-μl volume of sonicated chromatin were performed with antibodies against RFX5 (53), CIITA (38), the histone-specific antibodies indicated in the figures (Chemicon), or a TCR antibody (nonspecific control) at 4°C overnight. Chromatin–antibody complexes were incubated with 60 μl of protein A–sepharose beads in a final volume of 1 ml for another hour and washed eight times and prepared for PCR as previously described (13). All ChIP experiments were performed at least three times with independent siRNA-treated cell cultures.
To determine the amount of DNA associated with ChIP, quantitative real-time PCR was performed in a reaction containing SYBR green buffer (5% DMSO, 1× SYBR [BioWhittaker Molecular Applications], 0.04% gelatin, 0.3% Tween 20, 50 mM KCl, and 20 mM Tris [pH 8.3]), 3 mM MgCl2, 0.2 mM dNTP, and 100 nM of each primer using an iCycler with an optical assembly unit. PCR primers to analyze RFX5 and CIITA occupancy on the proximal promoter sequences of HLA-DRB1 and HLA-DQA1 genes were described previously (14). All of the primer sets used in this study produced a single specific amplicon. Quantitation of PCR products was determined by comparison to a standard curve produced with each primer set using a genomic DNA dilution series consisting of 500, 100, 20, 4, and 0.8 ng. A two-step PCR with denaturation at 95°C for 15 s and annealing and extension at 62°C for 1 min for 45 cycles was conducted. All real-time PCR reactions were performed in duplicate. Student's t tests were used to assess significance between samples.
Relative quantitative RT-PCR.
Total RNA was isolated using the RNeasy mini prep kit (QIAGEN). 2 μg RNA was DNase I treated and then reverse transcribed in a final volume of 20 μl using Superscript II RT (Invitrogen) and buffers from an RT-PCR kit (Applied Biosystems) according to the manufacturer's instructions. A negative control reaction lacking reverse transcription was performed with each RNA sample. After reverse transcription, sample volumes were increased to 200 μl with TE buffer, and 3 μl of the cDNA produced was used for subsequent quantitative PCR. PCR using primers for the GAPDH transcripts was conducted in each experiment for normalization. Primers used in these assays are listed in Fig. S5. Primers for CIITA and GAPDH were described previously (38). The threshold cycle values for all genes were normalized to the threshold cycle values determined for GAPDH. All real-time RT-PCR assays were repeated at least three times from independent RNA preparations. Student's t tests were used to assess significance differences between samples.
Coimmunoprecipitation and Western blotting.
For immunoprecipitation experiments, M-280 sheep anti–rabbit and anti–mouse magnetic beads (Invitrogen) were washed four times with the washing buffer containing 0.1% BSA in 1× PBS. 1.5 × 107 beads were incubated overnight with 3 μg of the appropriate antibody for each immunoprecipitation at 4°C. After two washes, beads were mixed with 100 μg of nuclear extract, which was prepared as previously described (54, 55), and the volume was adjusted 300 μl by addition of lysis buffer (50 mM Tris [pH 8], 150 mM NaCl, and 1% NP-40) plus protease inhibitors. Immunoprecipitation reactions were performed for 3 h at 4°C with gentle rotation. Protein-bound beads were washed four times with the described lysis buffer. After the last wash, the beads were resuspended in SDS-PAGE sample buffer. Precipitates were eluted from the beads by placement in a boiling bath for 5 min, separated by 6% SDS-PAGE, and analyzed by Western blotting. In some experiments, ethidium bromide or DNase I were used to determine whether the interactions were independent of DNA. 50 μg/ml ethidium bromide was added to the nuclear extract and incubated on ice for 30 min (30, 56). After incubation, the nuclear extract was cleared by centrifugation and used for coimmunoprecipitation. For DNase I treatment, the coimmunoprecipitation reaction was performed first and was followed by the addition of 30 U/ml DNase I (Roche) and a 5-min incubation at room temperature. The protein-bound beads were washed in lysis buffer and examined as described.
Western blots were performed according to standard protocols using 6 or 7.5% SDS-PAGE gels with transfer membranes (Immobilon-P; Millipore). After transfer, membranes were blocked for 1 h at room temperature in PBST (PBS, 0.05% Tween 20) and 5% nonfat dry milk. Membranes were incubated with the antibodies indicated in the figures diluted in PBST for 1 h. Anti-CTCF and β-actin were purchased from Millipore and Chemicon, respectively. After washing in blocking buffer, horseradish peroxidase–conjugated secondary antibodies were added in PBST containing 5% nonfat dry milk for another 1 h. Enhanced chemiluminescence substrate (Renaissance; DuPont) was added and the blots were exposed to film.
RNA FISH and immunofluorescence.
HLA-DRB1 and HLA-DQA1 direct-labeled FISH probes were prepared by nick translation of 1 μg of purified PCR products with either Spectrum orange or green dUTP (Abbott Laboratories) at 16°C for 2 h. Genomic DNA from Raji cells was amplified using the following PCR primers: HLA-DRB1, 5′-GCAGAGAAGCAGACACACAG-3′ and 5′-CCATACGGTTTAGGCAAAGGG-3′ (located in exons 3 and 6); and HLA-DQA1, 5′-ATGATTAAACGCTACAACTCTACCG-3′ and 5′-ATTTCCTTTATCCTAACTTATTTCTCTGTC-3′ (located in exons 2 and 5). Cells were seeded onto poly–l-lysine–coated microscope slides (Polysciences, Inc.). Approximately 106 cells in 1× PBS were applied per slide and allowed to settle for 10 min at room temperature. Slides were transferred to fixative (4% formaldehyde, 0.1% Triton X-100 in 1× PBS) for 10 min before washing twice for 2 min each in 1× PBS. Slides were dehydrated for 3 min each in 70 and 100% ethanol before air drying. Cells were washed twice for 2 min in 50% formamide, 2× SSC at room temperature, followed by two 3-min washes at 37°C in 50% formamide, 2× SSC. The probes were denatured and hybridized to the slides for 16 h at 37°C. The slides were washed twice in 50% formamide, 2× SSC at 42°C for 8 min each, followed by two washes in 2× SSC. The slides were treated in mounting media and counterstained with DAPI (Vector Laboratories). Images were collected using Axiovision software (version 4.0) on a microscope (Axiovert 200M; both from Carl Zeiss, Inc.).
Online supplemental material.
Figs. S1 and S2 extend the CTCF siRNA knockdown analysis, showing that the expression of several genes and that CIITA and RFX occupancy on MHC-II promoters were not affected by CTCF siRNA knockdown, respectively. Fig. S3 showed that the level of CIITA protein in the complemented RJ-CIITA cell line was similar to that in Raji cells. Fig. S4 displays the amplification efficiencies for the PCR amplicons used in the 3C assays from Fig. 4. Fig. S5 provides a table of primers used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071843/DC1.
Supplemental Material
Acknowledgments
We thank Drs. P. Wade, J. Lucchesi, D. Reines, C. Moran, B. Pollack, and M. Horne for helpful comments regarding this work.
This work was supported by National Institutes of Health grants to J.M. Boss (GM47310) and B.P. Chadwick (GM073120).
The authors have no conflicting financial interests.
Abbreviations used: 3C, chromatin conformation capture; BAC, bacterial artificial chromosome; ChIP, chromatin immunoprecipitation assay; CIITA, class II transactivator; CREB, cAMP response element binding protein; CTCF, CCCTC binding factor; FISH, fluorescence in situ hybridization; MHC-II, MHC class II; NF-Y, nuclear factor–Y; RFX, regulatory factor X; siRNA, small interfering RNA.
References
- 1.Germain, R.N., N.S. Braunstein, M.A. Brown, L.H. Glimcher, R.I. Lechler, J. McCluskey, D.H. Margulies, J. Miller, M.A. Norcross, W.E. Paul, et al. 1986. Structure and function of murine class II major histocompatibility complex genes. Mt. Sinai J. Med. 53:194–201. [PubMed] [Google Scholar]
- 2.Collins, T., A.J. Korman, C.T. Wake, J.M. Boss, D.J. Kappes, W. Fiers, K.A. Ault, M.A.J. Gimbrone, J.L. Strominger, and J.S. Pober. 1984. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc. Natl. Acad. Sci. USA. 81:4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reith, W., and B. Mach. 2001. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 19:331–373. [DOI] [PubMed] [Google Scholar]
- 4.Ting, J.P., and J. Trowsdale. 2002. Genetic control of MHC class II expression. Cell. 109(Suppl.):S21–S33. [DOI] [PubMed] [Google Scholar]
- 5.Boss, J.M., and P.E. Jensen. 2003. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr. Opin. Immunol. 15:105–111. [DOI] [PubMed] [Google Scholar]
- 6.Steimle, V., L.A. Otten, M. Zufferey, and B. Mach. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell. 75:135–146. [PubMed] [Google Scholar]
- 7.Steimle, V., C.-A. Siegrist, A. Mottet, B. Lisowska-Grospierre, and B. Mach. 1994. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 265:106–108. [DOI] [PubMed] [Google Scholar]
- 8.Leibundgut-Landmann, S., J.M. Waldburger, M. Krawczyk, L.A. Otten, T. Suter, A. Fontana, H. Acha-Orbea, and W. Reith. 2004. Mini-review: Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur. J. Immunol. 34:1513–1525. [DOI] [PubMed] [Google Scholar]
- 9.Van Ewijk, W., Y. Ron, J. Monaco, J. Kappler, P. Marrack, M. Le Meur, P. Gerlinger, B. Durand, C. Benoist, and D. Mathis. 1988. Compartmentalization of MHC class II gene expression in transgenic mice. Cell. 53:357–370. [DOI] [PubMed] [Google Scholar]
- 10.Dorn, A., H.J. Fehling, W. Koch, M. Le Meur, P. Gerlinger, C. Benoist, and D. Mathis. 1988. B-cell control region at the 5′ end of a major histocompatibility complex class II gene: sequences and factors. Mol. Cell. Biol. 8:3975–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masternak, K., N. Peyraud, M. Krawczyk, E. Barras, and W. Reith. 2003. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 4:132–137. [DOI] [PubMed] [Google Scholar]
- 12.Krawczyk, M., N. Peyraud, N. Rybtsova, K. Masternak, P. Bucher, E. Barras, and W. Reith. 2004. Long distance control of MHC class II expression by multiple distal enhancers regulated by regulatory factor X complex and CIITA. J. Immunol. 173:6200–6210. [DOI] [PubMed] [Google Scholar]
- 13.Gomez, J.A., P. Majumder, U.M. Nagarajan, and J.M. Boss. 2005. X box-like sequences in the MHC class II region maintain regulatory function. J. Immunol. 175:1030–1040. [DOI] [PubMed] [Google Scholar]
- 14.Majumder, P., J.A. Gomez, and J.M. Boss. 2006. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF binding enhancer-blocking element. J. Biol. Chem. 281:18435–18443. [DOI] [PubMed] [Google Scholar]
- 15.Kim, T.H., Z.K. Abdullaev, A.D. Smith, K.A. Ching, D.I. Loukinov, R.D. Green, M.Q. Zhang, V.V. Lobanenkov, and B. Ren. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 128:1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West, A.G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271–288. [DOI] [PubMed] [Google Scholar]
- 17.Hark, A.T., C.J. Schoenherr, D.J. Katz, R.S. Ingram, J.M. Levorse, and S.M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 405:486–489. [DOI] [PubMed] [Google Scholar]
- 18.Szabo, P.E., S.H. Tang, F.J. Silva, W.M. Tsark, and J.R. Mann. 2004. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol. Cell. Biol. 24:4791–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling, J.Q., T. Li, J.F. Hu, T.H. Vu, H.L. Chen, X.W. Qiu, A.M. Cherry, and A.R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 312:269–272. [DOI] [PubMed] [Google Scholar]
- 20.Burgess-Beusse, B., C. Farrell, M. Gaszner, M. Litt, V. Mutskov, F. Recillas-Targa, M. Simpson, A. West, and G. Felsenfeld. 2002. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. USA. 99(Suppl. 4):16433–16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi, C.F., A. Martensson, M. Mattioli, R. Dalla-Favera, V.V. Lobanenkov, and H.C. Morse III. 2003. CTCF functions as a critical regulator of cell-cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc. Natl. Acad. Sci. USA. 100:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasko, J.E., E.M. Klenova, J. Leon, G.N. Filippova, D.I. Loukinov, S. Vatolin, A.F. Robinson, Y.J. Hu, J. Ulmer, M.D. Ward, et al. 2001. Cell growth inhibition by the multifunctional multivalent zinc-finger factor CTCF. Cancer Res. 61:6002–6007. [PubMed] [Google Scholar]
- 23.Hamlyn, P.H., and T.H. Rabbitts. 1983. Translocation joins c-myc and immunoglobulin gamma 1 genes in a Burkitt lymphoma revealing a third exon in the c-myc oncogene. Nature. 304:135–139. [DOI] [PubMed] [Google Scholar]
- 24.Zhu, X.S., M.W. Linhoff, G. Li, K.C. Chin, S.N. Maity, and J.P. Ting. 2000. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell. Biol. 20:6051–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeSandro, A.M., U.M. Nagarajan, and J.M. Boss. 2000. Associations and interactions between bare lymphocyte syndrome factors. Mol. Cell. Biol. 20:6587–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Accolla, R.S. 1983. Human B cell variants immunoselected against a single Ia antigen subset have lost expression of several Ia antigen subsets. J. Exp. Med. 157:1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Accolla, R.S., L. Scarpellino, G. Carra, and J. Guardiola. 1985. Trans-acting element(s) operating across species barriers positively regulate expression of major histocompatibility complex class II genes. J. Exp. Med. 162:1117–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown, J.A., X.-F. He, S.D. Westerheide, and J.M. Boss. 1995. Characterization of the expressed CIITA allele in the class II MHC transcriptional mutant RJ2.2.5. Immunogenetics. 43:88–91. [DOI] [PubMed] [Google Scholar]
- 29.Steimle, V., B. Durand, B. Emmanuele, M. Zufferey, M.R. Hadam, B. Mach, and W. Reith. 1995. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome). Genes Dev. 9:1021–1032. [DOI] [PubMed] [Google Scholar]
- 30.Lai, J.S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA. 89:6958–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekker, J. 2006. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat. Methods. 3:17–21. [DOI] [PubMed] [Google Scholar]
- 32.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science. 295:1306–1311. [DOI] [PubMed] [Google Scholar]
- 33.Spilianakis, C.G., and R.A. Flavell. 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5:1017–1027. [DOI] [PubMed] [Google Scholar]
- 34.Tolhuis, B., R.J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 10:1453–1465. [DOI] [PubMed] [Google Scholar]
- 35.Spilianakis, C.G., M.D. Lalioti, T. Town, G.R. Lee, and R.A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature. 435:637–645. [DOI] [PubMed] [Google Scholar]
- 36.Lee, G.R., C.G. Spilianakis, and R.A. Flavell. 2005. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat. Immunol. 6:42–48. [DOI] [PubMed] [Google Scholar]
- 37.Kara, C.J., and L.H. Glimcher. 1991. In vivo footprinting of MHC class II genes: bare promoters in the bare lymphocyte syndrome. Science. 252:709–712. [DOI] [PubMed] [Google Scholar]
- 38.Beresford, G.W., and J.M. Boss. 2001. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat. Immunol. 2:652–657. [DOI] [PubMed] [Google Scholar]
- 39.Oestreich, K.J., R.M. Cobb, S. Pierce, J. Chen, P. Ferrier, and E.M. Oltz. 2006. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 24:381–391. [DOI] [PubMed] [Google Scholar]
- 40.Osborne, C.S., L. Chakalova, K.E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J.A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065–1071. [DOI] [PubMed] [Google Scholar]
- 41.Kara, C.J., and L.H. Glimcher. 1993. Developmental and cytokine-mediated regulation of MHC class II gene promoter occupancy in vivo. J. Immunol. 150:4934–4942. [PubMed] [Google Scholar]
- 42.Yusufzai, T.M., and G. Felsenfeld. 2004. The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA. 101:8620–8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yusufzai, T.M., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 13:291–298. [DOI] [PubMed] [Google Scholar]
- 44.Heng, H.H., S. Goetze, C.J. Ye, G. Liu, J.B. Stevens, S.W. Bremer, S.M. Wykes, J. Bode, and S.A. Krawetz. 2004. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J. Cell Sci. 117:999–1008. [DOI] [PubMed] [Google Scholar]
- 45.Jenuwein, T., and C.D. Allis. 2001. Translating the histone code. Science. 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 46.Lachner, M., R.J. O'Sullivan, and T. Jenuwein. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117–2124. [DOI] [PubMed] [Google Scholar]
- 47.Byrd, K., and V.G. Corces. 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerasimova, T.I., K. Byrd, and V.G. Corces. 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 6:1025–1035. [DOI] [PubMed] [Google Scholar]
- 49.Guy, K., A.S. Krajewski, and A.E. Dewar. 1986. Expression of MHC class II antigens in human B-cell leukaemia and non-Hodgkin's lymphoma. Br. J. Cancer. 53:161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, M.E., C.S. Holgate, J.M. Williamson, I. Grigor, P. Quirke, and C.C. Bird. 1987. Major histocompatibility complex class II antigen expression in B and T cell non-Hodgkin's lymphoma. J. Clin. Pathol. 40:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold, R., W. Maueler, G. Bassili, M. Lutz, L. Burke, T.J. Epplen, and R. Renkawitz. 2000. The insulator protein CTCF represses transcription on binding to the (gt)(22)(ga)(15) microsatellite in intron 2 of the HLA-DRB1(*)0401 gene. Gene. 253:209–214. [DOI] [PubMed] [Google Scholar]
- 52.Nagarajan, U.M., A. Bushey, and J.M. Boss. 2002. Modulation of gene expression by the MHC class II transactivator. J. Immunol. 169:5078–5088. [DOI] [PubMed] [Google Scholar]
- 53.Moreno, C.S., E.M. Rogers, J.A. Brown, and J.M. Boss. 1997. RFX, a bare lymphocyte syndrome transcription factor, is a multimeric phosphoprotein complex. J. Immunol. 158:5841–5848. [PubMed] [Google Scholar]
- 54.Shapiro, D.J., P.A. Sharp, W.W. Wahli, and M.J. Keller. 1988. A high-efficiency HeLa cell nuclear transcription extract. DNA. 7:47–55. [DOI] [PubMed] [Google Scholar]
- 55.Hasegawa, S.L., and J.M. Boss. 1991. Two distinct B cell factors bind the HLA-DRA X box region and recognize different subsets of MHC class II promoters. Nucleic Acids Res. 19:6269–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lochamy, J., E.M. Rogers, and J.M. Boss. 2007. CREB and phospho-CREB interact with RFX5 and CIITA to regulate MHC class II genes. Mol. Immunol. 44:837–847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.