Abstract
A whole blood peptide mapping intracellular cytokine staining (ICS) assay was developed that allows the direct comparison, at the individual peptide level, of CD4+ and CD8+ T-cell responses that span every encoded protein, in patients infected with HIV-1. Whole blood samples from HIV-1 infected patients were stimulated with overlapping synthetic peptides spanning nine subtype C HIV-1 gene regions (Gag, Pol, Nef, Env, Tat, Rev, Vif, Vpu, Vpr). Following stimulation and permeabilization, cells were stained with fluorochrome labelled antibodies to CD3, CD8 (CD4+ cells were defined as CD8 negative cells), and IL-2 and IFN-γ. A total of 396 overlapping peptides were arranged in pools with a matrix design which allowed the identification of individual peptide responses from multiple pool responses. HIV-1 infected patients screened using this method showed a broad range of peptide responses across the entire HIV-1 genome with CD8 T-cell responses being higher in frequency in magnitude than CD4+ T-cell responses. The advantages of this whole blood ICS assay include the following: (1) the response to all potential HIV-1 epitopes across the genome can be examined, (2) the responding cell type can be monitored in the same reaction, and (3) considerably less blood is required than would be necessary if peripheral blood mononuclear cells (PBMC) were first isolated prior to peptide stimulation.
Keywords: Intracellular cytokine staining assay, CD4+ and CD8+ T-cell responses, Peptide mapping, HIV-1
1. Introduction
Cellular immune responses appear to play an important role in control of viral replication and thereby may influence the clinical outcome of HIV-1 infected individuals. CD4+ T-cells coordinate effector immune responses (Kalams and Walker, 1998; Rosenberg and Walker, 1998), which involves both the induction and maintenance of B-cell functions (Letvin, 1998) as well as CD8+ cytotoxic T-lymphocyte (CTL) responses (Matloubian et al., 1994; Kalams and Walker, 1998). Vigorous HIV-1-specific CD4+ T-helper cell responses have been observed among long-term non-progressors (Rosenberg et al., 1997; Pitcher et al., 1999; Pontesilli et al., 1999; Alatrakchi et al., 2002), and have been found to be inversely correlated with viral load (Rosenberg et al., 1997; Musey et al., 1999; Pitcher et al., 1999), suggesting that CD4+ T-cells are critical in mediating an effective antiviral defence. HIV-1-specific CTL responses also play a central role in controlling viral replication (Koup et al., 1994; Koenig et al., 1995; Goulder et al., 1997; Price et al., 1997), by direct cytolysis of infected cells and by suppression of viral replication by secreted factors such as perforin, interferon-γ (IFN-γ), and the CC chemokines CCL3, CCL4, and CCL5 (Walker et al., 1986; Cocchi et al., 1995).
The quantification of antigen-specific T-cell responses to pathogens or immunogens has been greatly facilitated in recent years by the development of methods based on the detection of various cytokines, including assays such as enzyme-linked immunospot (ELISPOT) and intracellular cytokine staining (ICS) (Lalvani et al., 1997; Larsson et al., 1999; Betts et al., 2000; Gea-Banacloche et al., 2000). ELISPOT and ICS assays use short term in vitro stimulations to determine the frequency and cytokine profiles of responding cells. In addition, the tetramer assay was developed to directly stain CTL with a defined T-cell receptor (Altman et al., 1996). The ELISPOT assay was originally developed to detect antibody-producing cells (Klinman, 1992), but has subsequently been adapted to quantify viral-specific T-cell responses. Although the ELISPOT assay is valuable for detecting low-frequency T-cell responses, the enumeration of spots can be a subjective process due to different operators. While computer-aided counting apparatus has improved the efficiency and accuracy of spot counting, the definition of a positive spot (correct size and shape) has to still be made by the individual. In addition, the different cell types secreting the cytokine of interest in an ELISPOT assay are not easily discernable, although magnetic bead depletions of peripheral blood mononuclear cells (PBMC) prior to stimulation allows the testing of specific cell populations. The ICS assay allows rapid quantitation of a large number of cytokine-producing cells while the multi-parameter capability of flow cytometry allows determination of cell-specific production of individual cytokines. Methods aimed at mapping T-cell responses at the peptide level usually involve screening with an ELISPOT assay, with ICS used to confirm responses and identify responding cell types. The aim of this study was to develop an assay that allows the direct comparison, at the individual peptide level, of HIV-1-specific CD4+ and CD8+ T-cell responses that span every encoded protein, using as small a volume of blood as possible. We therefore evaluated a whole blood ICS assay using overlapping peptides arranged in a pool and matrix design to measure the breadth of T-cell responses across the genome in HIV-1 infected patients. This approach allows significant dissection of cellular immune responses in HIV-1 infected patients, and could help in understanding protective correlates in HIV-1 infection.
2. Materials and methods
2.1. Patient samples
To evaluate the whole blood intracellular cytokine staining assay, peripheral blood samples were collected from HIV-1 infected women (CD4 T-cell count > 200 cells/μl) at Chris Hani Baragwanath Hospital in Soweto, South Africa. Approximately 16 ml of blood was drawn into Sodium Heparin vacutainers and processed within 6 h of collection. Written informed consent was obtained from all study participants and the study was approved by the Institutional Review Boards of the investigators.
2.2. Synthetic HIV-1 peptides
A total of 396 overlapping synthetic peptides spanning nine HIV-1 subtype C gene regions (Gag, Pol, Nef, Env, Tat, Rev, Vif, Vpu, Vpr) were used in this study, arranged in pools with a matrix design which allowed identification of individual peptide responses, as described previously for ELISPOT (Masemola et al., 2004). The peptides were supplied by the South African AIDS Vaccine Initiative (SAAVI) Peptide Repository (Immunology Lab., AIDS Virus Research Unit, National Institute for Communicable Diseases, Johannesburg, South Africa). The peptides were synthesized using 9-fluorenylmethoxy carbonyl-based solid phase chemistry at the Natural and Medical Sciences Institute (University of Tübingen, Tübingen, Germany), and checked for correct molecular weight by Elektrospray quadruple time-of-flight (QTOF)-mass spectrometry. Table 1 shows details of the pools for each of the nine gene regions while Table 2 shows the arrangement of the peptides in each matrix. Some of the peptides, including Pol, Nef, Env (gp160), Tat, and Rev were designed to match regions that were selected for inclusion in clade C HIV-1 vaccine candidates (Du151 and Du179) (Williamson et al., 2003), while Gag, Vif, Vpu, and Vpr peptides were based on a consensus HIV-1 subtype C sequence. Gag, Pol, Env, Tat, Rev, Vif, Vpu, Vpr peptides varied from 15- to 18-mers overlapping at 10 amino acids, while Nef peptides were synthesized as 15-mers overlapping at 11 amino acids. The synthetic peptides were originally dissolved in 100% dimethyl sulphoxide (DMSO) at 10 mg/ml, and were then aliquoted into the pools and matrices at 40 μg/ml in phosphate-buffered saline (PBS).
Table 1.
Pool arrangement for each of the nine HIV-1 subtype C gene regions and the arrangement of peptides in each matrix
| Gene region | Pool 1 | Pool 2 | Pool 3 | Pool 4 | Pool 5 |
|---|---|---|---|---|---|
| Gag | 1–14 | 15–28 | 29–42 | 43–56 | 57–70 |
| Pol | 1–24 | 25–48 | 49–72 | 73–92 | |
| Nef | 1–10 | 11–20 | 21–30 | 31–40 | 41–50 |
| Env | 1–24 | 25–48 | 49–72 | 73–96 | 97–114 |
| Tat | 1–12 | ||||
| Rev | 1–14 | ||||
| Vif | 1–12 | 13–24 | |||
| Vpr | 1–11 | ||||
| Vpu | 1–9 | ||||
| Matrix | Peptides in matrix | Matrix | Peptides in matrix | ||
|
| |||||
| 1 | Pol 1, 25, 49, 73; Rev 5; Vpu 3; Vif 18; Env 7, 31, 55, 79, 103 | 25 | Gag 1, 15, 29, 43, 57 | ||
| 2 | Pol 2, 26, 50, 74; Rev 6; Vpu 4; Vif 19; Env 8, 32, 56, 80, 104 | 26 | Gag 2, 16, 30, 44, 58 | ||
| 3 | Pol 3, 27, 51, 75; Rev 7; Vpu 5; Vif 20; Env 9, 33, 57, 81, 105 | 27 | Gag 3, 17, 31, 45, 59 | ||
| 4 | Pol 4, 28, 52, 76; Rev 8; Vpu 6; Vif 21; Env 10, 34, 58, 82, 106 | 28 | Gag 4, 18, 32, 46, 60 | ||
| 5 | Pol 5, 29, 53, 77; Rev 9; Vpu 7; Vif 22; Env 11, 35, 59, 83, 107 | 29 | Gag 5, 19, 33, 47, 61 | ||
| 6 | Pol 6, 30 54, 78; Rev 10; Vpu 8; Vif 23; Env 12, 36, 60, 84, 108 | 30 | Gag 6, 20, 34, 48, 62 | ||
| 7 | Pol 7, 31, 55, 79; Rev 11; Vpu 9; Vif 24; Env 13, 37, 61, 85, 109 | 31 | Gag 7, 21, 35, 49, 63 | ||
| 8 | Pol 8, 32, 56, 80; Rev 12; Vif 1; Vpr 1; Env 14, 38, 62, 86, 110 | 32 | Gag 8, 22, 36, 50, 64 | ||
| 9 | Pol 9, 33, 57, 81; Rev 13; Vif 2; Vpr 2; Env 15, 39, 63, 87, 111 | 33 | Gag 9, 23, 37, 51, 65 | ||
| 10 | Pol 10, 34, 58, 82; Rev 14; Vif 3; Vpr 3; Env 16, 40, 64, 88, 112 | 34 | Gag 10, 24, 38, 52, 66 | ||
| 11 | Pol 11, 35, 59, 83; Tat 1; Vif 4; Vpr 4; Env 17, 41, 65, 89, 113 | 35 | Gag 11, 25, 39, 53, 67 | ||
| 12 | Pol 12, 36, 60, 84; Tat 2; Vif 5; Vpr 5; Env 18, 42, 66, 90, 114 | 36 | Gag 12, 26, 40, 54, 68 | ||
| 13 | Pol 13, 37, 61, 85; Tat 3; Vif 6; Vpr 6; Env 19, 43, 67, 91 | 37 | Gag 12, 27, 41, 55, 69 | ||
| 14 | Pol 14, 38, 62, 86; Tat 4; Vif 7; Vpr 7; Env 20, 44, 68, 92 | 38 | Gag 14, 28, 42, 56, 70 | ||
| 15 | Pol 15, 39, 63, 87; Tat 5; Vif 8; Vpr 8; Env 21, 45, 69, 93 | 39 | Nef 1, 11, 21, 31, 41 | ||
| 16 | Pol 16, 40, 64, 88; Tat 6; Vif 9; Vpr 9; Env 22, 46, 70, 94 | 40 | Nef 2, 12, 22, 32, 42 | ||
| 17 | Pol 17, 41, 65, 89; Tat 7; Vif 10; Vpr 10; Env 23, 47, 71, 95 | 41 | Nef 3, 13, 23, 33, 43 | ||
| 18 | Pol 18, 42, 66, 90; Tat 8; Vif 11; Vpr 11; Env 24, 48, 72, 96 | 42 | Nef 4, 14, 24, 34, 44 | ||
| 19 | Pol 19, 43, 67, 91; Tat 9; Vif 12; Env 1, 25, 49, 73, 97 | 43 | Nef 5, 15, 25, 35, 45 | ||
| 20 | Pol 20, 44, 68, 92; Tat 10; Vif 13; Env 2, 26, 50, 74, 98 | 44 | Nef 6, 16, 26, 36, 46 | ||
| 21 | Pol 21, 45, 69; Rev 1; Tat 11; Vif 14; Env 3, 27, 51, 75, 99 | 45 | Nef 7, 17, 27, 37, 47 | ||
| 22 | Pol 22, 46, 70; Rev 2; Tat 12; Vif 15; Env 4, 28, 52, 76, 100 | 46 | Nef 8, 18, 28, 38, 48 | ||
| 23 | Pol 23, 47, 71; Rev 3; Vpu 1; Vif 16; Env 5, 29, 53, 77, 101 | 47 | Nef 9, 19, 29, 39, 49 | ||
| 24 | Pol 24, 48, 72; Rev 4; Vpu 2; Vif 17; Env 6, 30, 54, 78, 102 | 48 | Nef 10, 20, 30, 40, 50 | ||
Table 2.
Representative patient data showing the positive pools and matrices, identified peptides and spanned amino acid regions
| Positive response
|
Identified peptide | Amino acid region spanned | |
|---|---|---|---|
| Pool | Matrix | ||
| CD4+ responses | |||
| Gag 4 | 31 | Gag 49 | SHKARVLAEAMSQANSA |
| Env 2 | 11–13 | Env 41, 42, 43 | IRIGPGQAFYTNHIIGDIRQAYCNISKQEWNK |
| Env 2 | 16, 17 | Env 46, 47 | KKLQEHFPNKTIKFNSSSGGDLEI |
| Nef 2 | 45 | Nef 17 | EVGFPVRPQVPLRPM |
| Tat | 11–13 | Tat 1, 2, 3 | MEPIDPNLEPWNHPGSQPNTPCNNCYCKHCSYH |
| Tat | 16, 17 | Tat 6, 7 | TKGLGISYGRKKRRQRRSTPPSSEDH |
| CD8+ responses | |||
| Gag 1 | 27 | Gag 3 | EKIRLRPGGKKHYMLKHL |
| Gag 1 | 34, 35 | Gag 10, 11 | QLQPALQTGTEELRSLYNTVATLY |
| Gag 1 | 38 | Gag 14 | IEVRDTKEALDKIEEEQNK |
| Gag 2 | 27 | Gag 17 | KAAADKGKVSQNYPIV |
| Gag 2 | 34, 35 | Gag 24, 25 | PMFTALSEGATPQDLNTMLNTVGGH |
| Gag 2 | 38 | Gag 28 | LKDTINEEAAEWDRLHPV |
| Pol 2 | 6, 7 | Pol 30, 31 | FWEVQLGIPHPAGLKKKKSVTVLDV |
| Pol 2 | 12–17 | Pol 36, 37, 38, 39, 40, 41 | PSINNETPGIRYQYNVLPQGWKGSPAIFQASMTKILEPFRAKNPEIVIY |
| Pol 4 | 6, 7 | Pol 78, 79 | TFYVDGAANRETKIGKAGYVTDRGR |
| Pol 4 | 12–17 | Pol 84, 85, 86, 87, 88, 89 | LALQDSESEVNIVTDSQYALGIIQAQPDRSESELVNQIIEQLIKKERAYLSWVPAHK |
| Env 2 | 6, 7 | Env 36, 37 | IRSENLTNNVKTIIVHLNESIGIV |
| Env 2 | 12–17 | Env 42, 43, 44, 45, 46, 47 | FYTNHIIGDIRQAYCNISKQEWNKTLEEVRKKLQEHFPNKTIKFNSSSGGDLEI |
| Env 4 | 6, 7 | Env 84, 85 | MQWDREINNYTNIIYQLLEDSQI |
| Env 4 | 12–17 | Env 90, 91, 92, 93, 94, 95 | WFSITNWLWYIKIFIMIVGGLIGLRIIFAVLSIVNRVRQGYSPLSFQTLTPNPR |
| Nef 2 | 40 | Nef 12 | SNTAHNNPDCAWLQA |
| Nef 2 | 44–46 | Nef 16, 17, 18 | EEEEEVGFPVRPQVPLRPMTYKA |
| Vif 1 | 12–17 | Vif 5, 6, 7, 8, 9, 10 | RRANGWFYRHHYESRHPKVSSEVHIPLGEARLVIKTYWGLQTGERDWHLGHGVSIEW |
| Tat | 12–17 | Tat 2, 3, 4, 5, 6, 7 | LEPWNHPGSQPNTPCNNCYCKHCSYHCLVCFQTKGLGISYGRKKRRQRRSTPPSSEDH |
2.3. Whole blood stimulation and intracellular cytokine staining
To map CD4+ and CD8+ T-cell responses in HIV-1 infected individuals, 200 μl of whole blood from HIV-1 patients were aliquoted into a total of 75 1.5 ml Eppendorf tubes together with 1 μg each of the costimulatory antibodies against CD28 and CD49d (BD Biosciences, San Jose, CA, USA). Each of the 25 HIV-1 peptide pools and the 48 peptide matrices were then added to individual tubes containing the whole blood at a final concentration of 10 μg/ml. A negative control tube (anti-CD28 and anti-CD49d) to control for non-specific production of intracellular IFN-γ and IL-2, as well as a positive control of Staphylococcus enterotoxin B (SEB); final concentration 1 μg/ml were also used for each experiment. In addition, the secretion inhibitor Brefeldin A (Sigma–Aldrich Corp., St. Louis, MI, USA) was added to each tube at 10 μg/ml final concentration. Samples were then gently mixed and incubated at 37 °C for 6 h.
Following stimulation, EDTA was added to all samples to arrest activation and to remove adherent cells. Samples were then transferred to FACS tubes and erythrocytes lysed by adding 2 ml 1× FACS lysing solution to each tube and allowed to incubate for 10 min (BD Biosciences). Samples were then centrifuged at 2000 rpm for 5 min, supernatants decanted, and the cells permeabilized using 500 μl 1× FACS Permeabilizing solution 2 (BD Biosciences) with a 10 min incubation at room temperature. Following permeabilization, cells were washed twice with 2 ml wash buffer (PBS containing 1% bovine serum albumin (BSA)) (Sigma–Aldrich Corp.) and 0.1% sodium azide (Sigma–Aldrich Corp.) (2000 rpm centrifugation for 5 min). A cocktail of fluorescent antibodies including CD3 allophycocyanin (APC), CD8 peridinin chlorophyll (PerCP), and interleukin-2 (IL-2) and interferon-γ (IFN-γ) phycoerythrin (PE) (BD Biosciences) were then added to each tube and incubated for 60 min at room temperature in the dark. After staining, the samples were washed with 2 ml wash buffer (2000 rpm centrifugation for 5 min), and resuspended in PBS containing 1% paraformaldehyde (Electron Microscopy Sciences, Pretoria, South Africa).
2.4. Flow cytometric acquisition and analysis of samples
Samples were acquired using a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems) within 24 h of staining with fluorescent antibodies. Where possible, 70 000 CD3+ cells were acquired per sample. Fig. 1 shows the gating procedure used in this study. The lymphocyte gate was based on the forward and side scatter characteristics of each sample. Since cells were not stained with a specific CD4 marker, CD4+ cells were defined as CD3+ CD8− cells, while CD8+ T-cells were identified as CD3+ CD8+ cells within the lymphocyte gate. All data was analyzed using FlowJo software (Tree Star, San Carlos, CA, USA).
Fig. 1.
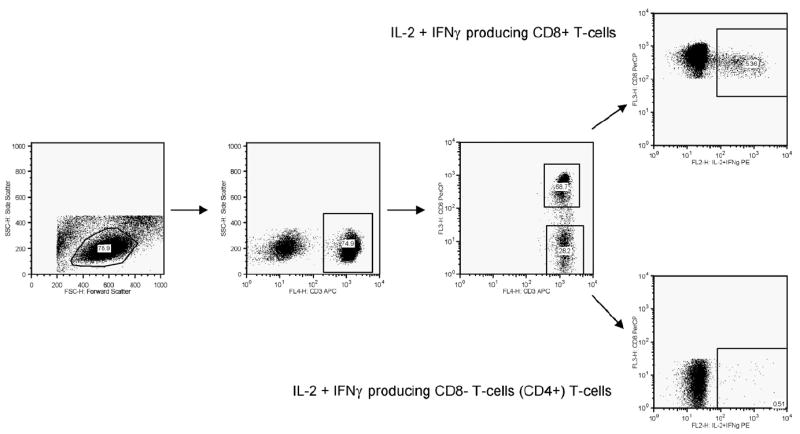
Representative data illustrating the gating strategy used in this study.
3. Results
The percentage of CD4+ and CD8+ T-cells responding to the peptides in a particular pool or matrix was determined using the whole blood ICS assay. Five HIV-1 infected individuals were screened for CD4+ and CD8+ T-cell responses across the HIV-1 genome. A positive response was considered as ≥0.1% IL-2 and IFN-γ producing cells after subtracting the background staining from cells stimulated with anti-CD28 and anti-CD49d in the absence of peptides. Detectable peptide responses identified in any of the major pools could be cross-referenced to a response in one of the matrix pools. In so doing, multiple peptide responses could be narrowed down to a single peptide response. In previous studies conducted in our laboratory measuring CD4+ and CD8+ T-cell responses to peptide pools representing entire HIV-1 genome regions, we have evaluated the production of different cytokines (IL-2, IFN-γ, TNF-α, MIP-1α/CCL3, MIP-1β/CCL4 and different combinations of these cytokines) in an attempt to determine optimal cytokine measurements to detect the strongest possible cellular responses. The combination of IL-2 and IFN-γ provided the greatest magnitude of CD4+ and CD8+ T-cell responses compared to their individual use, and provided the best cocktail of cytokine antibodies to use for the whole blood ICS assay (Shalekoff, unpublished data).
Fig. 2 shows representative data for one of the patients tested showing a comparison of individual CD4+ and CD8+ peptide responses across the entire HIV-1 genome. As an example of determining individual peptide responses, this individual showed a detectable CD8+ response in Gag pool 2 (Gag peptides 15–28) (Table 1) and in Matrix 38 (Gag peptides 14, 28, 42, 56 and 70) (Table 1), with the common peptide between Gag pool 2 and Matrix 38 being Gag 28. Table 2 shows the CD4+ and CD8+ peptide responses obtained for this patient, showing all positive pools and matrices, the individual peptides identified from these combinations, and the amino acid regions spanned by these individual peptides or groups of peptides. Upon searching the Los Alamos HIV Molecular Immunology database (http://www.hiv.lanl.gov/content/immunology/index.html), a number of the peptides identified in this patient contained previously described CTL (CD8+) and T-helper (CD4+) T-cell epitopes. Overall, all patients screened showed a broad range of peptide responses across the entire HIV-1 genome with CD8+ T-cell responses being higher in magnitude and frequency than CD4+ T-cell responses.
Fig. 2.
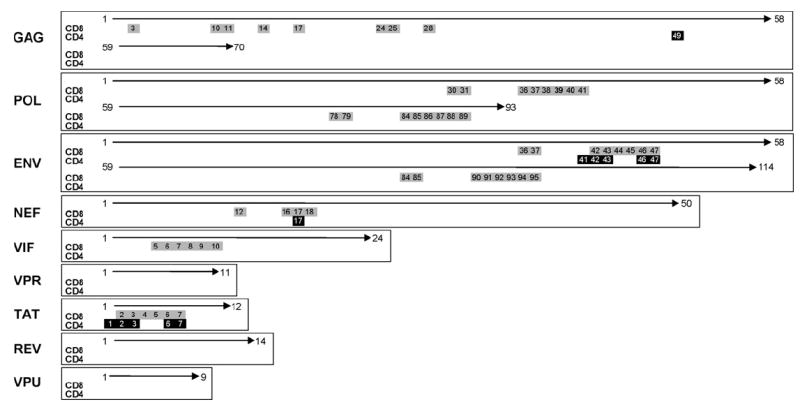
Representative patient data showing individual CD4+ and CD8+ peptide responses across the entire HIV-1 genome. Genome regions are shown on the left hand side of the figure with positive CD4+ responses (black boxes) and CD8+ responses (grey boxes) with peptide numbers indicated.
4. Discussion
The application of assays such as ELISPOT and ICS has greatly enhanced the understanding of the role cellular immune responses in HIV-1 infected individuals. Although certain reports have demonstrated inverse relationships between CD4+ (Rosenberg et al., 1997) and CD8+ (Ogg et al., 1998) T-cell responses and HIV-1 viral load, later studies evaluating HIV-1-specific T-cell responses to a variety of potential epitopes have failed to confirm these associations (Pitcher et al., 1999; Gea-Banacloche et al., 2000). Many studies to date using the ELISPOT and ICS assays to evaluate associations between cellular immune responses and viral parameters in HIV-1 infected individuals have however been restricted to responses against whole HIV-1 proteins (Gea-Banacloche et al., 2000), pools of peptides (Dalod et al., 1999), or single epitopes (Ogg et al., 1998). These approaches imply that certain responses to selected epitopes provide an illustration of the entire HIV-1-specific immune response, although findings by Betts et al. (2000) showing the correlation of particular restricting HLA alleles and epitope immunodominance is not absolute, contradict this assumption. In addition, the use of optimized epitopes will typically underestimate the total response, since only a few well-defined epitopes are used. In this study we developed a whole blood peptide mapping ICS assay that allows the direct comparison, at the individual peptide level, of CD4+ and CD8+ T-cell responses that span every encoded protein, in patients infected with HIV-1 to provide an appreciation of the overall immune response to HIV-1.
A total of 396 overlapping synthetic peptides spanning nine HIV-1 subtype C gene regions (Gag, Pol, Nef, Env, Tat, Rev, Vif, Vpu, Vpr) were used in this study. Peptides were arranged in pools with a matrix design, where detectable peptide responses identified in any of the major pools could be cross-referenced to a response in one of the matrices. In so doing, multiple peptide responses could be narrowed down to a single peptide response. Previously, virus or protein lysates, presented by MHC class II, have been used to detect CD4+-specific responses, while viral vector constructs expressing viral proteins (Nixon et al., 1988) and 8–11-mer peptides that mimic optimal epitopes have been used to measure CD8+-specific responses. Overlapping 15-mer peptides have been shown to stimulate both CD4+ and CD8+ T-cell responses (Kern et al., 2000; Maecker et al., 2001), circumventing the need to use separate antigens to detect CD4+ and CD8+ responses.
For detection of peptide-specific responses in this study, we have chosen to measure cells producing both IL-2 and IFN-γ, as opposed to measuring IFN-γ alone. De Rosa et al. (2004), using 12-colour flow cytometry, have illustrated significant limitations in using single functional measurements to evaluate immune responses, and have proposed that sensitive evaluation of antigen-specific T-cells requires the coordinated measurement of several cytokines. It has been shown that non-progressive HIV-1 infection is characterized by polyfunctional IL-2 and IFN-γ secreting CD4+ and CD8+ T-cell responses (Harari et al., 2004), while chronic, progressive HIV-1 infection is associated with a monofunctional T-cell response characterized by HIV-1-specific CD4+ and CD8+ T-cells that secrete IFN-γ (Champagne et al., 2001; Harari et al., 2004). Previous studies in our laboratory have shown that the combination of IL-2 and IFN-γ provided the greatest magnitude of CD4+ and CD8+ T-cell responses compared to using individual cytokine markers, which is in agreement with other studies showing IFN-γ secretion is typical of the early phase of immune response generation, but which decreases with antigen clearance, while IL-2 secretion is indicative of the long-term memory (predominantly CD4+) T-cell response.
Another important advantage of the whole blood ICS assay is that smaller blood volumes are required than would be necessary if PBMC were first isolated prior to peptide stimulation, as is the method of choice for ICS as cells can be stored. This is particularly advantageous in infant studies where blood volumes represent an important limiting factor to consider. The nature of the whole blood ICS assay also allows it to be modified in instances where blood volumes are limited, in order to assess cellular responses in specific regions of interest. Hanekom et al. (2004) have previously optimized an ICS assay to characterize BCG vaccination-induced CD4+ and CD8+ T-cell responses in infants using small volumes of whole blood, which they found to be sensitive and specific for detecting mycobacteria-specific immunity. We have previously used another whole blood assay to measure IFN-γ production in response to Mycobacterium tuberculosis antigens and BCG in infants born to HIV-1 infected mothers (Van Rie et al., 2006). In this assay, which was selected due to its relative simplicity and small blood volumes required, whole blood infants samples were stimulated with different antigens, following which IFN-γ release was measured in culture supernatants using ELISA. However, one major disadvantage of this whole blood assay was that the different cell types secreting the IFN-γ in response to the stimuli could not be identified. This is averted by the multi-parameter ability of an ICS assay, and with the development of flow cytometers which have the capacity to detect large numbers of different fluorochromes, flow cytometry is becoming a very powerful tool in measuring not only different responding cell types, but different cytokines and molecules produced in response to stimuli and the functional capacity of cells in a single reaction tube.
HIV-1 infected patients screened using the method developed in this study showed a broad range of peptide responses across the entire HIV-1 genome with CD8+ T-cell responses being higher in frequency in magnitude than CD4+ T-cell responses. The advantages of the whole blood ICS assay described here for peptide mapping include the following: (1) the response to potential HIV-1 epitopes across the genome can be examined, (2) the responding cell type can be monitored in the same reaction, and (3) that considerably less blood is required than would be necessary if peripheral blood mononuclear cells were first isolated prior to peptide stimulation. A disadvantage however of this method is that it is expensive to perform compared to other assays such as ELISPOT, due to the cost of the flow cytometer, fluorescent antibodies and other required reagents. In addition, although this assay is very useful in screening responses across the entire HIV-1 genome and allows the detection of stretches of targeted amino acids, the identification of optimal T-cell epitopes would require the patient to be re-bled for further testing which may not always be possible. Although the time taken to complete the stimulation, staining and acquisition of samples for an ICS peptide screening assay would be longer than completing an ELISPOT assay, time is saved on not having to isolate PBMC from whole blood. The ICS assay could also be adapted to use flow cytometric deep-well plates which would reduce the workload in performing this assay.
Overall, results indicate that the whole blood ICS peptide screening assay we have developed represents a useful approach to measuring HIV-1-specific T-cell responses across the entire HIV-1 genome, and could be a useful tool in dissecting immune parameters related to protective immunity in HIV-1 infected subjects. This whole blood ICS assay could also be adapted to monitoring immune responses (innate or adaptive) in defined cell subsets and studying immune responses to other viruses or organisms.
Acknowledgments
This study was supported by funding from SAAVI and by grants from NICHD (HD 42402, HD 36177) and the Wellcome Trust. CTT is a Wellcome Trust International Senior Research Fellow (076352/Z/05/Z). We would like to thank the medical and nursing staff at Chris Hani Baragwanath Hospital for patient recruitment and care.
References
- Alatrakchi N, Di Martino V, Thibault V, Autran B. Strong CD4 Th1 responses to HIV and hepatitis C virus in HIV-infected long-term non-progressors co-infected with hepatitis C virus. AIDS. 2002;16:713–717. doi: 10.1097/00002030-200203290-00006. [DOI] [PubMed] [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Betts MR, Casazza JP, Patterson BA, Waldrop S, Trigona W, Fu TM, Kern F, Picker LJ, Koup RA. Putative immunodominant human immunodeficiency virus-specific CD8(+) T-cell responses cannot be predicted by major histocompatibility complex class I haplotype. J Virol. 2000;74:9144–9151. doi: 10.1128/jvi.74.19.9144-9151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne P, Ogg GS, King AS, Knabenhans C, Ellefsen K, Nobile M, Appay V, Rizzardi GP, Fleury S, Lipp M, Forster R, Rowland-Jones S, Sekaly RP, McMichael AJ, Pantaleo G. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410:106–111. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- Dalod M, Dupuis M, Deschemin JC, Sicard D, Salmon D, Delfraissy JF, Venet A, Sinet M, Guillet JG. Broad, intense anti-human immunodeficiency virus (HIV) ex vivo CD8(+) responses in HIV type 1-infected patients: comparison with anti-Epstein-Barr virus responses and changes during antiretroviral therapy. J Virol. 1999;73:7108–7116. doi: 10.1128/jvi.73.9.7108-7116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa SC, Lu FX, Yu J, Perfetto SP, Falloon J, Moser S, Evans TG, Koup R, Miller CJ, Roederer M. Vaccination in humans generates broad T cell cytokine responses. J Immunol. 2004;173:5372–5380. doi: 10.4049/jimmunol.173.9.5372. [DOI] [PubMed] [Google Scholar]
- Gea-Banacloche JC, Migueles SA, Martino L, Shupert WL, McNeil AC, Sabbaghian MS, Ehler L, Prussin C, Stevens R, Lambert L, Altman J, Hallahan CW, de Quiros JC, Connors M. Maintenance of large numbers of virus-specific CD8+ T cells in HIV-infected progressors and long-term nonprogressors. J Immunol. 2000;165:1082–1092. doi: 10.4049/jimmunol.165.2.1082. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Hanekom WA, Hughes J, Mavinkurve M, Mendillo M, Watkins M, Gamieldien H, Gelderbloem SJ, Sidibana M, Mansoor N, Davids V, Murray RA, Hawkridge A, Haslett PA, Ress S, Hussey GD, Kaplan G. Novel application of a whole blood intracellular cytokine detection assay to quantitate specific T-cell frequency in field studies. J Immunol Methods. 2004;291:185–195. doi: 10.1016/j.jim.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Harari A, Petitpierre S, Vallelian F, Pantaleo G. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood. 2004;103:966–972. doi: 10.1182/blood-2003-04-1203. [DOI] [PubMed] [Google Scholar]
- Kalams SA, Walker BD. The critical need for CD4 help in maintaining effective cytotoxic T-lymphocyte responses. J Exp Med. 1998;188:2199–2204. doi: 10.1084/jem.188.12.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern F, Faulhaber N, Frommel C, Khatamzas E, Prosch S, Schonemann C, Kretzschmar I, Volkmer-Engert R, Volk HD, Reinke P. Analysis of CD8 T cell reactivity to cytomegalovirus using protein-spanning pools of overlapping pentadecapeptides. Eur J Immunol. 2000;30:1676–1682. doi: 10.1002/1521-4141(200006)30:6<1676::AID-IMMU1676>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Klinman DM. Analysis of B lymphocyte cross-reactivity at the single cell level. J Immunol Methods. 1992;152:217–225. doi: 10.1016/0022-1759(92)90143-h. [DOI] [PubMed] [Google Scholar]
- Koenig S, Conley AJ, Brewah YA, Jones GM, Leath S, Boots LJ, Davey V, Pantaleo G, Demarest JF, Carter C. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat Med. 1995;1:330–336. doi: 10.1038/nm0495-330. [DOI] [PubMed] [Google Scholar]
- Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalvani A, Brookes R, Hambleton S, Britton WJ, Hill AV, McMichael AJ. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson M, Jin X, Ramratnam B, Ogg GS, Engelmayer J, Demoitie MA, McMichael AJ, Cox WI, Steinman RM, Nixon D, Bhardwaj N. A recombinant vaccinia virus based ELISPOT assay detects high frequencies of Pol-specific CD8 T cells in HIV-1-positive individuals. AIDS. 1999;13:767–777. doi: 10.1097/00002030-199905070-00005. [DOI] [PubMed] [Google Scholar]
- Letvin NL. What immunity can protect against HIV infection? J Clin Invest. 1998;102:1643–1644. doi: 10.1172/JCI5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, Persaud N, Trigona W, Fu TM, Sinclair E, Bredt BM, McCune JM, Maino VC, Kern F, Picker LJ. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27–40. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- Masemola A, Mashishi T, Khoury G, Mohube P, Mokgotho P, Vardas E, Colvin M, Zijenah L, Katzenstein D, Musonda R, Allen S, Kumwenda N, Taha T, Gray G, McIntyre J, Karim SA, Sheppard HW, Gray CM the HIVNET 028 Study Team. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J Virol. 2004;78:3233–3243. doi: 10.1128/JVI.78.7.3233-3243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musey LK, Krieger JN, Hughes JP, Schacker TW, Corey L, McElrath MJ. Early and persistent human immunodeficiency virus type 1 (HIV-1)-specific T helper dysfunction in blood and lymph nodes following acute HIV-1 infection. J Infect Dis. 1999;180:278–284. doi: 10.1086/314868. [DOI] [PubMed] [Google Scholar]
- Nixon DF, Townsend AR, Elvin JG, Rizza CR, Gallwey J, McMichael AJ. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature. 1988;336:484–487. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon DF, McMichael AJ. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- Pitcher CJ, Quittner C, Peterson DM, Connors M, Koup RA, Maino VC, Picker LJ. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- Pontesilli O, Carotenuto P, Kerkhof-Garde SR, Roos MT, Keet IP, Coutinho RA, Goudsmit J, Miedema F. Lymphoproliferative response to HIV type 1 p24 in long-term survivors of HIV type 1 infection is predictive of persistent AIDS-free infection. AIDS Res Hum Retroviruses. 1999;15:973–981. doi: 10.1089/088922299310485. [DOI] [PubMed] [Google Scholar]
- Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg ES, Walker BD. HIV type 1-specific helper T cells: a critical host defence. AIDS Res Hum Retroviruses. 1998;14(Suppl 2):S143–S147. [PubMed] [Google Scholar]
- Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. Vigorous HIV-1-specific CD4+ T-cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- Van Rie A, Madhi SA, Heera JR, Meddows-Taylor S, Wendelboe AM, Anthony F, Violari A, Tiemessen CT. Gamma interferon production in response to Mycobacterium bovis BCG and Mycobacterium tuberculosis antigens in infants born to human immunodeficiency virus-infected mothers. Clin Vaccine Immunol. 2006;13:246–252. doi: 10.1128/CVI.13.2.246-252.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CM, Moody DJ, Stites DP, Levy JA. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- Williamson C, Morris L, Maughan MF, Ping LH, Dryga SA, Thomas R, Reap EA, Cilliers T, van Harmelen J, Pascual A, Ramjee G, Gray G, Johnston R, Karim SA, Swanstrom R. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res Hum Retroviruses. 2003;19:133–144. doi: 10.1089/088922203762688649. [DOI] [PubMed] [Google Scholar]


