Abstract
The membrane protein bacteriorhodopsin (BR) can be kept soluble in its native state for months in the absence of detergent by amphipol (APol) A8-35, an amphiphilic polymer. After an actinic flash, A8-35-complexed BR undergoes a complete photocycle, with kinetics intermediate between that in detergent solution and that in its native membrane. BR/APol complexes form well defined, globular particles comprising a monomer of BR, a complete set of purple membrane lipids, and, in a peripheral distribution, ∼2 g APol/g BR, arranged in a compact layer. In the absence of free APol, BR/APol particles can autoassociate into small or large ordered fibrils.
INTRODUCTION
In vitro study of the function and structure of membrane proteins (MPs) remains difficult. It is remarkable, for instance, that although they constitute ∼30% of coding sequences in genomes and 70% of drug targets, MPs still represent <1% of solved high-resolution structures (1,2). This is largely due to the challenge of obtaining stable preparations of MPs in their native state in sufficient amounts. Detergents are generally used to keep MPs soluble in water, but they tend to inactivate them. Among several new classes of surfactants that have been proposed to circumvent this difficulty, amphiphilic polymers named amphipols (APols) appear to be particularly promising in view of their universality and versatility (3,4). APols are comprised of a strongly hydrophilic backbone grafted with numerous hydrophobic chains that provide for multipoint attachment to MP transmembrane, hydrophobic surfaces. APols can substitute for detergents in keeping MPs soluble and in their native form in aqueous solution (4–6 and references therein). As a rule, APols, as compared to detergent solutions, enhance the stability of MPs (4,5), and they can be used to fold MPs to their active form (7), to immobilize them onto solid supports (D. Charvolin, J.-B . Perez, F. Rouvière, A. Abdine, F. Giusti, K. Martinez, and J.-L. Popot, unpublished data), and to study them by solution NMR (8) or cryoelectron microscopy (9). However, relatively scant information is available regarding the size, composition, structure, and solution properties of MP/APol complexes.
We report here on the functional and solution properties of APol-solubilized bacteriorhodopsin (BR), a light-driven proton pump from the purple membrane (PM) of the archebacterium Halobacterium salinarum. High-resolution 3D structures show BR to be folded into a bundle of seven transmembrane α-helices that surrounds the cofactor retinal and to which lipids are bound (10). The native state and functionality of BR can be asserted from the strongly red-shifted visible spectrum of retinal, which gives the protein its characteristic purple color, and from the photocycle it undergoes upon illumination. The APol used for this study, A8-35 (3), is a short polyacrylate grafted with octyle and isopropyle chains (Fig. 1). In aqueous solution, ∼4 molecules of A8-35 self-associate into ∼40-kDa globular particles with a hydrodynamic radius of ∼3 nm (11). BR was extracted from PM with octylthioglucoside (OTG) and transferred to either hydrogenated or partially deuterated (12) A8-35 (HAPol and DAPol, respectively). We have studied the functionality, composition, and structure of BR/A8-35 complexes, and the factors that control their homogeneity, using flash spectroscopy, chemical analysis, size-exclusion chromatography (SEC), analytical ultracentrifugation (AUC)—sedimentation velocity (SV) and sedimentation equilibrium (SE)—small angle neutron scattering (SANS), and electron microscopy (EM).
FIGURE 1.
Structure of amphipol A8-35, according to Tribet et al. (3). Molar fraction of each type of unit: x = 29–34%, y = 25–28%, and z = 39–44%.
EXPERIMENTAL PROCEDURES
Buffers
The buffers used were TpH (100 mM NaCl, 20 mM NaH2PO4/Na2HPO4, pH 7.1) and TpD. A known volume of TpH was lyophilized and rehydrated with the same volume of D2O.  (99% D, 75% 18O) was from Interchim (Montluçon, France). Its optical quality was poor (A280 = 0.25). Solubilization buffer contained 20 mM sodium phosphate, pH 7.1 and no salt, except in protocol 3: 20 mM potassium buffer, pH 7.1 (TpK).
(99% D, 75% 18O) was from Interchim (Montluçon, France). Its optical quality was poor (A280 = 0.25). Solubilization buffer contained 20 mM sodium phosphate, pH 7.1 and no salt, except in protocol 3: 20 mM potassium buffer, pH 7.1 (TpK).
Chemicals
OTG (n-octyl-β-d-thioglucopyranoside) was from Calbiochem (EMD Chemicals, San Diego, CA), and Bio-Beads SM2 Adsorbent from Bio-Rad (Marnes-la-Coquette, France). All organic solvents and acetic acid were from Carlo Erba (Val-de-Reuil, France), RPE grade.
Amphipols
The synthesis and characterization of HAPol (batches CT-961020H and FGH20) and DAPol (batches CT-980225D and CP-990520D) were described previously (11). In DAPol, octyle and isopropyle chains are perdeuterated. The synthesis of tritiated A8-35 is described in Supplementary Material (supporting information (S.I.)) 1.
Purple membrane purification and BR solubilization
H. salinarum cells (S9 strain, a gift of G. Zaccaï, IBS, Grenoble, France) were grown under illumination at 37°C in a liquid growth medium containing peptone (LP0037, Oxoid, Dardilly, France) prepared as in Oesterhelt and Stoeckenius (13). PM was isolated as in Lobasso et al. (14) and stored at −80°C. The next steps were performed at 4°C in the dark. Typically, a PM suspension containing 15–20 mg BR at 7 g L−1 was sonicated for 5 min and incubated for 40 h under constant magnetic stirring with 100 mM (30 g L−1) OTG (final concentration equivalent to >10 OTG micelles/BR monomer) in solubilization buffer. Solubilized BR was then briefly ultracentrifuged (20 min at 200,000 × g (TL100 ultracentrifuge, Beckman Coulter France, Roissy, France)). The concentration of solubilized BR was estimated using ɛ554 = 47 mM−1 cm−1 and ɛ280 = 81 mM−1 cm−1 (15,16).
Preparation of BR/A8-35 complexes.
Protocol 1
A8-35 from a 10% w/w stock solution in water was added to solubilized BR at 5:1 w/w APol/BR (1–2 g L−1 in 8–10 g L−1 OTG in solubilization buffer). After 15 min, detergent adsorption onto Bio-Beads SM2 (10 g/g OTG) was carried out for 2 h under gentle stirring. The Bio-Beads were removed and the ionic strength adjusted to 100 mM NaCl (TpH).
Protocol 2
The first steps were identical to Protocol 1 until after Bio-Bead removal. The BR/APol solution was then diluted 10× with TpH and concentrated by centrifugation for 24 h at 100,000 × g (10-mL tubes, 70.1 Ti Beckman rotor). The procedure was repeated by dilution of the concentrate to the initial volume and centrifugation for 17 h at 300,000 × g.
Protocol 3
After the solubilization step, BR in OTG was diluted three times with TpK to lower the OTG concentration, and then concentrated by centrifuging 18 h at 310,000 × g in a 70 Ti rotor (25-mL tubes), leading to [BR] ≥ 10 g L−1. After addition of APol and adsorption of the detergent onto Bio-Beads as in Protocol 1, the solution was diluted five times with TpK and centrifuged as before to concentrate the complexes and eliminate residual molecules of OTG and most of the unbound APol. The concentrate was diluted five times in either TpD or TpH and centrifuged (15 h at 370,000 × g) in 10-mL tubes (Beckman 70.1 Ti). The procedure was repeated twice in a TL100 ultracentrifuge (TLS 100.3 rotor, 3-mL tubes). The final samples (200 and 600 μL of BR/DAPol at ∼10 g L−1 in TpH and TpD, respectively) were used for SANS. After being stored frozen (−80°C), samples were concentrated and their buffer exchanged by ultrafiltration (Centricon 30, Amicon, Saint-Quentin-en-Yvelines, France) for a second set of measurements. We assume the amount of the detergent in the final samples to be insignificant, based on 1), parallel experiments in which the removal of OTG, in the absence of protein, was followed spectroscopically (not shown); 2), the absence of detectable traces of OTG in thin-layer chromatography (TLC) analyses of lipid content; 3), the good agreement of the contrast matching point (CMP) of the particles with that calculated assuming the complete absence of detergent (see Results); and 4), previous observations in which the removal of detergent was followed directly by either radiolabeling (3) or NMR (8). For instance, [14C]dodecylmaltoside, a detergent that is more difficult to remove than OTG due to its much lower critical micellar concentration, was eliminated to a level of <12 molecules per 228 kDa of cytochrome b6f complex using a much less drastic procedure than those used here in Protocol 2 or 3 (3).
Time-resolved absorption spectroscopy
The procedure has been described in a previous work (7). The samples ([BR] ≈ 0.12 g L−1), preilluminated for 4 min at 4°C to populate the light-adapted state, were excited at 640 nm by a 5-ns laser flash. Flash-induced transient absorption changes were monitored with a time resolution of 5 ns from 410 to 610 nm. The kinetics recorded at each wavelength were globally fitted with a sum of five exponential components, as described previously (17,18). The fitting procedure yields the halftime of each of the five components, and the amplitude for each wavelength. Plotting these amplitudes as a function of wavelength yields the spectra, commonly called decay-associated spectra (DAS), of the absorption changes specifically associated with each of these five components. From a mechanistic point of view, a sum of five exponential components corresponds to five successive irreversible reactions according to the scheme A0 → A1 → A2 → A3 → A4 → A5. Thus, DASi reflects the absorption changes resulting from the formation of Ai+1 at the expense of Ai, or, in other words, the (Ai − Ai+1) difference spectrum. For this reason, a DAS reflecting the formation of an absorbing species at the expense of a nonabsorbing species is expected to have negative amplitude. Since this is rather counterintuitive, the sign of the DASs plotted in Fig. 2 has been inverted with respect to the spectra yielded by the global fit analysis.
FIGURE 2.
Time-resolved absorption spectroscopy of BR in purple membrane (PM), solubilized in OTG, and trapped in A8-35. (A) UV-visible spectra of dark-adapted BR in purple membrane, in OTG solution, and after transfer to A8-35 (HAPol CT-961020H, protocol 1). (B) Scheme of BR photocycle (reprinted from Neutze et al. (53)). (C) Spectra of BR intermediates in PM (reprinted from Váró (54)). (D and E) Decomposition of the photocycles of PM, BR/OTG, and BR/A8-35 (HAPol batch FGH20, protocol 1) into five exponential components, each of them characterized by a decay-associated spectrum (DAS1 to DAS5 (D)) and a halftime (E).
Size-exclusion chromatography
SEC analysis was performed at 4°C on an Äkta Explorer 100 FPLC system (Pharmacia, GE Healthcare Europe, Orsay, France) equipped with a Superose 12 HR 10/30 column (Pharmacia) and a UV-visible detection system. A calibration curve (not shown) of the position of elution versus the Stokes radius (RS) was established as described previously (19), using a five-order polynomial fit. Standards, from Sigma Aldrich Chemie (Saint-Quentin Fallavier, France), were thyroglobulin (RS = 8.6 nm), β-galactosidase (6.9 nm), catalase (5.2 nm), aldolase (4.6 nm), apotransferrin (3.6 nm), albumin (3.5 nm), ovalbumin (2.8 nm), peroxidase (3.0 nm), trypsin inhibitor (2.2 nm), myoglobin (1.9 nm), and cytochrome c (1.7 nm). Samples of 60 μL containing 0.1–2 g L−1 protein were loaded onto the column equilibrated with TpH. For standards, concentrations were in the 2–5 g L−1 range with the same injection volume. The uncertainty of RS determinations is estimated to be ∼0.15 nm.
Lipid extraction and analysis
To separate BR complexes from free APol particles, which could contain unbound lipids, three 330-μL BR/HAPol samples ([BR] ≈ 4 g L−1) were layered onto 12–20% (w/w) 11-mL sucrose gradients (SW 41 rotor, Beckman) in TpH buffer and centrifuged for 91 h at 200,000 × g. The purple bands containing BR/APol complexes were collected and the buffer exchanged for 0.1 M NaCl by ultrafiltration on Centricon 10. Lipid extraction was performed using the Bligh and Dyer protocol modified for extreme halophiles (20,21). For reference, lipids were extracted in the same way from PM (4 mg BR). For TLC analyses, aliquots of each lipid extract were layered onto 12 cm × 10 cm silica gel plates (Kieselgel 60/Kieselgur F254, Merck, VWR International, Fontenay-sous-Bois, France) in chloroform/methanol/90% acetic acid (65:4:35 v/v/v). Assuming ∼10 lipids extracted per BR molecule, 2–20 nmol of lipids were applied per spot. Phospholipids were detected with molybdenum blue spray reagent (Sigma Aldrich), glycolipids with 0.5% orcinol/sulfuric acid, and total lipids with 10% sulfuric acid in ethanol, followed by charring at 120°C (21). Phospholipids, typically 10 nmol, were quantified by complete mineralization as described previously (22).
Analytical ultracentrifugation
AUC was carried out in XLA or XLI Beckman ultracentrifuges with Ti-60 or Ti-50 rotors. SV experiments used 3-mm or 12-mm optical path, two-channel centerpieces at 42,000, 50,000, or 60,000 rpm. SV profiles were typically recorded at 278 and 555 nm and, with the XLI, using interference optics. The solvent density, ρ0 (g mL−1), and viscosity, η0 (Pa s−1), were typically measured at 20°C with a DMA 5000 density meter and AMVn viscosity meter (Paar, Graz, Austria). Temperature corrections for viscosity were made using tabulated data for water. For an ideal solution, the sedimentation coefficient, s, in Svedberg (1 S = 10−13 s), is related to the buoyant molar mass, Mb, the Stokes radius, RS, and Avogadro's number NA by the Svedberg equation:
 |
(1) |
Mb depends on the particle molar mass, M, and partial specific volume,  in mL g−1:
in mL g−1:
 |
(2) |
A review of AUC typical methods of analysis can be found, e.g., in a previous work (23). For complex macromolecular systems, operational parameters for  φ′, and φ′R, defined previously (12), can be obtained from the chemical composition and from measurements in H2O and D2O solvents; corrected values of s in H2O and at 20°C, s20,w, were calculated using φ′ (instead of
φ′, and φ′R, defined previously (12), can be obtained from the chemical composition and from measurements in H2O and D2O solvents; corrected values of s in H2O and at 20°C, s20,w, were calculated using φ′ (instead of  in the usual formulae). SV profiles were globally analyzed with the program Sedfit (24,25). The analysis of SV profiles in terms of c(s) distribution allows the characterization of the sedimentation coefficients of the different types of particles in solution. The c(s) distributions are obtained with high resolution, because the effects of diffusion broadening are considered. Concentrations were derived from the area under the peaks of the c(s) distributions from interference data, using the increment of index of refraction ∂n/∂c = 0.186 g mL−1 for BR and 0.15 g mL−1 for lipid and APol (12). Modeling of s for bead assemblies was done with HYDRO (26).
in the usual formulae). SV profiles were globally analyzed with the program Sedfit (24,25). The analysis of SV profiles in terms of c(s) distribution allows the characterization of the sedimentation coefficients of the different types of particles in solution. The c(s) distributions are obtained with high resolution, because the effects of diffusion broadening are considered. Concentrations were derived from the area under the peaks of the c(s) distributions from interference data, using the increment of index of refraction ∂n/∂c = 0.186 g mL−1 for BR and 0.15 g mL−1 for lipid and APol (12). Modeling of s for bead assemblies was done with HYDRO (26).
SE profiles for 110-μL samples of BR/HAPol complexes at 1, 0.5, and 0.25 g L−1 in H2O, D2O, and  buffers were recorded at 560 nm with six-channel, 12-mm optical path centerpieces after centrifuging at 20°C for, typically, 1 day at each of four different speeds (8,000, 14,000, 22,000, and 31,000 rpm). A global analysis with the program Sedphat (24), including all the data for each solvent condition, gave estimates of Mb. For a single noninteracting species, the absorbance, a(r), as a function of the radial position, r in cm, is given by
buffers were recorded at 560 nm with six-channel, 12-mm optical path centerpieces after centrifuging at 20°C for, typically, 1 day at each of four different speeds (8,000, 14,000, 22,000, and 31,000 rpm). A global analysis with the program Sedphat (24), including all the data for each solvent condition, gave estimates of Mb. For a single noninteracting species, the absorbance, a(r), as a function of the radial position, r in cm, is given by
 |
(3) |
where a(r0) is the concentration—or signal proportional to concentration—at an arbitrary reference radial position r0; ω is the angular velocity (s−1); R is the gas constant; and T is the absolute temperature. For heterogeneous samples, Mb is a mean buoyant molar mass, weighed by weight concentration and extinction coefficients at the wavelength of the analysis of the different types of particles. Because SE profiles are obtained by measuring the absorbance at 560 nm, Mb estimates are insensitive to the distribution of free APol.
Mb for BR/APol particles, which also comprise lipids, can be expressed as a function of the mass density increment related to the protein concentration (∂ρ/∂cBR)μ, without units, μ expressing the conditions of constant chemical potential of the solvent component (27,28):
 |
(4) |
where MBR and cBR are the molecular mass and the concentration (w/v) of BR, respectively. (∂ρ/∂cBR)μ will be described in detail below.
If we consider a heterogeneous sample comprised of a main species and of dimers and trimers thereof with the same composition, i.e., of molar masses M1, 2M1, 3M1, respectively, each present in the weight fractions f1, f2, and f3, respectively,
 |
(5) |
 |
(6) |
The numerical values used in the calculations are given below.
Small-angle neutron scattering
SANS measurements were performed at the Institut Laue Langevin (ILL, Grenoble, France) on beam lines D22 and D11. Settings and data normalization are as described previously (11). The scattering curve represents the scattered intensity, I, as a function of the scattering vector, Q (nm−1). Q is related to the scattering angle θ and wavelength λ by Q = (4π/λ)sin(θ/2). The Guinier approximation,
 |
(7) |
was used to extrapolate the forward intensity I(0) and determine the radius of gyration Rg (nm). Given the presence of two species of particles with very different size (Rg ≈ 10.0 nm and Rg ≈ 3.0 nm, respectively), linear regression analysis was performed in two angular regions, namely at very low Q (below Q2 ≈ 0.001 Å−2) and between Q2 ≈ 0.001 and Q2 ≈ 0.003 Å−2 (11). I(0) is related to the signal of the different macromolecular species, i, each at concentration ci (g mL−1), of molar mass Mi (mol g−1), and neutron scattering length density increment (∂ρN/∂ci)μ (cm g−1) (27,28):
 |
(8) |
In our samples, a very small weight fraction of large APol particles contributes to scattering at the smallest angles, whereas for the wider angle region, the main contribution results from the presence of complexes of BR containing one molecule of BR, lipids, and A8-35. These complexes are present as monomers, dimers, and trimers, in weight fractions f1, f2, and f3. In addition, there are free APol particles comprised, on average, of nAPol polymer chains of mean molar mass MAPol per chain. The contribution of BR/APol complexes and small APol particles to the intensity at zero angle, I(0), when normalized to BR concentration, cBR, and the molar mass of monomeric BR, MBR, can be written as
 |
(9) |
where (∂ρN/∂cBR)μ and (∂ρNAPol/∂cAPol)μ are the neutron scattering length density increments (cm g−1) of BR/APol and free APol, respectively. The numerical values used in the calculations are given below.
Particle composition, mass density increments, and neutron scattering length density increments
The increments (∂ρ/∂cBR)μ and (∂ρN/∂cBR)μ can be expressed as the sum of the contributions of the protein, lipids, and polymer present in the complex, denoted by the indices BR, L, or APol (referring to either HAPol or DAPol), respectively). NaCl appears as a fourth component interacting with the polyelectrolyte APol, with ξi,NaCl the preferential binding parameter in g NaCl/g APol. Mi designs the molar mass of component i (g mol−1),  its partial specific volume (mL g−1), bi its neutron scattering length (cm g−1), δi the amount of bound lipid and APol (g/g protein), and z the effective number of charges of the polymer. Under our experimental conditions, ξNaCl = (MAPol/MNaCl)z/2 (12). (∂ρ/∂cBR)μ and (∂ρN/∂cBR)μ depend on these parameters and on the solvent density, ρ0 (g mL−1), and neutron scattering length density, ρN0 (cm−2). When the solvent contains D2O, a number of H atoms of the components exchange for D, which modifies Mi and bi to MiD and biD. The latter values depend on the percentage of D2O in the solvent. For each component, this can be expressed as a function of M*iD and b*iD, corresponding to full exchange, and of the percentage, EXCHi, of exchangeable H atoms that are indeed exchanged after a given time in 100% D2O (see, e.g., Gohon et al. (12)).
its partial specific volume (mL g−1), bi its neutron scattering length (cm g−1), δi the amount of bound lipid and APol (g/g protein), and z the effective number of charges of the polymer. Under our experimental conditions, ξNaCl = (MAPol/MNaCl)z/2 (12). (∂ρ/∂cBR)μ and (∂ρN/∂cBR)μ depend on these parameters and on the solvent density, ρ0 (g mL−1), and neutron scattering length density, ρN0 (cm−2). When the solvent contains D2O, a number of H atoms of the components exchange for D, which modifies Mi and bi to MiD and biD. The latter values depend on the percentage of D2O in the solvent. For each component, this can be expressed as a function of M*iD and b*iD, corresponding to full exchange, and of the percentage, EXCHi, of exchangeable H atoms that are indeed exchanged after a given time in 100% D2O (see, e.g., Gohon et al. (12)).
 |
(10) |
 |
(11) |
cBR is the product of BR molar concentration and its (hydrogenated) molar mass, MBR; the values of bi are in centimeters per gram of hydrogenated component. The numerical values used in the calculations are given below.
Numerical values used for the analysis of AUC and SANS results
The contribution of lipids was estimated on the basis of the lipid composition of PM, assuming all lipids to be quantitatively retained. The exact composition of PM lipids is still debated (29). We have assumed that for each BR molecule, PM contains 2.4 molecules of phosphatidylglycerophosphate methyl ester, three of sulfo-triglycosyl-diether-1, one of glycocardiolipin, two of squalene, and two of phosphatidylglycerol, i.e., ∼0.38 g lipid/g BR. For each lipid,  was calculated from the ratio of its estimated volume and molecular mass, using Tanford's estimates for the volume of saturated alkyl chains (30), 40 Å3 per methyl group, 120 Å3 per glycerol group (31), and 508 Å3 per glycoside headgroup (32). The characteristics of APols have been published previously (11,12). We used MBR = 27,067.1 g mol−1 (monomer of BR and its retinal); ML (mass of bound lipids) = 10,184 g mol−1; MHAPol = 12,391 g mol−1; MDAPol = 13,096 g mol−1 (mean values for 100 monomers, an arbitrary figure used solely for the sake of calculations; the average molecular mass of A8-35 is actually 9–10 kDa (11)); nDAPolMDAPol = 40,000 g mol−1; M*BRD − MBR = 360; M*LD − ML = 53.2; M*APolD − MAPol = 65; EXCHBR = 75%; EXCHL = EXCHAPol = 100%; zHAPol = zDAPol = 35, determining ξHAPol,NaCl = 0.0825 g g−1; ξDAPol,NaCl = 0.0781 g g−1;
was calculated from the ratio of its estimated volume and molecular mass, using Tanford's estimates for the volume of saturated alkyl chains (30), 40 Å3 per methyl group, 120 Å3 per glycerol group (31), and 508 Å3 per glycoside headgroup (32). The characteristics of APols have been published previously (11,12). We used MBR = 27,067.1 g mol−1 (monomer of BR and its retinal); ML (mass of bound lipids) = 10,184 g mol−1; MHAPol = 12,391 g mol−1; MDAPol = 13,096 g mol−1 (mean values for 100 monomers, an arbitrary figure used solely for the sake of calculations; the average molecular mass of A8-35 is actually 9–10 kDa (11)); nDAPolMDAPol = 40,000 g mol−1; M*BRD − MBR = 360; M*LD − ML = 53.2; M*APolD − MAPol = 65; EXCHBR = 75%; EXCHL = EXCHAPol = 100%; zHAPol = zDAPol = 35, determining ξHAPol,NaCl = 0.0825 g g−1; ξDAPol,NaCl = 0.0781 g g−1;  bBR = 1.30 1010 cm g−1 (monomer); and bL = 2.52 109 cm g−1. For the term ((bAPolD + ξAPol,NaCl) − ρN0 (
bBR = 1.30 1010 cm g−1 (monomer); and bL = 2.52 109 cm g−1. For the term ((bAPolD + ξAPol,NaCl) − ρN0 ( )) of Eq. 10, which is also (∂ρN/∂cAPol)μ of Eq. 8, we used the experimental values determined in Gohon et al. (12), namely, for DAPol in 100% H2O and 100% D2O buffers, 4.1 × 1010 and −7.2 × 109 cm g−1, respectively, and a linear combination of these two values at intermediate D2O contents. The ratio of the effective molar masses in D2O and H2O solvents, MD/MH = 1.007, was used for the determination of φ′R of BR/APol particles from SV measurements in H2O and D2O buffers.
)) of Eq. 10, which is also (∂ρN/∂cAPol)μ of Eq. 8, we used the experimental values determined in Gohon et al. (12), namely, for DAPol in 100% H2O and 100% D2O buffers, 4.1 × 1010 and −7.2 × 109 cm g−1, respectively, and a linear combination of these two values at intermediate D2O contents. The ratio of the effective molar masses in D2O and H2O solvents, MD/MH = 1.007, was used for the determination of φ′R of BR/APol particles from SV measurements in H2O and D2O buffers.
Analysis of Rg
The Rg of a two-component particle varies as a function of the inverse contrast according to
 |
(12) |
where ΔρN is the difference between the scattering length density (cm−2), ρN, of the solvent at a given D2O content and that of the particle, ρN°, at its CMP (33). The general form of this expression is a parabola where the term β is a measure of the separation between the centers of mass of the two components. β equals zero for a particle where the two components have the same center of mass (e.g., a spherical virus). In such a case, the plot becomes a straight line whose slope, α (positive or negative), determines whether the component of higher or lower scattering length density is located at the periphery of the particle. Rg°, the interpolated value of Rg at infinite contrast, i.e., for 1/ΔρN = 0, corresponds to the Rg of a homogeneous particle with the same envelope. Rg°—as the Rg at the match point of one component—corresponds only approximately to the Rg of the visible components, since the Rg is influenced by density fluctuations within the “invisible” component even when, on average, it is matched out (34).
3D model of the monomeric BR/A8-35 particle
A set of (x,y,z) positions surrounding the protein was generated using the VOIDOO program available from Uppsala Software Company (35): using probes of various radii, surfaces described by grids of dots were built at distances varying by steps of 0.15 nm, from 0.15 to 1.5 nm from the surface of the complex of BR and lipid tails (PDB code 1QHJ (10)). Output files were adapted to PDB format and a program was written to fill them with atoms of A8-35 with the following constraints: 1), a mass ratio APol/BR of 2:1; 2), a half-height of 2.2 nm from the midmembrane plane (only the transmembrane part of BR being assumed to bind APols; cf. (8)); 3), an overall thickness of the APol layer in the membrane plane of 1.7 nm; and 4), octyl chain atoms distributed preferentially close to the transmembrane surface of BR. Conditions 1–3 result in the density of the APol layer surrounding the protein being similar to that of A8-35 in pure APol particles. The output file can be read as a usual PDB file made of heteroatom positions.
Model-based estimates of Rg
Theoretical evaluations of Rg based on the above model were calculated using CRYSON, version 2.5 (36,37), and adapted PDB files. For BR, C-atoms from lipids and retinal were replaced by composite protein “atoms” such as CA from ALA, CB from ARG, and CD1 from LEU, according to their nature. In the case of A8-35, a new PDB file was built with CH, CH2, and CH3 groups being respectively described as CA from ALA, CB from ARG, and CD1 from LEU, NH groups as NH, C and O as C and O from ALA, and deuterium atoms as C from ALA. Lipid headgroups were not included in the calculations, introducing an error on Rg estimated, on the basis of geometrical considerations, not to exceed ∼5%.
Electron microscopy
Specimens of BR/APol complexes at ∼0.15 g L−1 in TpH were adsorbed to the clean side of a carbon film formed on mica (the carbon-mica interface) and negatively stained using 1% uranyl acetate. Photographs were taken under low-dose conditions with a JEOL 1200 EXII electron microscope operating at 100 kV at a nominal magnification of 40,000×.
Supporting information
See the online Supplementary Material: (S.I. 1.) Estimation of APol binding by BR using tritiated A8-35. (S.I. 2) Sedimentation velocity analyses of BR/HAPol complexes at 0.13 g L−1 in H2O buffer. (S.I. 3) Equilibrium sedimentation measurements in medium-length cells. (S.I. 4) BR/DAPol preparations contain a very small amount of pure DAPol aggregates. (S.I.5) Sedimentation velocity analyses of BR/HAPol complexes after 6-month storage at 4°C.
RESULTS
Transfer of BR from OTG to A8-35
Typically, OTG-solubilized BR was supplemented with A8-35 and the detergent removed by adsorption on Bio-Beads. This was followed by dilution (protocol 1) or centrifugation (protocol 2). The preparation for SANS of concentrated BR/DAPol solutions with a limited excess of APol (protocol 3) included centrifugation steps before and after the exchange (see Experimental Procedures). UV-visible absorbance spectra of BR in OTG solution and after trapping by A8-35 were strictly identical (Fig. 2 A), indicating that the protein remains in its native state and that the environment of retinal is not detectably affected by the exchange of surfactants.
Time-resolved absorption spectroscopy
The kinetics of the absorption changes associated with the rise and decay of intermediates during the photocycle (Fig. 2, B and C) were recorded in the 410–600 nm range from 5 ns to 100 ms after a 5-ns actinic flash. Time-resolved spectral changes were globally fitted with five exponentials (see Experimental Procedures). This procedure yields so-called decay-associated spectra (DAS), each of them characterized by its halftime, t1/2 (Fig. 2, D and E). Particularly worth noting are that
The rise of M intermediates is characterized by an increase of absorbance at 420 nm (Fig. 2 D), due to the deprotonation of the Schiff base. This rise is slower in PM (a low-amplitude phase, DAS2, with t1/2 ≈ 21 μs, and a larger, slower one, DAS3, with t1/2 ≈ 84 μs) than in OTG (major phase, t1/2 ≈ 4.3 μs, very minor one, t1/2 ≈ 23 μs; note that the contribution of M is already visible in DAS1, t1/2 ≈ 0.4 μs). This effect of solubilization is comparable to that reported by Milder et al. (38) upon transfer of delipidated BR to any of six other detergents (halftime for the rise of M, 9–15 μs). After transfer to A8-35, the rise of M follows intermediate kinetics, with ∼2/3 of it almost as rapid as in OTG (DAS2; t1/2 ≈ 5.8 μs) and ∼⅓ more akin to that in PM (DAS3; t1/2 ≈ 53 μs). The contribution of M to DAS1 (t1/2 ≈ 0.5 μs) is present, but barely detectable.
In PM, M decays in two phases (DAS4 and DAS5) with halftimes of 1.4 ms and 4.7 ms. After transfer to A8-35, M's decay resembles that in PM spectroscopically and kinetically (t1/2 ≈ 1.0 and 6.3 ms). In both samples, DAS4 shows no indication of the accumulation of the N state (no peak at 560 nm). We thus assign DAS4 to the M → O transition. DAS5 would then correspond to the formation of the ground state (peak at 570 nm) at the expense of O. In OTG, M's decay is split into an accelerated phase, with t1/2 ≈ 0.5 ms, and a slow one, with t1/2 ≈ 9 ms, with distinctly different spectra: DAS4 likely reflects the formation of N (peak at ∼560 nm), whereas DAS5 would correspond to the decay of N to the ground state, a transition that is not expected to yield very specific features, the N and BR absorption spectra being similar (Fig. 2 C). Thus, the N → O transition seems significantly slowed down in OTG, so that the N state transiently accumulates, whereas it does not in the PM and BR/A8-35 samples.
In brief, the photocycle of BR is complete whatever the environment. After trapping with A8-35, its kinetics feature characters intermediate between those in PM and those in detergent.
Size-exclusion chromatography
Upon SEC, BR/HAPol complexes typically elute as a well-defined peak (Fig. 3), with an apparent Stokes radius RS = 5.0 ± 0.15 nm. This is significantly larger than that observed in OTG (RS = 2.9 ± 0.15 nm (16)). The latter value is consistent with that of 2.8 ± 0.5 reported in Dencher and Heyn (39) for BR/OG complexes, but quite distinct from that of 3.6 ± 0.2 nm previously reported for BR/OTG ones (40). The latter samples, however, had been obtained by a multistep solubilization procedure very different from that used here. The half-height width for BR/HAPol complexes compares well with that observed for catalase, suggesting homogeneity. That for BR/DAPol complexes is slightly broader (by ∼4%), indicating the presence of a minor fraction of larger particles, consistent with AUC analyses (see below). SEC fractionation of BR/DAPol samples yielded purified fractions that, as judged by SEC or SV analyses, were similar to those shown in Fig. 4 C′ (see below), or even closer to homogeneity (∼99%, not shown).
FIGURE 3.
Size-exclusion chromatography of BR/A8-35 complexes. Arrows indicate the void (V0) and total (Vt) volumes. The elution profiles of BR/HAPol complexes (batch CT-961020H, protocol 1, 3.3 g L−1 BR), BR/DAPol complexes (batch CT-980225D, protocol 3, 1.2 g L−1 BR), and catalase (220 kDa, 2 g L−1) were scaled to the same maximum.
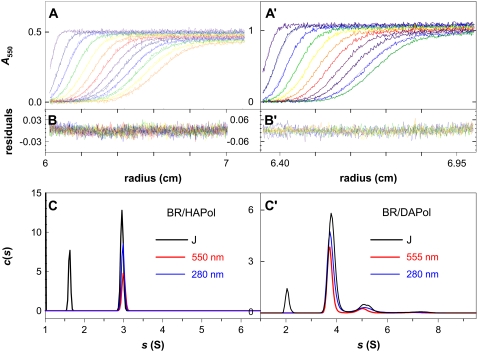
FIGURE 4.
Sedimentation velocity analysis of BR/A8-35 complexes. (A and A′) Superimposition of selected experimental and modeled profiles obtained at 20°C in 3-mm optical path cells over 270 min at 42,000 rpm for a BR/HAPol preparation (batch FGH20, protocol 1) at 1 g L−1 (A) and over 90 min at 50,000 rpm for a BR/DAPol preparation (batch CT-980225D, protocol 3) at 2.3 g L−1 (A′). (B and B′) Corresponding residuals. (C and C′) c(s) distributions determined using different optics. For clarity, the c(s) scale for interference data is in arbitrary units.
Lipid composition of BR/APol particles
TLC analyses revealed no differences between lipids extracted from either PM or purified BR/HAPol complexes and were similar to literature profiles (41) (not shown). Phosphate determination performed on two different preparations of BR/APol complexes indicated the presence of 5.2 and 4.5 mol phospholipid/mol BR versus 4.6 mol/mol in PM. These values are within experimental error of each other and of those previously reported for PM: 6.4, 5, and 4.3 mol/mol (29 and references therein). This suggests that BR/APol complexes retain all or almost all PM lipids, i.e., ∼0.38 g lipid/g BR (see Experimental Procedures, Numerical values).
Sedimentation velocity analysis of BR/APol complexes
Fig. 4 shows two sets of typical SV profiles and the corresponding distributions (c(s)) of sedimentation coefficients (s), according to measurements of A280 (protein), A550 (native BR), and the interference fringe shift, J (all species). Whether for BR/HAPol or BR/DAPol complexes, the position of the main peaks does not depend on the protocol used for sample preparation. Their relative intensities are not significantly modified when diluting the complexes from 2.3 to 0.1 g L−1, nor when exchanging the solvent for D2O buffer (as an example, a SV analysis of BR/HApol complexes at 0.13 g L−1 in H2O buffer is shown in S.I. 2). The equilibrium between species in solution—if any—is, therefore, slow on the timescale of these experiments (hours to days), and each peak corresponds to a distinct species (Table 1).
TABLE 1.
Sedimentation velocity analysis of two BR/APol preparations
| BR/HAPol | BR/DAPol | |
|---|---|---|
| Free APol | ||
| s20,w (S) | 1.6 | 2.2 |
| Free APol/BR(main species) (g g−1) | 1.6 | 0.2 |
| BR/APol complexes, major species | ||
| s20,w (S) | 3.2 | 4 |
| A280/A555 | 1.6 | 1.6 |
| (bound APol + lipids)/BR (g g−1) | 2.4 ± 0.2 | 2.1 ± 0.2 |
| bound APol/BR (g.g−1) | 2.0 ± 0.2 | 1.7 ± 0.2 |
| φ′ (mL g−1) | 0.868 | 0.824 |
| φ′R (mL g−1) | 0.84 | 0.80 |
| BR/APol complexes, minor species: | ||
| A1/Atot (at 555 nm) | 98% | 88% |
| s2/s1 (A2/Atot at 555 nm) | 1.37 (11%) | |
| s3/s1 (A3/Atot at 555 nm) | 1.87 (2%) | |
| Mw/M1 | 1.2 |
Samples are the same as in Fig. 4. Experimental errors are typically ±0.1 S for s, ±10% for APol/BR mass ratios, ±0.01 mL g−1 for φ′and φ′R, and ±0.05 for Mw/M1.
The slowest sedimenting species—detected only by interference optics—features the s value of pure HAPol or DAPol (11) and thus corresponds to particles of free A8-35. The relative concentrations of free APol and complexes depend on sample preparation, but the s values of the main components do not.
The major species (s20,w = 3.2 S for BR/HAPol, 4.0 S for BR/DAPol) presents a ratio A280/A555 = 1.6, characteristic of native BR (Fig. 2 A). From the ratio J/A555, one can estimate this species to comprise 2.2 ± 0.2 g of APol and lipids per g BR, i.e., ∼1.8 ± 0.2 g of APol, assuming the presence of a full complement of PM lipids (∼0.38 g g−1; see Experimental Procedures). As shown in Table 1, there is a rather good agreement between the values of two operational partial specific volumes, φ′, calculated from the composition of the complexes, and φ′R, deduced from complementary SV measurements in D2O buffers (not shown; note that, because the complex is a polyelectrolyte, φ′R is expected to be lower than φ′ by 0.01–0.02 mL g−1 (12)). Combining s20,w and φ′ with RS = 5 nm from SEC leads to molar mass M1 = 130 kDa for both BR/HAPol and BR/DAPol complexes, somewhat larger than the value (90 kDa) calculated from the composition, assuming the protein to be monomeric (see below). The difference originates from the overestimation (by ∼30%) of RS by SEC (see below). The values of s20,w and J/A are essentially invariant from sample to sample, indicating a fixed composition of the major species.
In the BR/DAPol sample, small amounts of larger complexes sediment with s values of 5.5 and 7.5 S. We consider them as multimers of the main BR/DAPol species (arguments are given below). In this particular sample, dimers represented ∼10% and trimers ∼2% of BR. The ratio of the mass-averaged molar mass of all BR/DAPol species to that of the monomeric one, Mw/M1, is 1.2. The relative proportion of the different species did not vary significantly upon decreasing the BR concentration from 1 g L−1 to 0.15 g L−1. In the BR/HAPol sample, in addition to free polymer and the major BR/APol complex, a small amount of larger species (scattered to up to ∼15 S) was detected (barely) with interference optics (not shown). These species are, very likely, aggregates of free HAPol, which were also present in the solution of pure polymer (∼5% in mass; not shown).
Sedimentation equilibrium analysis of Mb
SE analyses carried out in buffers prepared in either H2O, D2O, or  yielded information about the composition of BR/APol particles through the determination of buoyant molar masses (Mb), which depend on particle mass and density (Fig. 5). Data were acquired at 560 nm, so as to focus on the distribution of native BR. In
yielded information about the composition of BR/APol particles through the determination of buoyant molar masses (Mb), which depend on particle mass and density (Fig. 5). Data were acquired at 560 nm, so as to focus on the distribution of native BR. In  buffer, Mb for BR/HAPol complexes is close to null, the particles being only slightly redistributed even at the highest angular velocity. Despite the slight heterogeneity of the sample (∼9% dimer according to SV data), rather good fits were obtained assuming a single species of noninteracting particles (Fig. 5). Two other samples of BR/HAPol and BR/DAPol were investigated in H2O and D2O buffers, using longer columns (see S.I. 3). Plots of Mb from all SE experiments as a function of solvent density indicate a good consistency of the results (Fig. 5, D and E).
buffer, Mb for BR/HAPol complexes is close to null, the particles being only slightly redistributed even at the highest angular velocity. Despite the slight heterogeneity of the sample (∼9% dimer according to SV data), rather good fits were obtained assuming a single species of noninteracting particles (Fig. 5). Two other samples of BR/HAPol and BR/DAPol were investigated in H2O and D2O buffers, using longer columns (see S.I. 3). Plots of Mb from all SE experiments as a function of solvent density indicate a good consistency of the results (Fig. 5, D and E).
FIGURE 5.
Sedimentation equilibrium analysis of BR/A8-35 complexes. (A–C′) Short-column SE centrifugation of BR/HAPol complexes. The complexes (batch CT-961020H, protocol 2) were transferred by SEC in 0.1 M NaCl, 0.2 M sodium phosphate, pH 7, before concentration (Centricon 30, Amicon) and 20× dilution in 0.1 M NaCl in H2O, D2O, or  For each buffer, data were collected at 1, 0.5, and 0.25 g L−1 and analyzed globally assuming the presence of a single type of particles. Data at 0.5 g L−1 (black dots) and fits (colored lines) (A–C) are shown above the corresponding residuals (A′–C′). (D and E) Mb as a function of solvent density, ρ°, for BR/HAPol and BR/DAPol complexes. Solid squares, triangles, and diamonds refer to SE data in A–C, in S.I. 2, and not shown. Open symbols in E represent the buoyant mass Mb1 calculated for the monomeric BR/DAPol particle, derived from Mb considering Mw/M1 = 1.3. Lines represent theoretical contributions from monomeric BR (purple), a full complement of lipids (blue), APol at 1.8 g/g BR (green), and a complex with either 1.8 (solid red line) or 2.2 g APol/g BR (red dots).
For each buffer, data were collected at 1, 0.5, and 0.25 g L−1 and analyzed globally assuming the presence of a single type of particles. Data at 0.5 g L−1 (black dots) and fits (colored lines) (A–C) are shown above the corresponding residuals (A′–C′). (D and E) Mb as a function of solvent density, ρ°, for BR/HAPol and BR/DAPol complexes. Solid squares, triangles, and diamonds refer to SE data in A–C, in S.I. 2, and not shown. Open symbols in E represent the buoyant mass Mb1 calculated for the monomeric BR/DAPol particle, derived from Mb considering Mw/M1 = 1.3. Lines represent theoretical contributions from monomeric BR (purple), a full complement of lipids (blue), APol at 1.8 g/g BR (green), and a complex with either 1.8 (solid red line) or 2.2 g APol/g BR (red dots).
Fig. 5, D and E, shows the expected individual contributions to Mb of the different partners in a complex comprised of one monomer of BR and, per gram of protein, 0.38 g lipid and 1.8 g APol (as inferred from chemical analysis and SV experiments), as well as their sum (solid red lines). For BR/HAPol complexes, experimental values of Mb are slightly larger than calculated ones. This small discrepancy can be related to the slight heterogeneity of the sample and/or to an underevaluation of the amount of bound HAPol (assuming 2.2 g HAPol/g BR (dotted line) gives a better fit for purely monomeric BR), but the two phenomena of course can well contribute. For BR/DAPol complexes, a relatively good fit is obtained assuming the binding of 1.8 g DAPol/g BR and Mw/M1 = 1.3 (open symbols), as found for this sample using SV analysis (data not shown).
SANS contrast variation analysis of BR/DAPol complexes
SANS measurements yield information about the mass, size, composition, and internal structure of the complexes. A dilution series of a concentrated BR/DAPol sample (Fig. 6 A) indicated that a concentration of 1.9 g L−1 is a good compromise for contrast variation experiments, yielding a good signal/noise ratio without significant interparticle interactions. Guinier plots (Fig. 6 B) indicate that both scattered intensities and the shape of the scattering curves vary with D2O content. This is because the relative contributions of the complexes and of free DAPol particles depend on their respective contrast with the solvent. Increased scattering at very small angles (Q < 0.03 Å−1) reveals the presence of small amounts of very large aggregates of pure DAPol ((11), and see S.I. 4). At larger angles, these aggregates do not scatter significantly and the signal from BR/DAPol complexes and small DAPol particles is predominant. At ∼85% D2O, where DAPol is contrast-matched (11), Guinier plots are linear, consistent with the low polydispersity of BR/DAPol complexes already observed by SEC and AUC. Fig. 6, C–E, shows the derived values for the normalized forward intensities (Fig. 6, C and D) and radii of gyration (Fig. 6 E) of the small particles.
FIGURE 6.
Small-angle neutron scattering by BR/DAPol samples. (A) Guinier plots of a BR/DAPol preparation in H2O buffer (batch CT-980225D, protocol 3), showing the effect of dilution (intensities are normalized by the concentration of BR; only half of the points are plotted for clarity). From bottom to top, concentrations of BR decrease from 10.0 g L−1 (diamonds) to 5.0 (squares), 2.5 (inverted triangles), and 1.9 g L−1 (circles). (B) Guinier plots for the same sample at 1.9 g L−1 BR at various D2O contents. Dashed vertical lines delimit the Q2-domain used to determine Rg and the forward intensity, I(0). From bottom to top, the D2O content varies from 65% (squares) to 40% (diamonds), 84.5% (inverted triangles), 0% (triangles), and 100% (circles). (C) Forward intensities as a function of D2O content. Crosses are experimental values for two sets of measurements at [BR] = 1.9 g L−1. Lines are calculations, assuming BR/DAPol complexes to comprise 75% monomers, 20% dimers, and 5% trimers, with a full complement of PM lipids and either 1.8 (model 1 solid black line) or 2.2 (model 2, dashed black line) g bound DAPol/g BR, along with 0.38 g L−1 free DAPol particles. The gray lines represent the individual contributions of the individual components of the monomeric BR/DAPol complex, namely, from bottom to top, PM lipids at 0.38 g g−1, BR, and DAPol at 1.8 g g−1. (D) Predicted effect on I(0) of increasing the free DAPol particle concentration from 0.38 g L−1 (solid line) to 1.9 (dashed line) and 3.8 g L−1 (dash-dotted line). (E) Stuhrmann plot, where ρN is the scattering length density of the buffer as a function of D2O content and ρN° that of BR/DAPol particles at their CMP (61.2% D2O). The dashed line represents the linear regression on  measurements (black circles).
measurements (black circles).
In Fig. 6, C and D, the value of ±√(NAI(0)/cBR) is plotted as a function of the percentage of D2O in the solvent. This term includes the contributions of both BR/DAPol complexes and free DAPol particles. The contribution of the latter is small (maximum ∼15% of I(0) in H2O), but, close to the CMP of the complexes, it prevents perfect matching (see Fig. 6 D). The composition of the BR/DAPol complexes determines their CMP (experimental interpolated value, 61 ± 2% D2O), whereas the proportion of multimers is reflected in the value of I(0). Predicted I(0) values were calculated for various models and compared to experimental ones. The BR/DAPol sample was assumed to contain a major species, comprised of one molecule of BR, all PM lipids, and 1.8 or 2.2 g DAPol/g BR (models 1 and 2, respectively), along with an adjustable number of multimers thereof. Scattering by free DAPol particles (0.4 g L−1, based on SV data) was taken to be identical to that of pure DAPol solutions (11). The calculated contribution of the different components is shown in Fig. 6 C. A good fit of the forward intensities is obtained, for this sample, for 75% monomers, 20% dimers, and 5% trimers (w/w), which is consistent with SE results and corresponds to Mw (average mass) = 1.3 times the mass of the monomeric BR/DAPol particle, M1.
DAPol distribution within BR/DAPol complexes
The square of the radius of gyration of BR/DAPol particles,  was found to vary linearly with the inverse of the contrast as seen in a Stuhrmann plot (Fig. 6 E). This indicates that BR and the DAPol moiety of the complexes have the same center of mass (33). The positive slope of the plot indicates that, as expected, the component of highest scattering density (DAPol) lies farther from the center of mass of the particle compared to BR. Fig. 7 shows a model built from the crystallographic structure of BR, including nine lipid (phytanyl) chains (10) to which 2 g g−1 DAPol have been added as a 1.7-nm thick belt surrounding the transmembrane surface. The Rg for monomeric BR calculated from the PDB file is 1.8 nm; the Rg° (Rg at infinite contrast) calculated for our model is 2.6 nm. As expected, given the slight heterogeneity of the samples (see above), experimental values are somewhat larger, namely, Rg ≈ 2.3 nm at the match point of DAPol (signal due to BR and lipids) and Rg° ≈ 3.0 nm (Fig. 6 E). The Rg at the match point of BR (where the scattering is due to DAPol and lipids) is 3.2 nm, which is, logically, larger than that measured for pure DAPol particles (2.4 nm) (11).
was found to vary linearly with the inverse of the contrast as seen in a Stuhrmann plot (Fig. 6 E). This indicates that BR and the DAPol moiety of the complexes have the same center of mass (33). The positive slope of the plot indicates that, as expected, the component of highest scattering density (DAPol) lies farther from the center of mass of the particle compared to BR. Fig. 7 shows a model built from the crystallographic structure of BR, including nine lipid (phytanyl) chains (10) to which 2 g g−1 DAPol have been added as a 1.7-nm thick belt surrounding the transmembrane surface. The Rg for monomeric BR calculated from the PDB file is 1.8 nm; the Rg° (Rg at infinite contrast) calculated for our model is 2.6 nm. As expected, given the slight heterogeneity of the samples (see above), experimental values are somewhat larger, namely, Rg ≈ 2.3 nm at the match point of DAPol (signal due to BR and lipids) and Rg° ≈ 3.0 nm (Fig. 6 E). The Rg at the match point of BR (where the scattering is due to DAPol and lipids) is 3.2 nm, which is, logically, larger than that measured for pure DAPol particles (2.4 nm) (11).
FIGURE 7.
A model of BR/A8-35 complexes. (A) Cross section in a plane normal to the membrane plane. (B) View along an axis normal to the membrane plane (see text).
SV analysis of heterogeneous samples
Much more heterogeneous samples of BR/HAPol and BR/DAPol complexes were occasionally obtained. Fig. 8, A–C, shows the SV analysis of such a sample, which comprised ∼40% of large species (8–25 S) and three to four well resolved small species in the 1–8 S range. The spectroscopic characteristics of BR (A278/A550) are the same for all species, as shown by the good superimposition of the normalized c(s) curves. The sedimentation coefficients of the second, third, and fourth species are in the ratios 1.36, 1.71, and 2.11, respectively, to that of the smallest, major one. The first two values are similar to those obtained for other slightly heterogeneous preparations of BR/HAPol (1.35 and 1.74) and BR/DAPol (1.37 and 1.87) complexes (Table 1). They fit, quite well, predictions for linear assemblies of two, three, and four beads (1.5, 1.8, and 2.04, respectively), whereas expected ratios for globular multimers would be somewhat larger (1.6, 2.1, and 2.5, respectively).
FIGURE 8.
SV analysis and transmission electron microscopy of homogeneous and heterogeneous BR/A8-35 samples. (A) SV analysis of a heterogeneous BR/HAPol preparation (batch CT-961020H, protocol 3); successive profiles obtained over 140 min at 60,000 rpm, 5°C. (B) c(s) analysis of data recorded at 561 nm (red) and 277 nm (blue) (same sample). (Inset) Analysis for the slowest species. (C) c(s) analysis of the same sample after 2 years of storage at 4°C, with, in addition, the c(s) distribution derived from interference data (inset, black line). Data were collected at 42,000 rpm, 20°C. Differences in the resolution of the c(s) data in B and C are related to instrumental and experimental factors. (D–H) EM images (in negative stain) of (D) a rather homogeneous BR/DAPol sample (batch CP-990520D, protocol 3 followed by SEC fractionation); (E) a slightly heterogeneous BR/HAPol sample (batch CT-961020H, protocol 2); and (F–H) the 2-year old BR/HAPol sample from C. Scale bars, 50 nm. (I and J) Size distribution of BR/A8-35 particles, measured from several EM images. Color bars refer to the same EM images in I and J. BR/HAPol and BR/DAPol measurements are from images with homogeneous or slightly heterogeneous particles; BR/HAPol filament and BR/DAPol filament measurements are from images such as that in D.
Electron microscopy of BR/A8-35 complexes
EM of negatively stained samples of homogeneous or moderately heterogeneous samples of BR/HAPol and BR/DAPol and of a strongly heterogeneous sample of BR/HAPol are presented in Fig. 8, D–H. All samples investigated showed, in different proportions, the same three kinds of objects, namely, 1), more or less spherical particles; 2), short rods; and 3), long filaments. All structures have the same width, 6.3 ± 0.8 nm, with a rather narrow distribution (Fig. 8 I). The distribution of lengths shows a first maximum at 8 nm, corresponding to the major, globular species, followed by maxima at 13, 19, 25, and 31 nm for the elongated particles (Fig. 8 J). Apparently, BR/APol monomers have a propensity to autoassemble in a linear way, as already suggested by SV data. The filaments showed a fairly regular longitudinal segmentation (Fig. 8 G). The length of the segments corresponds roughly to tetra- or pentamers of the globular species. Because the stain formed rather thick puddles in filament-rich regions (Fig. 8 F), however, it is difficult to decide from these micrographs whether the segmented appearance results from the lengthwise association of smaller oligomers or from the filaments adopting a helical structure that rises periodically above the carbon film.
Long-term colloidal and biochemical stability of BR/APol complexes
Spectroscopic and SV analyses of the BR/HAPol sample presented in Fig. 4 A ([BR] = 1 g L−1, 98% monomeric) were repeated after 6-month storage at 4°C. Absorbance spectra were identical, indicating the absence of denaturation, but the c(s) distribution revealed the presence of ∼20% aggregates, mostly distributed around s ≈ 25 S (see S.I. 5). A280/A550 and J/A280 ratios were identical, within experimental error, for the monomeric and larger species, indicating that BR was native and all particles had the same chemical composition. In a parallel sample in which the fresh preparation had been supplemented with 4 g L−1 HAPol, thereby increasing the free HAPol concentration from ∼1.6 to ∼5.6 g L−1, large aggregates were detected neither in the fresh sample nor after 6 months of storage at 4°C (see S.I. 5). Thus, free APol in excess prohibits the formation of large complexes. A related observation was made in the course of estimating the APol/BR ratio using a tritiated amphipol, where two successive sucrose gradients were used to separate free polymer from BR/APol complexes: there was no indication of aggregation in the first gradient, whereas a pellet was detected in the second one, most probably due to the low concentration of free APol (see S.I. 1).
The long-term stability of the very heterogeneous sample of BR/HAPol complexes shown in Fig. 8 B was evaluated after 2 years of storage at 4°C (Fig. 8 C). According to the protein distribution profile recorded at 280 nm, the proportion of very large species had not changed. The A280/A550 ratio (∼1.5) was unchanged for the large aggregates, but had increased to ∼2 for the smaller ones, indicating partial denaturation. There was also a significant decrease in the proportion of small oligomers. Examination of the SV profiles of slightly heterogeneous samples of BR/HAPol and BR/DAPol before and after 2-year freezing, followed by a few weeks at 4°C (not shown), also showed the small oligomers (in small amounts to start with) disappearing with time, and demonstrated the weaker capacity of dimers to keep BR in its native state over years (increasing A280/A550 ratio). On the other hand, the A280/A550 ratio for the monomer remained unchanged, a testimony to the preservation of the native state of BR.
DISCUSSION
The functionality of native BR in APol
A8-35-trapped BR features exactly the same blue-shifted visible spectrum as in detergent, and it accomplishes its entire photocycle. The kinetics of the cycle presents composite features. As in detergent, the early part of the building up of M is accelerated compared to that in PM (by ∼4.9 times in OTG, ∼3.6 times in A8-35). However, this step shows essentially no slow phase in OTG, whereas a slow phase is distinctly apparent in A8-35, with kinetics close to that in PM (53 vs. 84 μs). A straightforward interpretation is that deprotonation of the Schiff base, which yields M1, is accelerated in both OTG and A8-35, possibly by the same phenomenon that is proposed to be responsible for the characteristic blue shift of solubilized BR, namely, a shortening of the distance between the Schiff base and the proton acceptor Asp-85 (38). On the other hand, the completion of the formation of M is slower in PM and BR/A8-35 complexes than it is in OTG. It has been proposed that the formation of M2, which corresponds to the largest conformational departure from the ground state, with one end of helix F moving outward by ∼0.35 nm to open the cytoplasmic proton channel, is hindered in PM by the close packing of the lattice (42). Although not univocal, a simple interpretation of the contrasted effects of OTG and A8-35 on the M1 → M2 transition would be that solubilization releases the pressure from neighboring lipid and BR molecules, whereas transfer from OTG to A8-35 reinstates some degree of hindrance. During M's decay, A8-35-trapped BR again resembles PM more than the OTG-solubilized protein: the halftimes of the last two phases are more similar, and the O intermediate accumulates transiently, rather than the N one in detergent solution.
BR is the third MP whose functionality is studied in the presence of APols. Upon binding a fluorescent agonist, the nicotinic acetylcholine receptor exhibits similar allosteric transitions in postsynaptic membranes and after trapping with A8-35, whereas in detergent solution equilibria are strongly shifted toward the desensitized state (43). As regards the sarcoplasmic Ca2+-ATPase, APols slow down both the hydrolytic activity and Ca2+ dissociation (44). Crystallographic structures of the ATPase show that its transmembrane region undergoes large conformational transitions during its functional cycle (45), whereas that of the acetylcholine receptor does not (46,47). On the basis of these observations, we have proposed that multipoint attachment of APols to the transmembrane surface of MPs may damp those rearrangements of this surface that entail reorganization of the polymer, an effect that could also contribute to the protective effect of APols against MP denaturation (4,5). The slowing down of the M1 → M2 transition of BR by A8-35, as compared to OTG, is consistent with this proposal. Other causes, however, such as the rebinding of lipids to critical sites at the surface of BR upon replacement of OTG by A8-35, could equally well be considered.
BR is primarily trapped by A8-35 as a monomer
When OTG-solubilized BR is supplemented with A8-35 and the detergent removed by adsorption onto Bio-Beads, a unique type of well defined BR/APol particles is usually obtained. These particles have similar size and chemical composition regardless of the A8-35 type (HAPol or DAPol), batch and sample preparation (typical protocol followed, or not, by ultracentrifugation, SEC, dialysis, ultrafiltration, etc.), or conservation (fresh preparation or storage for months at 4°C or −80°C), and they are the major species in less homogeneous preparations. Each particle contains a monomer of BR and lipids, with a lipid composition and lipid/protein ratio similar to those in PM. Both features most likely reflect the protein's state in detergent solution, preserved upon trapping with APols. Similar observations have been made previously with the cytochrome b6f complex (48,49) and the nicotinic acetylcholine receptor (43). APols thus seem to preserve the oligomeric state and bound lipids of detergent-solubilized MPs, a useful property for biochemical and structural investigations. The preservation of protein/lipid interactions may contribute to the stabilizing effects of APols (4). In the case of BR, we show elsewhere that, even though lipids do contribute to stabilization, A8-35-trapped delipidated BR is also highly stable (50).
BR/A8-35 particles contain 2.0 ± 0.2 g APol/g BR. Upon increasing the free APol/BR ratio, the sedimentation coefficient of the complex does not increase (it actually decreases slightly, due to interparticle effects (not shown)). Binding of APols thus appears saturated under our conditions. Each monomeric complex comprises ∼54 kDa A8-35, or 110 octyle chains. The latter number is somewhat below the range of published values for detergent binding by BR, which, for C12E8, Triton X-100, and lauryl maltoside, ranges from ∼120 to ∼200 molecules/monomer (51). A similar trend has already been reported for photosynthetic reaction centers and b6f complexes trapped with radiolabeled A8-75 (a close analog of A8-35) (4,48).
For the most part, our data, gathered by a wide range of approaches, provide consistent estimates of the composition, size, and mass of monomeric BR/A8-35 complexes. Combining composition and s values yields RS ≈ 3.6 nm for both BR/HAPol and BR/DAPol complexes, whereas SE and SV analyses give RS ≈ 3.8 nm for BR/HAPol complexes. SANS yields an Rg° value for BR/DAPol complexes of ∼3.0 nm, which, for spherical particles, would correspond to RS = Rg° × (5/3)1/2 ≈ 3.8 nm. By EM, one observes halfwidths and halflengths of ∼3.2 and ∼4 nm for both BR/HAPol and BR/DAPol particles. The outlier is SEC, which provides significantly larger estimates (RS ≈ 5.0 ± 0.15 nm). Earlier observations have already suggested that, for reasons that are not clear yet, SEC tends to overestimate the hydrodynamic radius of MP/A8-35 complexes as compared to MP/detergent ones. Indeed, gel filtration (6) yields much larger apparent RS differences between tOmpA/A8-35 complexes (RS ≈ 4.3–3.7 nm) and tOmpA/dihexanoylphosphatidylcholine complexes (RS ≈ 2.6 nm) than is indicated by an NMR study of their respective rotational correlation times, τc (8). The NMR data suggest that SEC overestimates the RS of tOmpA/A8-35 particles by as much as 30–50% (6), a difference close to that observed here for BR (∼30%). Electrostatic repulsion between the complexes and resin-bound A8-35 does not seem to account for this effect (for a discussion, see Zoonens et al. (6)).
Except for SEC, all experimental approaches therefore indicate that the monomeric BR/A8-35 complex is a compact, globular particle with a hydrodynamic radius of ∼3.8 nm. This value fits well with that obtained by combining the particle composition with a frictional ratio of 1.25, typical of globular compact particles, namely RS ≈ 3.9 nm. As expected, SANS data show the polymer to be in a peripheral position with respect to the protein. To build the model shown in Fig. 7, we further assumed, based on NMR observations on tOmpA/A8-35 complexes (8), that MP/APol contacts are restricted to the transmembrane surface of the protein. The compact arrangement of the polymer around the protein evidenced in this work—a layer 1.5–2 nm thick—is a favorable feature for such structural approaches as solution NMR or—more hypothetically—crystallography.
Factors that control the dispersity of BR/A8-35 complexes
In several preparations, small assemblies (dimers, trimers, etc.) and larger aggregates of BR/APol complexes were detected in addition to the major monomeric particle. Two factors leading to polydispersity have been identified. First, polydispersity is related to the solution behavior of the APol itself. We have shown previously that some batches of A8-35 with nonstandard composition exhibit marked polydispersity (11). Trapping BR with such batches always led to the formation of very heterogeneous complexes. Similar observations have been made with lipid-free preparations of tOmpA, a β-barrel MP domain (6).
Two observations indicate that decreasing the concentration of free APol is a second cause of heterogeneity: 1), a procedure designed to eliminate unbound polymer (protocol 3) consistently yielded heterogeneous samples (estimated final free APol/BR mass ratio, 0.2–0.4/1); and 2), supplementing BR/APol preparations with excess free APol prevented the long-term formation of large aggregates. We recently reported that the polydispersity of tOmpA/A8-35 complexes increases markedly after elimination of free APol by immobilized metal affinity chromatography, an effect that is reversed by adding back free APol (6). All these observations converge to indicate that complete elimination of unbound polymer can promote the reversible association of MP/APol complexes.
In addition to these two factors, it is worth recalling that the solubility and monodispersity of A8-35, and, therefore, that of the complexes it forms with MPs, diminish upon lowering the pH below ∼6.8 (8,11), adding Ca2+ ions (5), or increasing the salt (NaCl) concentration to 1 M (data not shown; cf. (11,52)). Each of these conditions most probably affects the solubility of the complexes by lowering the effective charge of A8-35, whose counterions, in our usual solvent conditions, are essentially fully dissociated (12), and/or the electrostatic repulsion between particles.
Altogether, these observations provide guidelines for the preparation of monodisperse MP/APol preparations for such applications as NMR, SANS, SAXS, EM, and chromatographic studies.
Aggregates of BR/APol are structured
Under such conditions as the use of a batch of polymer prone to aggregation or depletion of free APol, monomeric BR/A8-35 complexes exhibit a tendency to self-assemble. The aggregates, whatever their size and the cause of the polydispersity, are built by linear association of monomeric particles. Small assemblies—never abundant—consist of wormlike particles comprising four or five monomers. Those may be the building blocks of much longer fibers, which are sometimes present in a significant amount. According to SV and EM observations of preparations obtained with different batches of HAPol and DAPol, the structure of the small, medium-sized, and large assemblies does not seem to depend on the properties of the APol. Monomers incorporating native BR are physically and biochemically stable for months (marginal incorporation of monomers into large aggregates and limited denaturation of monomeric BR were each noted once over 6-month and 2-year periods, respectively, at 4°C). The small oligomers (dimers, etc.) are the least stable species (over months) in terms of colloidal and biochemical stability, suggesting that they are kinetically trapped. In our buffer conditions of dilute salt and pH 7, the fibers evolve only very slightly over months or years at 4°C, and the native state of BR is impressively stabilized, as compared to detergent solution (>2 years at 4°C, versus a couple of weeks in OTG). Currently available data do not permit a decision as to whether the formation of the fibers involves contact between the transmembrane regions of successive BR molecules, as their formation upon surfactant depletion may suggest, or between APol-free hydrophilic protein surfaces, which would naturally account for the linear mode of association. The width and incremental increase of length of the fibers (6.3 and 6 nm, respectively) are compatible with either model. The tendency of native BR/APol complexes to self-organize may provide clues to the identification of conditions promoting the formation of 3D crystals of MP/APol complexes.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Guy Schoehn and the CIBB joint electron microscopy platform for help with electron microscopy, Christophe Dugave (CEA, Saclay) for a gift of 3,4[3H]octylamine, Paulette Hervé (UMR 7099) for the synthesis of tritiated amphipol, Carla Prata (UMR 7615) for the synthesis of DAPol batch CP-990520D, and the referees of the MS for many useful remarks.
This work was supported by the Centre National de la Recherche Scientifique, the Université Paris-7, the Commissariat à l'Energie Atomique, and the Université Joseph Fourier, and by grants to J.-L.P. from the CNRS interdisciplinary program Physique et Chimie du Vivant, from the European Union (BIO4-CT98-0269), from the Human Frontier Science Program Organization (RG00223/2000-M), and from European Union Specific Targeted Research Project IMPS (Innovative tools for membrane protein structural proteomics). D.M.E. held a Chaire Blaise Pascal from the Région Île-de-France. Y.G. and T.D. were recipients of fellowships from the Ministère de la Recherche et de la Technologie.
Yann Gohon's present address is UMR 206, Chimie Biologique, Agro ParisTech, INRA, F-78850 Thiverval-Grignon, France.
Editor: Lukas K. Tamm.
References
- 1.http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
- 2.http://www.mpibp-frankfurt.mpg.de/michel/public/memprotstruct.html.
- 3.Tribet, C., R. Audebert, and J.-L. Popot. 1996. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. USA. 93:15047–15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popot, J.-L., E. A. Berry, D. Charvolin, C. Creuzenet, C. Ebel, D. M. Engelman, M. Flötenmeyer, F. Giusti, Y. Gohon, Q. Hong, J. L. Lakey, K. Leonard, H. A. Shuman, P. Timmins, D. E. Warschawski, F. Zito, M. Zoomens, B. Pucci, and C. Tribet. 2003. Amphipols: polymeric surfactants for membrane biology research. Cell. Mol. Life Sci. 60:1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard, M., T. Dahmane, M. Garrigos, C. Gauron, F. Giusti, M. le Maire, J.-L. Popot, and P. Champeil. 2006. Protective and inhibitory effects of various types of amphipols on the Ca2+-ATPase from sarcoplasmic reticulum: a comparative study. Biochemistry. 45:1861–1869. [DOI] [PubMed] [Google Scholar]
- 6.Zoonens, M., F. Giusti, F. Zito, and J.-L. Popot. 2007. Dynamics of membrane protein/amphipol association studied by Förster resonance energy transfer. Implications for in vitro studies of amphipol-stabilized membrane proteins. Biochemistry. 46:10392–10404. [DOI] [PubMed] [Google Scholar]
- 7.Pocanschi, C. L., T. Dahmane, Y. Gohon, F. Rappaport, H.-J. Apell, J. H. Kleinschmidt, and J.-L. Popot. 2006. Amphipathic polymers: tools to fold integral membrane proteins to their active form. Biochemistry. 45:13954–13961. [DOI] [PubMed] [Google Scholar]
- 8.Zoonens, M., L. J. Catoire, F. Giusti, and J.-L. Popot. 2005. NMR study of a membrane protein in detergent-free aqueous solution. Proc. Natl. Acad. Sci. USA. 102:8893–8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flötenmeyer, M., H. Weiss, C. Tribet, J.-L. Popot, and K. Leonard. 2007. The use of amphipathic polymers for cryo-electron microscopy of NADH:ubiquinone oxidoreductase (complex I). J. Microsc. 227:229–235. [DOI] [PubMed] [Google Scholar]
- 10.Belrhali, H., P. Nollert, A. Royant, C. Menzel, J. P. Rosenbusch, E. M. Landau, and E. Pebay-Peyroula. 1999. Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 Å resolution. Struct. Fold. Des. 7:909–917. [DOI] [PubMed] [Google Scholar]
- 11.Gohon, Y., F. Giusti, C. Prata, D. Charvolin, P. Timmins, C. Ebel, C. Tribet, and J.-L. Popot. 2006. Well-defined nanoparticles formed by hydrophobic assembly of a short and polydisperse random terpolymer, amphipol A8–35. Langmuir. 22:1281–1290. [DOI] [PubMed] [Google Scholar]
- 12.Gohon, Y., G. Pavlov, P. Timmins, C. Tribet, J.-L. Popot, and C. Ebel. 2004. Partial specific volume and solvent interactions of amphipol A8–35. Anal. Biochem. 334:318–334. [DOI] [PubMed] [Google Scholar]
- 13.Oesterhelt, D., and W. Stoeckenius. 1974. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 31:667–678. [DOI] [PubMed] [Google Scholar]
- 14.Lobasso, S., P. Lopalco, V. M. Lattanzio, and A. Corcelli. 2003. Osmotic shock induces the presence of glycocardiolipin in the purple membrane of Halobacterium salinarum. J. Lipid Res. 44:2120–2126. [DOI] [PubMed] [Google Scholar]
- 15.London, E., and H. G. Khorana. 1982. Denaturation and renaturation of bacteriorhodopsin in detergents and lipid-detergent mixtures. J. Biol. Chem. 257:7003–7011. [PubMed] [Google Scholar]
- 16.Gohon, Y. 2002. Structural and functional study of two membrane proteins, bacteriorhodopsin and the nicotinic acetylcholine receptor, kept soluble in detergent-free aqueous solutions by amphipathic polymers. PhD thesis. Université Paris-VI, Paris.
- 17.Baymann, F., and F. Rappaport. 1998. Electrostatic interactions at the donor side of the photosynthetic reaction center of Rhodopseudomonas viridis. Biochemistry. 37:15320–15326. [DOI] [PubMed] [Google Scholar]
- 18.Li, Y., A. van der Est, M. G. Lucas, V. M. Ramesh, F. Gu, A. Petrenko, S. Lin, A. N. Webber, F. Rappaport, and K. Redding. 2006. Directing electron transfer within Photosystem I by breaking H-bonds in the cofactor branches. Proc. Natl. Acad. Sci. USA. 103:2144–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harlan, J. E., D. Picot, P. J. Loll, and R. M. Garavito. 1995. Calibration of size-exclusion chromatography: use of a double Gaussian distribution function to describe pore sizes. Anal. Biochem. 224:557–563. [DOI] [PubMed] [Google Scholar]
- 20.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. 37:911–917. [DOI] [PubMed] [Google Scholar]
- 21.Kates, M., S. D. Kushawa, and G. D. Sprott. 1982. Lipids of purple membranes from extreme halophiles and of methanogenic bacteria. Methods Enzymol. 88:98–111. [Google Scholar]
- 22.Rouser, G., S. Fleischer, and A. Yamamoto. 1970. Two-dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids. 5:494–496. [DOI] [PubMed] [Google Scholar]
- 23.Ebel, C. 2007. Analytical ultracentrifugation. State of the art and perspectives. In Protein Structures: Methods in Protein Structure and Stability Analysis. V. Uversky and E. A. Permyakov, editors. Nova Science, New York. 229–260.
- 24.www.analyticalultracentrifugation.com.
- 25.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.http://leonardo.fcu.um.es/macromol/programs/hydro++/hydro++.htm.
- 27.Eisenberg, H. 1981. Forward scattering of light, X-rays and neutrons. Q. Rev. Biophys. 14:141–172. [DOI] [PubMed] [Google Scholar]
- 28.Ebel, C. 2007. Solvent mediated protein-protein interactions. In Protein Interactions: Biophysical Approaches for the Study of Complex Reversible Systems. P. Schuck, editor. Springer, Berlin. 255–287.
- 29.Renner, C., B. Kessler, and D. Oesterhelt. 2005. Lipid composition of integral purple membrane by 1H and 31P NMR. J. Lipid Res. 46:1755–1764. [DOI] [PubMed] [Google Scholar]
- 30.Tanford, C. 1972. Micelle shape and size. J. Phys. Chem. 76:3020–3024. [Google Scholar]
- 31.Maryadele J. O'Neil, editor. 2001. Merck Index, 13th ed. Monograph No. 4497. Merck Publishing, Whitehouse Station, N.J. 799.
- 32.Perkins, S. J. 1986. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280–nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur. J. Biochem. 157:169–180. [DOI] [PubMed] [Google Scholar]
- 33.Stuhrmann, H. B. 1970. Interpretation of small-angle scattering functions of dilute solutions and gases. A representation of the structures related to a one-particle scattering function. Acta Crystallogr. A. 26:297–306. [Google Scholar]
- 34.Witz, J. 1983. Contrast variation of the small-angle neutron scattering of globular particles: the influence of hydrogen exchange. Acta Crystallogr. A. 39:706–711. [Google Scholar]
- 35.Kleywegt, G. J., and T. A. Jones. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50:178–185. [DOI] [PubMed] [Google Scholar]
- 36.http://www.embl-hamburg.de/ExternalInfo/Research/Sax/cryson.html.
- 37.Svergun, D. I., S. Richard, M. H. Koch, Z. Sayers, S. Kuprin, and G. Zaccaï. 1998. Protein hydration in solution: experimental observation by X-ray and neutron scattering. Proc. Natl. Acad. Sci. USA. 95:2267–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milder, S. J., T. E. Thorgeirsson, L. J. Miercke, R. M. Stroud, and D. S. Kliger. 1991. Effects of detergent environments on the photocycle of purified monomeric bacteriorhodopsin. Biochemistry. 30:1751–1761. [DOI] [PubMed] [Google Scholar]
- 39.Dencher, N. A., and M. P. Heyn. 1978. Formation and properties of bacteriorhodopsin monomers in the non-ionic detergents octyl-β-D-glucoside and Triton X-100. FEBS Lett. 96:322–326. [DOI] [PubMed] [Google Scholar]
- 40.Seigneuret, M., J.-M. Neumann, and J.-L. Rigaud. 1991. Detergent delipidation and solubilization strategies for high-resolution NMR of the membrane protein bacteriorhodopsin. J. Biol. Chem. 266:10066–10069. [PubMed] [Google Scholar]
- 41.Corcelli, A., M. Colella, G. Mascolo, F. P. Fanizzi, and M. Kates. 2000. A novel glycolipid and phospholipid in the purple membrane. Biochemistry. 39:3318–3326. [DOI] [PubMed] [Google Scholar]
- 42.Vonck, J. 2000. Structure of the bacteriorhodopsin mutant F219L N intermediate revealed by electron crystallography. EMBO J. 19:2152–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez, K. L., Y. Gohon, P.-J. Corringer, C. Tribet, F. Mérola, J.-P. Changeux, and J.-L. Popot. 2002. Allosteric transitions of Torpedo acetylcholine receptor in lipids, detergent and amphipols: molecular interactions vs. physical constraints. FEBS Lett. 528:251–256. [DOI] [PubMed] [Google Scholar]
- 44.Champeil, P., T. Menguy, C. Tribet, J.-L. Popot, and M. le Maire. 2000. Interaction of amphipols with the sarcoplasmic Ca2+-ATPase. J. Biol. Chem. 275:18623–18637. [DOI] [PubMed] [Google Scholar]
- 45.Toyoshima, C., and H. Nomura. 2002. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 418:605–611. [DOI] [PubMed] [Google Scholar]
- 46.Miyazawa, A., Y. Fujiyoshi, and N. Unwin. 2003. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 423:949–955. [DOI] [PubMed] [Google Scholar]
- 47.Unwin, N. 2003. Structure and action of the nicotinic acetylcholine receptor explored by electron microscopy. FEBS Lett. 555:91–95. [DOI] [PubMed] [Google Scholar]
- 48.Tribet, C., R. Audebert, and J.-L. Popot. 1997. Stabilisation of hydrophobic colloidal dispersions in water with amphiphilic polymers: application to integral membrane proteins. Langmuir. 13:5570–5576. [Google Scholar]
- 49.Tribet, C., D. Mills, M. Haider, and J.-L. Popot. 1998. Scanning transmission electron microscopy study of the molecular mass of amphipol/cytochrome b6f complexes. Biochimie. 80:475–482. [DOI] [PubMed] [Google Scholar]
- 50.Dahmane, T. 2007. Amphipol-assisted renaturation of membrane proteins and development of sulfonated amphipols for solution NMR. PhD thesis. Université Paris-7, Paris.
- 51.Møller, J. V., and M. le Maire. 1993. Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. J. Biol. Chem. 268:18659–18672. [PubMed] [Google Scholar]
- 52.Diab, C., C. Tribet, Y. Gohon, J.-L. Popot, and F. M. Winnik. 2007. Complexation of integral membrane proteins by phosphorylcholine-based amphipols. Biochim. Biophys. Acta. 1768:2737–2747. [DOI] [PubMed] [Google Scholar]
- 53.Neutze, R., E. Pebay-Peyroula, K. Edman, A. Royant, J. Navarro, and E. M. Landau. 2002. Bacteriorhodopsin: a high-resolution structural view of vectorial proton transport. Biochim. Biophys. Acta. 1565:144–167. [DOI] [PubMed] [Google Scholar]
- 54.Váró, G. 2000. Analogies between halorhodopsin and bacteriorhodopsin. Biochim. Biophys. Acta. 1460:220–229. [DOI] [PubMed] [Google Scholar]