Abstract
Analysis of infrared polarized absorbance spectra and linear dichroism spectra of reconstituted melibiose permease from Escherichia coli shows that the oriented structures correspond mainly to tilted transmembrane α-helices, forming an average angle of ∼26° with the membrane normal in substrate-free medium. Examination of the deconvoluted linear dichroism spectra in H2O and D2O makes apparent two populations of α-helices differing by their tilt angle (helix types I and II). Moreover, the average helical tilt angle significantly varies upon substrate binding: it is increased upon Na+ binding, whereas it decreases upon subsequent melibiose binding in the presence of Na+. In contrast, melibiose binding in the presence of H+ causes virtually no change in the average tilt angle. The data also suggest that the two helix populations change their tilting and H/D exchange level in different ways depending on the bound substrate(s). Notably, cation binding essentially influences type I helices, whereas melibiose binding modifies the tilting of both helix populations.
INTRODUCTION
Melibiose permease (MelB) of Escherichia coli couples the uphill transport of α- or β-galactosides to the downhill inward movement of Na+, Li+, or H+. It is a representative member of the galactoside-pentose-hexuronide family. This integral membrane protein is encoded by melB gene (1) and consists of 473 amino acids, of which 70% are apolar. Although most bacterial transporters cotransport H+ with the solute, MelB constitutes an exception, since it has the ability to transport sugars using Na+, Li+, or H+. Each cation binds to the same site, enhancing the affinity of the cotransported sugar, with Na+ and Li+ being the best activators (2,3). The currently accepted topological model, consisting of 12 transmembrane (TM) domains and N- and C-termini located in the cytoplasm, is strongly supported by immunological studies (4,5), extensive melB-phoA fusion analyses (5,6), and proteolytic mapping (7). Moreover, Fourier transform infrared (FTIR) studies showed a high α-helix content (50%), giving support to the 12-TM segment topological model (8). Additionally, these studies and subsequent attenuated total reflection-Fourier transform infrared (ATR-FTIR) analyses suggest the existence of two different types of transmembrane α-helices (8,9). Photolabeling (10) and mutagenesis studies combined with fluorescence analysis (11) have highlighted the importance of the helix IV loop 4-5 domain for MelB transport function. Furthermore, fluorescence and FTIR spectroscopies have revealed substrate-induced conformational changes at different stages of the transport cycle (8,12–15). An electrophysiological approach revealed fast transient currents (20-ms range) triggered by either Na+ or melibiose binding, resulting from movements of charged amino acids and/or a reorientation of helix dipoles (16).
One of the most important aims of protein research is to study the relation between structure and function. Though MelB 3D structure determination would be an essential step in this regard, FTIR spectroscopy in some of its different applications (secondary-structure quantification (8), H/D exchange (15), and substrate-induced difference spectroscopy (9,17)) has provided relevant information about structural changes related to MelB function. Determination of the orientation of the secondary-structure components of proteins with polarized incident light is another of the important applications of FTIR spectroscopy (18–20). The method is based on the fact that infrared light absorption is maximal when the transition dipole moment is parallel to the electric field component of the incident light beam. The dipole orientation can be assessed by measuring the spectrum intensity on changing the incident light polarization (parallel, ‖, versus perpendicular, ⊥).
An electron crystallographic study of 2D MelB crystals (21) strongly suggest that at least 7 out of the 12 transmembrane (TM) domains (possibly α-helices), display significant albeit variable tilt with respect to the membrane normal. A recent reconstitution of 3D MelB structure, although of limited resolution, confirms the presence of such tilted transmembrane helical domains (22). Moreover, tilted membrane helices are observed in all the 3D structures of secondary transporters successfully crystallized (23–29). It is important to note that changes in the tilting of transmembrane helices have been proposed, almost systematically, to be involved in the alternating-access mechanisms (30,31).
In this study, we use polarized ATR-FTIR spectroscopy to quantitatively evaluate the average helix tilting of MelB with respect to the lipid membrane normal in proteoliposomes containing purified MelB transporters. Moreover, the possibility of independently manipulating the sodium and sugar substrates offers, additionally, the opportunity to investigate whether their interaction with MelB modifies the tilting of the helical domains of the transporter, in particular that of two hypothetical populations of α-helices that were previously identified on the basis of their different FTIR signatures (8).
MATERIALS AND METHODS
Materials
p-Nitrophenyl α-D-(6-3H) galactopyranoside ([3H] α-NPG) was synthesized in our department (Institut de Biologie et Technologies-Saclay, CEA, France), under the direction of Dr. B. Rousseau. Synthesis of LAPAO ((3-lauryl-amido)-N,N′-(dimethylamino)propylamine oxide) was performed as previously described (21). Dodecyl maltoside (DM) was obtained from Boehringer Mannheim (Cambridge, MA), and Ni-NTA resin was from Qiagen (Hilden, Germany). SM-2 Bio-Beads were obtained from Bio-Rad (Hercules, CA). Total E. coli lipids (acetone/ether precipitated) were purchased from Avanti Polar Lipids (Alabaster, AL). High-purity-grade salts or chemicals (Suprapur, Merck, Whitehouse Station, NJ) were used to prepare nominally Na+-free media containing <20 μM sodium salts. All other chemicals were obtained from commercial sources.
MelB overproduction and purification
As described previously (32), a RecA− derivative of E. coli DW2 (Δmel ΔlacZY) was transformed with pK95ΔAHB plasmid to overexpress a wild-type His-tagged MelB (8). Transformed cells were grown at 30°C in 200 L of M9 medium supplemented with appropriate carbon sources and ampicillin (100 mg/ml) at the Centre de Fermentation, Centre National de la Recherche Scientifique (Marseille, France), and used to prepare inverted membrane vesicles (IMVs), by means of a French press (American Instrument Exchange, Haverhill, MA). Purification of the His-tagged MelB was essentially carried out as previously described (33).
Preparation of MelB proteoliposomes
Purified MelB protein (1–1.5 mg/ml) solubilized in dodecyl maltoside (DM, 0.1% w/v) was mixed with E. coli lipids to give a protein/lipid ratio of 1:2 (w/w). DM was removed by overnight adsorption in SM-2 Bio-Beads (Bio-Rad) at 4°C (34). The proteoliposomes were then subjected to repeated freeze/thaw-sonication-wash cycles in nominally Na+-free buffer (20 mM MES, 10 mM KCl, pH 6.6) to eliminate the NaCl from the sample and medium.
MelB activity and protein assays
MelB activity in proteoliposomes or in the solubilized state was assessed by measuring [α-3H] NPG binding activity as previously described (2). The protein concentration was assayed using serum bovine albumin as standard.
ATR-FTIR measurements
Twenty-microliter aliquots of MelB proteoliposome suspension (155 μg protein), previously incubated with the desired substrates and briefly sonicated, were spread homogeneously on the upper surface of a germanium crystal (50 × 10 × 2 mm, yielding 12 internal reflections at the sample side, Harrick, Ossining, NY) under a stream of nitrogen. The film was left at room temperature for >1 h and the ATR device was covered with a stainless steel plate containing a gas input and output. All the spectra were recorded at 20°C with a thermostatic circulatory bath connected to the stainless plate, using a FT6000 Bio-Rad spectrometer equipped with a cooled liquid nitrogen mercury-cadmium-telluride detector (Kolmar Technologies, Newburyport, MA) at a nominal resolution of 2 cm−1.
Orientation experiments of MelB in H2O (before H/D exchange) were performed with the incident light polarized parallel and perpendicular with respect to the incidence plane with a KRS polarizer (Perkin-Elmer, Wellesley, MA). Polarized spectra were also acquired after H/D exchange. H/D exchange experiments were performed basically as described in detail elsewhere (15). A film was exposed to a continuous flow of N2 gas (220 ml/min), saturated with 2H2O by flushing it through nine closed 10-ml vials containing 2–3 ml of 2H2O each. After 900 min exposure, the film was dried by flushing N2, and polarized absorbance spectra were then acquired. Linear dichroism spectra were then computed by subtracting the perpendicular polarized spectrum from the parallel polarized spectrum with a suitable scaling factor (see below). Each polarized spectrum is the average of 2000 interferograms at a spectral resolution of 2 cm−1.
Four film conditions were analyzed: either no substrates were added (H+ is the only substrate) or samples were equilibrated for 12 h in the presence of 10 mM NaCl, 5 mM melibiose, or 5 mM melibiose plus 10 mM NaCl. All samples were sonicated shortly before use. In the rehydrated state (where H/D exchange was performed) and the dry state (where polarized spectra were recorded), the relative concentrations of the media salt and sugar constituents increase to their respective solution states, ensuring full occupancy of the protein binding sites.
Analysis of MelB secondary-structure tilting using ATR-FTIR dichroism spectra
Gaining information about structure orientation relies on the dependency of the infrared intensity on the angle between the transition dipole moment of a vibrational mode and the exciting electric field. By using polarized infrared radiation, these angles can be calculated, provided that the sample has a sufficient degree of orientation. The experimental quantity used to compute the structure orientation is the dichroic ratio RATR, defined as the ratio between the absorption of light polarized parallel and that polarized perpendicular to the incidence plane (35). On the other hand, the orientation is quantified by the experimentally accessible order parameter, Sexperimental (ϕ), which is a time- and space-averaged function of the angle between the transition dipole moment of a vibration and the z axis.
A perfect alignment with the z axis (ϕ = 0° or ϕ = 180°) gives Sexperimental = 1, whereas a perfect alignment in the xy plane (ϕ = 90°) yields Sexperimental = −1/2. Alignment at the magic angle (ϕ = 54.7°) gives Sexperimental = 0, but an isotropic arrangement (there is no order) also gives an Sexperimental equal to zero. Hence, these conditions cannot be discerned based solely on the Sexperimental value. In fact, Sexperimental can be considered as a segmental parameter, since it is the product of three suborder parameters: the order parameter of the absorbing vibration dipole with respect to a molecular or a structure axis, Sdipole (α); the order parameter of the molecular or structural axis with respect to the bilayer normal, Sstructure (β); and the order parameter of the bilayer normal with respect to the z axis, Smembrane (γ) (see Fig. 23 in Goormarghtigh et al. (35)):
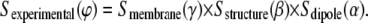 |
(1) |
Sexperimental can be estimated from polarized ATR experiments, using the relation (35)
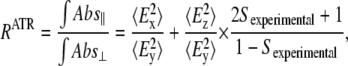 |
(2) |
where RATR is the experimentally determined dichroic ratio from an ATR experiment and 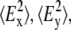 and
and 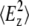 are the time-averaged electromagnetic field intensities along the x, y, and z directions of the ATR crystal (35). Their accurate values cannot usually be computed, since they depend on some parameters difficult to know, as the complex refraction index of the sample or the film thickness. However, if some experimental conditions are preserved, the thick film hypothesis provides a simple way for a simple calculation of
are the time-averaged electromagnetic field intensities along the x, y, and z directions of the ATR crystal (35). Their accurate values cannot usually be computed, since they depend on some parameters difficult to know, as the complex refraction index of the sample or the film thickness. However, if some experimental conditions are preserved, the thick film hypothesis provides a simple way for a simple calculation of 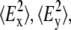 and
and 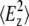 values (18,35,36). This hypothesis assumes that the sample is a weak absorber and the film thickness is much larger than the evanescent wave penetration for all the studied wavenumbers. These assumptions hold in our experiments, since all proteoliposome vibrations can be considered as weak absorbers and, even if completely dried, the amount of sample allows for at least 0.93-μm film thickness, which is higher than the evanescent wave penetration above 700 cm−1. Therefore, in the following we will use the thick-film hypothesis.
values (18,35,36). This hypothesis assumes that the sample is a weak absorber and the film thickness is much larger than the evanescent wave penetration for all the studied wavenumbers. These assumptions hold in our experiments, since all proteoliposome vibrations can be considered as weak absorbers and, even if completely dried, the amount of sample allows for at least 0.93-μm film thickness, which is higher than the evanescent wave penetration above 700 cm−1. Therefore, in the following we will use the thick-film hypothesis.
In general, the polarizers are not able to polarize the infrared light to 100%, i.e., they have some leak. We determined our polarization leak to be 0.024 from the experimentally determined isotropic ratio of 1.93 (see below). This leaking is consistent with typical reported values (37). If we take into account the estimated leak of the polarizer in Eq. 2, then
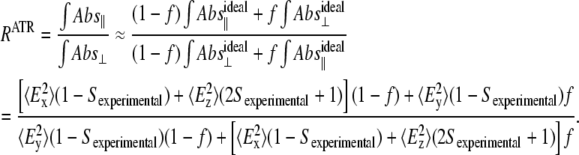 |
(3) |
Using the expressions for 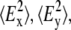 and
and 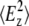 under the thick film/weak absorber approximation (35), taking into account our experimental setup (angle of the infrared (IR) beam with the ATR surface normal θ = 45°, refractive index of the ATR internal reflection element n1 = 4), and simplifying, we obtain
under the thick film/weak absorber approximation (35), taking into account our experimental setup (angle of the infrared (IR) beam with the ATR surface normal θ = 45°, refractive index of the ATR internal reflection element n1 = 4), and simplifying, we obtain
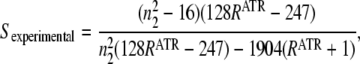 |
(4) |
where n2 is the real refraction index of the sample, which is usually assumed to be close to 1.5 for proteoliposomes (35). From Eq. 4, we obtain an experimental order parameter using the RATR of the polarized infrared spectra, which in turn can be used in Eq. 1 to estimate a structure order parameter or any other order parameter. To calculate RATR for different vibrations, the following integration intervals were used: 3400–3200 cm−1 (amide A); 1696–1605 cm−1 (amide I and amide I–I′); 1750–1720 cm−1 (lipid C=O stretching); and 2931–2912 cm−1 for lipid CH2 asymmetrical stretching. With the aim of obtaining a RATR value for the amide I as accurately as possible, the amino acid side-chain contribution was subtracted before RATR was calculated. The amino acid side-chain contribution synthetic data in H2O and D2O were constructed from published data (38,39). For the lipid CH2 stretching, the areas used to calculate RATR were measured from the Fourier deconvoluted spectra, to reduce the contribution of nearby bands. All errors reported in Table 1 and throughout this article correspond to standard errors obtained from at least three repetitions.
TABLE 1.
Order-parameter values for vibrations and tilt angles of some molecular structures in the MelB proteoliposomes for different substrate conditions
| H+ | Na+ | H+·mel | Na+·mel | |
|---|---|---|---|---|
| Order parameters | ||||
| Protein amide I* | 0.15 ± 0.01 | 0.10 ± 0.01 | 0.15 ± 0.01 | 0.13 ± 0.01 |
| H/D resistant protein amide A† | 0.44 ± 0.03 | 0.32 ± 0.01 | 0.40 ± 0.01 | 0.40 ± 0.04 |
| Lipid C=O | −0.26 ± 0.01 | −0.24 ± 0.01 | −0.25 ± 0.01 | −0.24 ± 0.01 |
| Lipid C-H‡ | −0.33 ± 0.01 | −0.32 ± 0.01 | −0.33 ± 0.01 | −0.32 ± 0.01 |
| Lipid C-H§ | −0.27 | −0.27 | −0.27 | −0.27 |
| Tilt angles¶ | ||||
| Helices | 26 ± 2 | 36 ± 1 | 25 ± 1 | 30 ± 1 |
| H/D resistant helices | 28 ± 2 | 36 ± 1 | 31 ± 1 | 31 ± 3 |
All order parameters and tilt angles are given with respect to the lipid membrane normal.
Calculated from the amide I in both H2O and D2O. Roughly corresponding to the order parameter of the amide C=O bond.
Calculated from the amide A in D2O. Corresponds to the order parameter of the amide N-H bond.
From the lipid asymmetric CH2 stretching.
From the lipid symmetric CH2 stretching, used as an fixed internal standard to correct for differences in membrane orientation over the ATR crystal (see Materials & Methods).
Details about the assumptions involved can be found in Materials & Methods and Results.
It should be taken into account that when RATR is obtained from a band area with overlapped subcomponent bands (such as the amide I band of proteins), the experimental order parameter should better be interpreted as a global experimental order parameter, 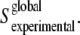 Once the order-parameter properties are given, the global order parameter will be an average of the subcomponent bands order parameters, weighed by their area (35). If the amide I subcomponents are assembled in two groups, one will correspond to the averaged order parameter of the amide groups located in transmembrane helices, with a fractional contribution to the amide I total area, m; the other will correspond to the averaged order parameter of the remainder of structures contributing to the amide I:
Once the order-parameter properties are given, the global order parameter will be an average of the subcomponent bands order parameters, weighed by their area (35). If the amide I subcomponents are assembled in two groups, one will correspond to the averaged order parameter of the amide groups located in transmembrane helices, with a fractional contribution to the amide I total area, m; the other will correspond to the averaged order parameter of the remainder of structures contributing to the amide I:  In our case, the amide I area fraction corresponding to transmembrane helices has been estimated to be ∼0.5 (8). If we consider the averaged order parameter of amide I vibrations of the structures other than helices to be close to zero, then
In our case, the amide I area fraction corresponding to transmembrane helices has been estimated to be ∼0.5 (8). If we consider the averaged order parameter of amide I vibrations of the structures other than helices to be close to zero, then 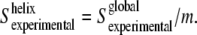 Once
Once 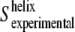 is obtained, the order parameter of the molecular axis of the helices,
is obtained, the order parameter of the molecular axis of the helices, 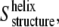 can be determined as
can be determined as
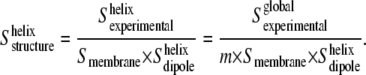 |
(5) |
This equation allows us to estimate the averaged order parameter (and tilt angle) of the transmembrane helices from the amide I band. The above assumption, considering the order parameter of nonhelical structures to be zero for amide I vibrations, does not mean that they have no orientation. Instead, we are assuming that the amide I vibration (roughly equivalent to the direction of the amide C=O bond) in nonhelical structures will have no net preferential orientation, and so their order parameter will average out to nearly zero. This assumption is strongly supported when the deconvoluted amide I absorbance and the linear dichroism spectra are compared (compare Dave et al. (8), Fig. 5, with this article (see Fig. 3)). In the absorbance spectrum, many bands are observed, and only about half of the area is assignable to transmembrane helices (8). In contrast, fewer bands are observed in the linear dichroism spectrum, and >80–85% of its area is assignable to transmembrane helices. The 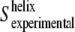 value obtained from Eq. 5 will be subjected to some uncertainties, related to the use of the m value and the assumption that
value obtained from Eq. 5 will be subjected to some uncertainties, related to the use of the m value and the assumption that  However, even if there is a 20% relative increase in m (from m = 0.5 to m = 0.6), the calculated tilt angle for the transmembrane helices will increase only ∼3–5°. Moreover, since these corrections are the same for all the substrate conditions, the tilt differences between substrate conditions will remain basically unaffected, as well as our conclusions about substrate effects on transmembrane helix orientation.
However, even if there is a 20% relative increase in m (from m = 0.5 to m = 0.6), the calculated tilt angle for the transmembrane helices will increase only ∼3–5°. Moreover, since these corrections are the same for all the substrate conditions, the tilt differences between substrate conditions will remain basically unaffected, as well as our conclusions about substrate effects on transmembrane helix orientation.
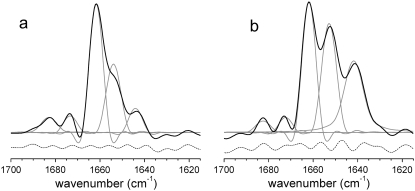
FIGURE 5.
(a) Deconvoluted linear dichroism spectra for a dry film of MelB proteoliposomes in buffer (20 mM MES and 10 mM KCl, pH 6.6, with no added substrates (black solid line)). Curve-fitted bands are represented by gray lines. The fitting residual (short-dashed line) is shifted down for a clear visualization. (b) Same as a, but for a sample incubated for 900 min in D2O.
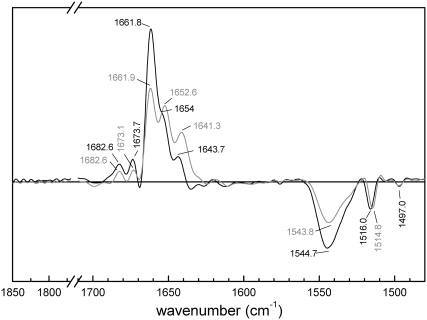
FIGURE 3.
Deconvoluted linear dichroism spectra for a dry film of MelB proteoliposomes in buffer (20 mM MES and 10 mM KCl, pH 6.6; no substrates added) before (black) and after (gray) 900 min incubation in D2O.
Once the Sstructure value is determined for the different molecular axes, we obtain the corresponding angle β:
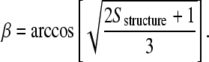 |
(6) |
This equation assumes a uniaxial orientation for the molecular axis. The molecular angle β will henceforth be referred to in this article as the average tilt angle of the structure.
Reliability on structure tilting changes
Knowing Sexperimental, Sdipole, and Smembrane and applying Eq. 1 (or the modified Eq. 5 for the amide I) enables the computation of the molecular structures order parameter. The actual values of Sdipole for the amide vibrations in α-helices have been controversial, with important discrepancies between authors (see, e.g., Table IIIB in Goormaghtigh et al. (40)). Recently, Marsh et al. experimentally determined the Sdipole of the amide vibrations (amides A, I, and II) using an α-helix model polypeptide (41). The Sdipole values obtained were in very good agreement with values estimated later from the correlation of orientation experiments of bacteriorhodopsin with its x-ray crystallographic structure (42). Following Marsh et al., we used Sdipole 0.65 for the amide A vibration mode, 0.43 for the amide I, and −0.37 for the amide II (41). It should be noted that the use of different values for Sdipole would affect the tilt angles estimated in this work, but the substrate-induced tilt angle differences should remain basically unaffected. A more serious problem could be the possibility that helices having different structures also could have a different Sdipole, as suggested by a theoretical work by Reisdorf and Krimm on αI and αII helices (43). In such a case, a difference in Sdipole between two types of helices could be artifactually interpreted as a difference in their Sstructure, and so as having a different tilt angle. However, it should be pointed out that the relative differences in Sdipole calculated by Reisdorf and Krimm for αI and αII helices were only in the order of 10% despite their significant structural differences, and no significant difference in Sdipole as a function of the helix length was observed. Moreover, we should note that the work of Marsh and Pali suggests no significant differences in Sdipole for the different helices of bacteriorhodopsin (42). Therefore, we will make the reasonable assumption that all the helices in MelB have the same Sdipole independent of their actual conformation, length, etc. For phospholipids, the order parameter of the CH2 stretching and scissoring with respect to the lipid chain axis, is Sdipole = −0.5 (44). On the other hand, to determine accurate values and be able to reliably detect changes of Sstructure, it is important to have a good estimate of Smembrane for each sample. For this purpose, it would be interesting to know Sstructure for E. coli lipids to obtain Smembrane, since the Sstructure for E. coli lipids is little affected by the presence of the protein and substrates. This last conclusion is based on the invariance of the lipid CH2 stretching band maxima under different MelB substrate conditions, which is a very sensitive marker to detect changes on the lipid Sstructure (45).
Unfortunately, no Sstructure values for E. coli lipids are found in the literature, so we estimated it in the following way. The order parameter for a lipid chain of DMPC in the crystalline state is Sstructure ≈ 0.93 (46). We obtained Sexperimental = −0.356 ± 0.03 for the CH2 stretching of DMPC in a dry film (pseudocrystalline state), which is roughly equivalent to a crystalline state. Applying Eq. 1, we obtained Smembrane = 0.77 ± 0.04 for the DMPC films. We assumed that the same Smembrane value will be valid for other lipid films prepared in the same way, such as E. coli lipids. In this way, we determined, for E. coli lipids, Sstructure ≈ 0.54. Using this value, together with Sexperimental and Sdipole of the lipid CH2 stretching we were able to obtain an accurate estimation of Smembrane for each MelB film.
Linear dichroism spectra
Linear dichroism spectra were obtained as Abs‖ − Riso × Abs⊥. The Riso is the dichroic ratio for a band with an order parameter equal to zero (not oriented or tilted at the magic angle). We experimentally determined this ratio from the dichroic ratio of pure liquid water (a nonordered substance), as 1.93. The band intensity in a linear dichroism spectra is proportional to its absorbance × its experimental order parameter. Therefore, the dichroism spectra properties are 1), bands showing Sexperimental ≈ 0 cancelled; 2), bands with Sexperimental > 0 appearing as positive bands; and 3), bands with Sexperimental < 0 appearing as negative bands. Due to the disappearance of bands with Sexperimental ≈ 0, the resolution of bands arising from oriented structures is enhanced. Furthermore, the resolution of these oriented bands is further enhanced by using Fourier self-deconvolution (FSD) (47,48), using slightly different parameters depending on the region. In the 1800–1580 cm−1 region, FSD was applied with a full width at half-height (γ) of 18 cm−1, a band-narrowing factor (k) of 2.5 and a Bessel filter. In the 1590–1450 cm−1 region, where some narrow bands are present, FSD was performed with γ = 18 cm−1, k = 2.2, and a Gaussian filter. For the purpose of quantifying amide I and amide I-I′, FSD spectra were curve-fitted using Fourier deconvoluted Lorentzian bands with probabilistic constraints in the positions (±2 cm−1) and bandwidths (±8 cm−1), as described in detail elsewhere (49,50).
Obtaining the order-parameter spectra
The structural order parameter for the amide I as a function of wavenumber was obtained in the following way. First, deconvoluted parallel and perpendicular polarized absorbance spectra (with the amino acid side chain contribution subtracted) were rationed, giving RiATR versus wavenumber. This dichroic ratio spectrum was converted to a structural order parameter spectrum by applying Eq. 4 and then Eq. 1, and using the Sdipole corresponding to the amide I of an α-helix. FSD (γ = 18 cm−1, k = 2.2, and a Gaussian filter) was used to decrease the band overlapping, as well as improving the accuracy of the obtained order parameters. Further details about the interpretation of order parameter spectra are given as Supplementary Material.
RESULTS
To obtain information on MelB secondary-structure orientation and analyze the changes in tilting caused by substrate interaction, experiments were carried out on dry proteoliposomes prepared with purified MelB. Proteoliposome samples were fully equilibrated with media differing in substrate composition, favoring the formation of different MelB intermediates of the catalytic cycle, namely H+·MelB, Na+·MelB, H+·mel·MelB, and Na+·mel·MelB complexes.
As detailed in Materials and Methods, the order parameters directly obtained from the experimental data are relative to the ATR crystal normal, whereas the order parameter and especially the tilting of any of the MelB secondary-structure components are of interest if expressed with respect to the bilayer normal. To convert the reference frame, the experimental order parameter, Sexperimental, must be corrected by the order parameter of the mosaic spread, Smembrane (order parameter of the membrane normal with respect to the ATR crystal normal). For each sample, Smembrane was determined from Eq. 1, using the Sexperimental of the CH2 symmetric stretching from the lipids, and Sstructure = 0.54 and Sdipole = −0.5 (see Materials and Methods for details). The average mosaic spread order parameter obtained for the MelB proteoliposomes under all conditions examined was 0.69 ± 0.09. By comparing this value with that obtained for pure DMPC lipid (Smembrane ≈ 0.77) it can be inferred that the presence of the protein reduces, to some extent, the film order over the ATR crystal, as we expected. Using the Smembrane value estimated for each MelB film, we could express the vibration order parameters and tilt angles of molecular structures in MelB with respect to the lipid bilayer normal, no longer to the ATR crystal normal.
Polarized and dichroism spectra of MelB in H2O medium without substrates
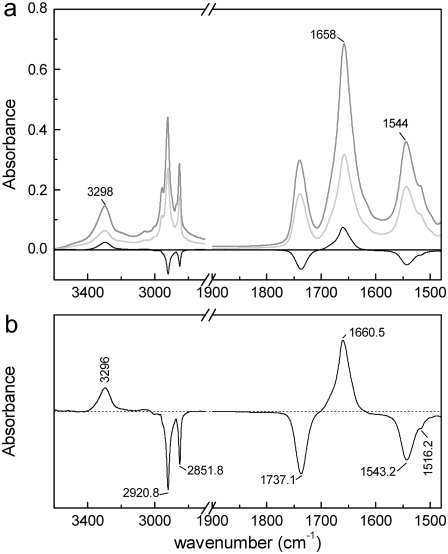
Preliminary information on the presence of oriented secondary structures in MelB proteoliposomes equilibrated with Na+- and sugar-free aqueous buffer (H+·MelB species) was obtained by recording absorption spectra with perpendicular and parallel polarized IR radiations (Fig. 1 a). The amide I, II, and A maxima wavenumbers (1658, 1544, and 3298 cm−1, respectively) are typical of membrane proteins with a main contribution of helical structures (40). However, the small shoulder in the amide A at 3200 cm−1, as well as a previous quantitative analysis of the amide I (8), indicate some significant contribution of β-sheet structures, as well as turns.
FIGURE 1.
(a) Absorbance spectra of a dry film of MelB proteoliposomes in buffer (20 mM MES and 10 mM KCl, pH 6.6; no substrates added) for perpendicular (gray) and parallel (light gray) polarized light, together with the corresponding linear dichroism spectrum (black line). (b) Expanded linear dichroism spectrum.
The dichroism spectrum, enlarged in Fig. 1 b, was calculated by subtracting the perpendicular (⊥) spectrum from the parallel (‖) spectrum, corrected by a subtraction factor of 1.93 (see Materials and Methods). From the negative sign of the CH2 and C=O lipid bands in the dichroism spectrum, we can deduce that the film is correctly oriented over the ATR crystal. The presence of signals at wavenumber intervals corresponding to the amide A (3296 cm−1, positive), amide I (1660.5 cm−1, positive), and amide II (1543.2 cm−1, negative) bands clearly indicates that MelB contains a significant amount of oriented secondary-structural components. Moreover, the amide A, I, and II wavenumber maxima are all assignable to α-helices, indicating that the major part of the oriented structures corresponds essentially to transmembrane helices. One may note the coherence of the amide A, I, and II signal signs, because the major vibration contributing to the amide I normal mode is due to the C=O stretching, which is parallel to NH stretching (amide A vibration), and perpendicular to NH bending, the major contribution to the amide II normal mode.
Polarized and dichroism spectra of MelB in D2O medium without substrates
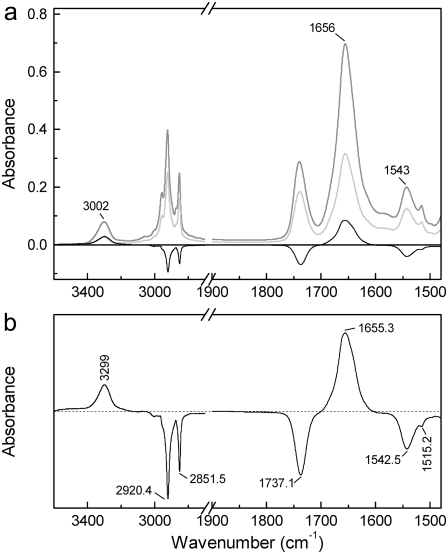
Similar analysis was carried out on MelB film previously fluxed for 900 min with D2O vapor (Fig. 2 a). In this situation, and in agreement with previous measurements (15), 40% of the amide group hydrogens appeared to be resistant to the H/D exchange, as determined from the amide II area decrease and after amino acid side-chain corrections. From the amide A and II bands maxima (3302 and 1543 cm−1, respectively) and the symmetric shape of the amide A, we can deduce that the H/D-resistant amide hydrogens are mostly located in helices.
FIGURE 2.
(a) Absorbance spectra of a dry film of MelB proteoliposomes in buffer (20 mM MES and 10 mM KCl, pH 6.6; no substrates added) for perpendicular (gray) and parallel (light gray) polarized light after 900 min incubation in D2O, together with the corresponding linear dichroism spectrum (black line). (b) Expanded linear dichroism spectrum.
The enlarged dichroism spectrum is presented in Fig. 2 b. Amide A and amide II correspond to H/D-resistant structures, which in addition are oriented. From the signal intensity and signs arising in the amide A and II bands (maxima at 3299.0 and 1542.5 cm−1, respectively) and the symmetric shape of the amide A, we can deduce that the oriented structures resistant to H/D exchange are most likely transmembrane helices (52), although there exists a certain amount of other oriented structures. On the other hand, the analysis of amide A′ and amide II′ (potential characterizers of the structure adopted by the H/D-exchanged amide hydrogens) presented unworkable difficulties. The amide A′ gave two bands in the dichroism spectrum, at 2590 and 2470 cm−1 (data not shown). However, since no amide A′ assignments are currently available in the literature it was not possible to assign the corresponding secondary structures. The amide II′, totally overlapped with a lipid band at 1460 cm−1, is therefore not experimentally observable. Finally, the analysis and interpretation of the highly overlapped amide I and I′ signals (amide I-I′) is challenging (35,53), since it represents a mixture of the nonexchanged and exchanged oriented structures. This problem was solved by combining deconvolution and curve-fitting, as described below.
Deconvoluted dichroism spectra in H2O or D2O medium without substrates
To improve the band resolution and to identify the oriented secondary-structural components, the MelB dichroism spectra were deconvoluted. In Fig. 3, the black trace shows a deconvoluted spectrum in H2O medium devoid of substrates (i.e., in the presence of H+). The most salient feature is the amide I showing an intense maximum at 1661.8 cm−1 and a shoulder at 1654.0 cm−1. According to theoretical studies, an ideal transmembrane α-helix gives two amide I vibrations with opposite orientation: a first main component parallel to the helix axis (A mode), and a second one, around five times less intense, perpendicular to the helix axis (E mode) (43). Hence, the band observed at 1654.0 cm−1 cannot be assigned to the lower component of the band at 1661.8 cm−1, since both bands show positive intensity in the dichroism spectrum and comparable intensity in the absorbance spectrum (8). From this, we infer that the two individualized bands correspond to dichroic signals arising from two different types of transmembrane α-helices. It is worth noting that their respective positions coincide with those of the previously assigned type I and type II α-helices (8). For amide II, only one band at 1544.7 cm−1 is observed, meaning a major contribution of oriented α-helices (52). Further examination of Fig. 3 suggests that other MelB secondary-structure components may also display a significant degree of orientation. Thus, the bands at 1682.6 and 1673.7 cm−1 may represent open loops, contributed by the first amino acids belonging to the loops linking the transmembrane segments, which can adopt reverse turns structure (54–56) and should show orientation. Also, the band at 1643.7 cm−1 may correspond to 310 helices or to α-helices with strong hydrogen bonds (8). This band could be assigned to oriented β-sheet structures if the corresponding amide II signal at ∼1535 cm−1 was observed (52). The assignment could still be possible if the amide II component was hidden by the more intense helix band, but the amide II envelope does not give a clear support for the presence of an unresolved component at ∼1535 cm−1. Furthermore, no band is observed in the 1640–1620 cm−1 amide I region, a typical interval for β-sheet absorption (52). Finally, typical bands from tyrosine and phenylalanine side chains appear at 1516.0 cm−1 and 1497.0 cm−1, respectively (38). Not surprisingly, about half of the tyrosines (13 out of 25) and phenylalanines (19 out of 33) are presumably located in the transmembrane segments (5).
In Fig. 3, the gray trace shows the deconvoluted dichroism spectrum of MelB after equilibration with a D2O saturated atmosphere for 900 min . In addition to the three main bands observed in the amide I′ region (1661.9, 1652.6, and 1641.3 cm−1), two smaller bands are observed in the spectrum at 1682.6 and 1673.1 cm−1. In the amide II, a single band at 1543.8 cm−1 appears with a very similar shape to that in H2O although with lower intensity with respect to the amide I-I′ area, which works as an internal standard. The band at 1661.9 cm−1 in D2O should correspond to the nonexchanged structure at 1661.8 cm−1 of amide I in H2O. As mentioned above, precise assignment of the other dichroic signals to a given secondary structure is often complicated by possible overlapping of exchanged (amide I′) with nonexchanged (amide I) oriented structures. Thus, the 1652.6 cm−1 band is probably due to contributions of the exchanged 1661.8 cm−1 band and the nonexchanged 1654 cm−1 band. Finally, the band at 1641.3 cm−1 should be considered the result of an exchanged structure from the 1654 cm−1 band plus the nonexchanged structure of the band at 1643.7 cm−1. In any instance, all these bands arise from α-helical components. Taken together, the data in D2O medium therefore suggest that the predominant oriented structures in MelB are, in substance, mostly α-helical. Incidentally, the band corresponding to tyrosine shifts from 1516.0 in H2O to 1514.8 cm−1 in D2O, meaning that, despite their putative transmembrane location, the oriented tyrosines have the −OH group H/D-exchangeable (39).
Effect of substrate on MelB secondary-structure tilting
Table 1 compares the order-parameter values of several protein and lipid vibrations with respect to the membrane normal, obtained as Sexperimental/Smembrane, for MelB proteoliposomes equilibrated in the four different substrate conditions (H+, Na+, H+/melibiose, or Na+/melibiose). A comparison of the linear dichroism spectra under different substrate conditions is included as Supplementary Material (Fig. S1), in both H2O and D2O. This allows the interested reader to evaluate the substrate effects independently of the way the dichroic ratios are converted in structural order parameters and tilt angles.
One first important observation from Table 1 is that the lipid vibrations do not change their order significantly as a result of MelB substrate presence, in contrast to the changes clearly exhibited by the protein vibrations. The protein vibrations considered are the amide I (roughly equivalent to the direction of the amide C=O bonds), and the amide A after 900 min incubation in D2O (direction of the amide NH bond with hydrogens resistant to H/D exchange). These vibration order parameters are positive, which means that both the amide C=O and NH bonds show a preferential orientation toward the membrane normal in MelB, as expected for a protein containing a high percentage of amide groups involved in transmembrane helices.
To convert the order parameter of the amide I vibration into a structural order parameter of the helix axis, we need to take into account that we have other structures besides helices contributing to the amide I area, and thus to its experimental order parameter. Taking into account that the helix contribution to the amide I area is roughly 50%, we obtained the average tilt of the helices for each substrate condition, shown in Table 1. Note that in this calculus we also assumed that the order parameter of the amide I vibration averages to nearly zero, except for the amide groups located in helices. This assumption becomes justified after observing that although helix contribution to the amide I area in the absorbance spectra is ∼50% (8), the area contribution in a linear dichroism is 85–90% (see Figs. 3 and 5). This implies that the average order parameter of an amide I vibration located in helices is more than eight times higher than the average order parameter of the amide I vibration located in other structures, i.e., the assumptions used to obtain the tilted angles in Table 1 are completely appropriate.
As deduced by the amide A and II maxima after D2O incubation (Fig. 2 a), the H/D-exchange-resistant amide NH bonds correspond to helices. Making the approximation that all the H/D-exchange-resistant amide groups contributing to the amide A area (3400–3200 cm−1) are located in helices, we estimated the tilt axis of the corresponding H/D-exchange-resistant helices. The calculated tilt angles for different substrates conditions are shown in Table 1.
Note that the estimated tilt angles for the MelB helices (from amide I in H2O and D2O) and those for the H/D-exchange-resistant helices (from amide A in D2O) are in good agreement, although the assumptions involved in the two cases were completely different. This confirms that H/D-exchange-resistant structures correspond mainly to transmembrane segments. More important, these values indicate that the transmembrane helices reorient upon substrate binding. The averaged angle calculated for H+·MelB and H+·mel·MelB complexes is ∼26°, whereas the Na+·MelB complex has an angle around 36°, and finally, the Na+·mel·MelB complex shows an angle of 30°.
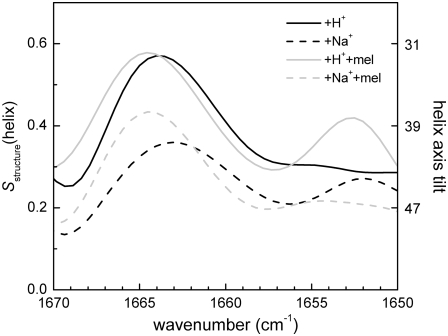
One of our aims was to get more specific information about the tilting of type I and II helices and their variation as a function of Na+ and melibiose interaction. To this end, we evaluated the order parameter of the helices, Sstructure, as a function of wavenumber (order-parameter spectrum) for amide I in the 1670–1650 cm−1 region, since it includes the signals arising from the two helix types (Fig. 4). The deconvoluted polarized absorbance spectral data were used for the calculation to improve structural resolution. Details about how Fig. 4 was obtained and how it should be interpreted are given as Supplementary Material (Fig. S2).
FIGURE 4.
Estimated structure order parameter Sstructure of helices in MelB versus the wavenumber for different substrate conditions: (black solid line) no added substrates, (black dashed line) addition of Na+, (light gray solid line) addition of melibiose, and (light gray dashed line) addition of melibiose plus Na+. The right axis shows the helix tilt angle (°) corresponding to the helix structure order parameter. The maxima values around 1664 and 1652 cm−1 give an approximate lower bond to the tilt for helices I and II, respectively. For more detail, see text and Supplementary Material.
A clear maximum in the region at 1663–1665 cm−1 appears in all substrate conditions, whereas a second, smaller maximum is only observed for two conditions at ∼1653 cm−1 (Fig. 4). It should first be noted that according to simulations using synthetic data, when two oriented (e.g., type I and II helices) and one nonoriented bands overlap, two maxima will usually appear in the order-parameter spectrum, at frequencies close (but not equal) to the absorbance maxima of each of the overlapping species (see Supplementary Material). Moreover, the order parameter at the band maxima will be representative of the oriented-band order parameter, giving in this case an indication of the tilt angle of the two types of helices. We should note, however, that these order parameters and tilt angles should be regarded as approximated lower bounds for the actual order parameters, and approximated higher bounds for the tilt angles.
Therefore, from the order-parameter spectra in Fig. 4, we can conclude that the tilting of both helical bands change upon substrate binding (see the scale on the right). The highest maxima, at around 1664 cm−1, represent the less tilted helix with respect to the membrane normal, whereas the small maxima around 1653 cm−1 correspond to more tilted helices. It is evident from Fig. 4 that the tilting of both types of helices is influenced by substrate interaction with the transporter.
H/D exchange properties of α-helices type I and type II
Fig. 5 shows a representative curve-fitting of deconvoluted linear dichroism spectra of MelB without added substrates (Fig. 5 a in H2O, and Fig. 5 b in D2O). Comparison of the fitted-band areas in H2O and D2O provides a means to evaluate the percentage of exchange of the structures giving rise to the two bands at 1661.8 and 1654.0 cm−1 in H2O, assigned to oriented transmembrane α-helix types I and II, respectively (Table 2). H/D exchange for the 1661.7 cm−1 band amounts to 36%, whereas that of the 1653.9 cm−1 band is 53%. This calculation was repeated for diverse substrate conditions, and the results are collected in Table 2. The data suggest that either Na+ or melibiose binding to MelB decreases the H/D-exchange level of type I α-helices, whereas the H/D exchange of type II α-helices clearly decreases in the presence of melibiose.
TABLE 2.
Estimated positions and percentage of H/D exchange for helix types I and II in MelB from the curve-fitting of deconvoluted linear dichroism spectra
| H+ | Na+ | H+·mel | Na+·mel | |
|---|---|---|---|---|
| Type I α-helices | ||||
| Position (cm−1) | 1661.7 | 1661.5 | 1662.5 | 1663.1 |
| H/D exchange (%) | 36 | 22 | 19 | 22 |
| Type II α-helices | ||||
| Position (cm−1) | 1653.9 | 1653.4 | 1654.1 | 1655.0 |
| H/D exchange (%) | 53 | 46 | 37 | 37 |
The uncertainties in the H/D exchange quantification are approximately ±3 for type I helices and ±5 for type II helices.
DISCUSSION
The results reported in this study provide evidence for the presence of an oriented structure that essentially corresponds to transmembrane helical domains, including, in particular, two previously identified populations of helices with different IR signatures (∼1661 and ∼1653 cm−1). The data also suggest that both the average tilt angle and the orientation of each of these two particular tilted helices vary to a different extent according to the bound substrate(s).
Oriented components in the MelB secondary structure
Analysis of MelB structure properties using IR polarized light indicates MelB contains a significant amount of tilted structural components, forming an average angle of ∼26° with the membrane normal in a substrate-free medium. The dichroic bands in H2O or D2O media arise at ∼1660 and ∼1655 cm−1 (amide I interval), ∼1554 cm−1 (amide II interval), and ∼3296 and ∼3299 cm−1 (amide A interval). These frequencies strongly suggest that the oriented structural components of MelB are essentially α-helices. Furthermore, the deconvoluted linear dichroism spectra in H2O and D2O make it apparent that three individualized α-helical populations, differing in their absorbance frequency (∼1661, ∼1653, and ∼1644 cm−1), contribute for most of the oriented protein amide I vibrations. Their positive sign in the linear dichroism spectra probes a preferential orientation in the direction of the membrane normal, strongly suggesting that these helical structures correspond to transmembrane domains. It is noteworthy that the existence of a significant number of tilted transmembrane domains in the MelB structure is strongly suggested by the projection map or the surface-rendered 3D map derived from 2D crystals (21). From the available resolved 3D structures, we learn that large proportions of helices are transmembrane and tilted. This general feature is found in 12 TM transporters belonging either to the MSF transporter family, namely, the LacY symporter (24), the oxalate OxlT, and the Glycerol-3-P GltP antiporters (23,26), to the Na+-H+ antiporter family (NhaA, (27)), or, finally, to the neurotransmitter sodium symporter family (the Na+-leucine symporter LeuTaa, (28)).
The average angle value for the H+·MelB binary complex, estimated as 26°, is of the same order of magnitude as those reported for the H+·lacY symporter, the K+ tetrameric channel of Streptomyces lividans (33°) (19,36), or the EmrE transporter (27°) (18). However, this comparison is only qualitative, as different factors can affect the average tilt angle value. For example, the lipid/protein ratio used for protein reconstitution, which is known to modify the average tilt angle (36), may be different in these reconstituted proteins. We should note that in all these works the orientation was estimated from dry films of the reconstituted proteins, and the possibility exists that the orientation (and the tilt changes on substrate binding) could be different in solution. However, it is important to keep in mind that water is present in dry films of proteins, and the hydration level may be not very different from the hydration level in the protein crystals used in x-ray diffraction (35).
Presence of two major populations of oriented transmembrane helices
Distinction between α-helical signals at 1661 cm−1 and at 1653 cm−1 has already been inferred from the Fourier self-deconvoluted amide I (8) and ATR-FTIR difference spectra of MelB (9,16). The corresponding α-helices were termed, respectively, types I and II. The detection of structural components at similar frequencies in the linear dichroism spectra clearly indicates that type I and II helices are oriented. They account for a large proportion of the MelB oriented structures. It is noteworthy that they differ by their orientation with respect to the membrane normal, type I helices being proportionally less tilted than type II helices. In addition, and as will be discussed in more detail below, the tilt of type I and type II helices are modified in a different manner by the substrates. The physicochemical factors leading to the different maximal absorption frequency at which the two helix types are recorded is a complex issue. Theoretical and experimental studies show that the amide I frequency for an α-helix increases as the helix length decreases (57,58), decreases as the distortions and irregularities of the peptide bond dihedral angles increase (59), decreases as the helix hydrations increase (57), and increases as the peptide backbone i→i + 4 hydrogen bond length increases (60). For type I and II helices, one could invoke, for example, a difference in their length. If this were the case for MelB, the type I α-helix signal (1661 cm−1), or part of it, would arise from short helices, whereas the type II α-helix signal (1653 cm−1), or—again—part of it, would be generated by longer helices. However, one might expect that all these distortions of the canonical structure lead not only to a wavenumber maximum shift, but also to a change in the dipole orientation of the amide vibrations with respect to the helix axis. Therefore, it is conceptually possible that the differences in the dichroic ratio between type I and type II helices may originate not from tilt differences, but just from structural differences (43). However, we consider that reasonable differences in the dipole orientation caused by structural changes are insufficient to explain the observed dichroic differences (see Materials and Methods), and the observed differences in tilt between type I and type II helices are highly likely to be real. Nevertheless, it is important to recognize that structural changes leading to a change in the dipole orientation, but not affecting the helix tilt angle, can be misleadingly interpreted as a change in the helix tilt.
Changes of helical tilting upon substrate interaction
Undoubtedly, a most interesting result is the observed change of tilting of the transmembrane α-helical components resulting from the interaction of each cosubstrate with MelB. These changes are not only substrate-specific, but are also different for each of the two major type I and II α-helix components (Fig. 4). A general trend is that type I helices are systematically less tilted than type II helices in all MelB intermediates.
The average MelB tilt increases when shifting from the H+·MelB to the Na+·MelB intermediate. This is associated with a significant increase in the tilt of type I α-helices, and not—or at most only modestly—in that of type II α-helices (Fig. 4). Associating the quite selective Na+-dependent change in tilt angle of type I α-helices and the possibility that they might correspond to short MelB helices raises the interesting possibility of a structural analogy between the helices lining the Na+-binding site of the Na+-H+ antiporter NhaA or of the Na+-leucine LeuTaa symporter and that of MelB (27,28). It should also be stressed that MelB and NhaA have comparable projection maps at medium resolution, showing two regions with different shape and helical arrangements (21,22). It is striking that the determined Na+ binding site of NhaA and that predicted for MelB are both located in the corresponding compact regions. As learned from the NhaA and LeuTaa structures, the structural determinant of their respective Na+ binding site (Na1 site in the case of LeuTaa) includes, in addition to a long helix (HV for NhaA and H7 for LeuTaa), two helices (H4 and HXI for NhaA and HI and H6 for LeuTaa) interrupted by an unwound stretch, generating two sets of two short helices. The stretch flanking regions harbor the side-chain or main-chain carbonyls expected to be involved in Na+ coordination. From an IR spectroscopic point of view, the resulting inner or outer “half-helices” of any interrupted helix would behave as short helices. In view of the polarization data presented in this study, we hypothesize that one or more helices surrounding the ion binding site of MelB may also be interrupted helices. Tilting of some of these “half-helices” to accommodate the bound Na+ may lead to a reduction of their accessibility to the solvent, in agreement with our H/D measurements. Finally, based on biochemical evidence suggesting that helices I, II, and IV of the N-terminal half of MelB harbor Asp residues that are critical for Na+ recognition or binding (32,61), it is suggested that one (or more) of these particular helices may contain an unwound stretch. It should finally be recalled that Na+ cooperatively enhances the affinity of MelB for the sugar. One cannot a priori exclude that part of the tilt variations of type I α-helices, and perhaps also of the modest tilt change of type II α-helices, also reflect structural change involved in the cooperative effect. This would be consistent with a recent suggestion by Gouaux and co-workers (62) that ion binding to Na+-coupled secondary transporters causes not only local but also long-distance conformational changes through the bending and twisting of the associated helical elements.
As illustrated in Table 1 and Fig. 4, interaction of melibiose with H+·MelB does not modify the MelB average angle. This result is similar to that reported for the interaction of lactose with the H+-coupled LacY transporter (36). However, melibiose interaction with Na+·MelB decreases the average angle. When MelB operates according to the H+-coupled mode, melibiose significantly reduces the tilt of type II α-helices, whereas no modification of tilting of type I α-helices is observed. In the Na+-coupled mode, melibiose binding accentuates tilting of type II α-helices but reduces that of type I α-helices. The large diversity in the observed effects could be due in large part to the utilization of a different coupling ion species for the sugar transport reaction. This diversity is most likely related to the large variations of kinetic constants (substrate affinity and rate of transport), cosubstrate binding, and cooperative properties existing between MelB functioning as H+- or an Na+-coupled sugar transporter (8,12,13,63). It is also significant that melibiose seems to affect the tilt of type II α-helices proportionally more than that of type I α-helices, whereas Na+ has the opposite impact on the tilt of these two helix populations, as discussed above. Finally, it must be stressed that the sugar-induced structural signals reported here may not be limited to the sugar-binding reaction, but may be mixed with (or replaced by) structural events associated with cooperative phenomena and/or to subsequent reorientation of the ternary MelB complex. With the sole information currently at hand, it appears unrealistic to look for any detailed structural explanation for the effect of the sugar on various helix tilt variations of MelB operating as H+- and/or Na+-coupled transport reaction. At a minimum, however, our study with polarized IR light provides additional support for a previous suggestion that conformational changes occur at different stages of the MelB transport cycle (12,13,16,63) and involve tilt variations of the two major helical populations of the transporter structure (9,17).
Clearly, improvement of the resolution of MelB structure will be essential to critically assess the validity of some of the proposed interpretations and/or speculations. It is also evident that examination of the fate of these signals in the many available mutants of MelB with altered substrate selectivity or with impaired coupling and/or translocation capacity should be useful to unravel the structure-function relationships of the Na+-coupled MelB transporter in particular and of Na-coupled symporters in general.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Raymonde Lemonnier for preparing the MelB samples of this study and Elodia Serrano and Neus Ontiveros for technical help.
This work was supported by Dirección General de Investigación (MEC) grant BFU2006-04656/BMC.
Víctor A. Lórenz-Fonfría's present address is Department of Materials Science and Engineering, Nagoya Institute of Technology, Nagoya, Japan.
Editor: Brian R. Dyer.
References
- 1.Yazyu, H., S. Shiota-Niiya, T. Shimamoto, H. Kanazawa, M. Futai, and T. Tsuchiya. 1984. Nucleotide sequence of the melB gene and characteristics of deduced amino acid sequence of the melibiose carrier in Escherichia coli. J. Biol. Chem. 259:4320–4326. [PubMed] [Google Scholar]
- 2.Damiano-Forano, E., M. Bassilana, and G. Leblanc. 1986. Sugar binding properties of the melibiose permease in Escherichia coli membrane vesicles. Effects of Na+ and H+ concentrations. J. Biol. Chem. 261:6893–6899. [PubMed] [Google Scholar]
- 3.Bassilana, M., T. Pourcher, and G. Leblanc. 1988. Melibiose permease of Escherichia coli. Characteristics of co-substrate release during facilitated diffusion reactions. J. Biol. Chem. 263:9663–9667. [PubMed] [Google Scholar]
- 4.Botfield, M. C., and T. H. Wilson. 1989. Peptide-specific antibody for the melibiose carrier of Escherichia coli localizes the carboxyl terminus to the cytoplasmic face of the membrane. J. Biol. Chem. 264:11649–11652. [PubMed] [Google Scholar]
- 5.Pourcher, T., E. Bibi, H. R. Kaback, and G. Leblanc. 1996. Membrane topology of the melibiose permease of Escherichia coli studied by melB-phoA fusion analysis. Biochemistry. 35:4161–4168. [DOI] [PubMed] [Google Scholar]
- 6.Botfield, M. C., K. Naguchi, T. Tsuchiya, and T. H. Wilson. 1992. Membrane topology of the melibiose carrier of Escherichia coli. J. Biol. Chem. 267:1818–1822. [PubMed] [Google Scholar]
- 7.Gwizdek, C., G. Leblanc, and M. Bassilana. 1997. Proteolytic mapping and substrate protection of the Escherichia coli melibiose permease. Biochemistry. 36:8522–8529. [DOI] [PubMed] [Google Scholar]
- 8.Dave, N., A. Troullier, I. Mus-Veteau, M. Dunach, G. Leblanc, and E. Padros. 2000. Secondary structure components and properties of the melibiose permease from Escherichia coli: a Fourier transform infrared spectroscopy analysis. Biophys. J. 79:747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.León, X., V. A. Lórenz-Fonfría, R. Lemonnier, G. Leblanc, and E. Padrós. 2005. Substrate-induced conformational changes of melibiose permease from Escherichia coli studied by infrared difference spectroscopy. Biochemistry. 44:3506–3514. [DOI] [PubMed] [Google Scholar]
- 10.Ambroise, Y., G. Leblanc, and B. Rousseau. 2000. Active-site-directed photolabeling of the melibiose permease of Escherichia coli. Biochemistry. 39:1338–1345. [DOI] [PubMed] [Google Scholar]
- 11.Cordat, E., G. Leblanc, and I. Mus-Veteau. 2000. Evidence for a role of helix IV in connecting cation- and sugar-binding sites of Escherichia coli melibiose permease. Biochemistry. 39:4493–4499. [DOI] [PubMed] [Google Scholar]
- 12.Mus-Veteau, I., T. Pourcher, and G. Leblanc. 1995. Melibiose permease of Escherichia coli: substrate-induced conformational changes monitored by tryptophan fluorescence spectroscopy. Biochemistry. 34:6775–6783. [DOI] [PubMed] [Google Scholar]
- 13.Maehrel, C., E. Cordat, I. Mus-Veteau, and G. Leblanc. 1998. Structural studies of the melibiose permease of Escherichia coli by fluorescence resonance energy transfer. I. Evidence for ion-induced conformational change. J. Biol. Chem. 273:33192–33197. [DOI] [PubMed] [Google Scholar]
- 14.Cordat, E., I. Mus-Veteau, and G. Leblanc. 1998. Structural studies of the melibiose permease of Escherichia coli by fluorescence resonance energy transfer. II. Identification of the tryptophan residues acting as energy donors. J. Biol. Chem. 273:33198–33202. [DOI] [PubMed] [Google Scholar]
- 15.Dave, N., V. A. Lórenz-Fonfría, J. Villaverde, R. Lemonnier, G. Leblanc, and E. Padrós. 2002. Study of amide-proton exchange of Escherichia coli melibiose permease by attenuated total reflection-Fourier transform infrared spectroscopy: evidence of structure modulation by substrate binding. J. Biol. Chem. 277:3380–3387. [DOI] [PubMed] [Google Scholar]
- 16.Meyer-Lipp, K., C. Ganea, T. Pourcher, G. Leblanc, and K. Fendler. 2004. Sugar binding induced charge translocation in the melibiose permease from Escherichia coli. Biochemistry. 43:12606–12613. [DOI] [PubMed] [Google Scholar]
- 17.León, X., R. Lemonnier, G. Leblanc, and E. Padrós. 2006. Changes in secondary structures and acidic side chains of melibiose permease upon cosubstrates binding. Biophys. J. 91:4440–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arkin, I. T., W. P. Russ, M. Lebendiker, and S. Schuldiner. 1996. Determining the secondary structure and orientation of EmrE, a multi-drug transporter, indicates a transmembrane four-helix bundle. Biochemistry. 35:7233–7238. [DOI] [PubMed] [Google Scholar]
- 19.le Coutre, J., H. R. Kaback, C. K. Patel, L. Heginbotham, and C. Miller. 1998. Fourier transform infrared spectroscopy reveals a rigid α-helical assembly for the tetrameric Streptomyces lividans K+ channel. Proc. Natl. Acad. Sci. USA. 95:6114–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimard, V., C. Vigano, A. Margolles, R. Wattiez, H. W. van Veen, W. N. Konings, J. M. Ruysschaert, and E. Goormaghtigh. 2001. Structure and dynamics of the membrane-embedded domain of LmrA investigated by coupling polarized ATR-FTIR spectroscopy and 1H/2H exchange. Biochemistry. 40:11876–11886. [DOI] [PubMed] [Google Scholar]
- 21.Hacksell, I., J. L. Rigaud, P. Purhonen, T. Pourcher, H. Hebert, and G. Leblanc. 2002. Projection structure at 8 Å resolution of the melibiose permease, an Na-sugar co-transporter from Escherichia coli. EMBO J. 21:3569–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purhonen, P., A. K. Lundback, R. Lemonnier, G. Leblanc, and H. Hebert. 2005. Three-dimensional structure of the sugar symporter melibiose permease from cryo-electron microscopy. J. Struct. Biol. 152:76–83. [DOI] [PubMed] [Google Scholar]
- 23.Hirai, T., J. A. Heymann, D. Shi, R. Sarker, P. C. Maloney, and S. Subramaniam. 2002. Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol. 9:597–600. [DOI] [PubMed] [Google Scholar]
- 24.Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science. 301:610–615. [DOI] [PubMed] [Google Scholar]
- 25.Chang, C. I., K. Ihara, Y. Chelliah, D. Mengin-Lecreulx, S. Wakatsuki, and J. Deisenhofer. 2005. Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition. Proc. Natl. Acad. Sci. USA. 102:10279–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y., M. J. Lemieux, J. Song, M. Auer, and D. N. Wang. 2003. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 301:616–620. [DOI] [PubMed] [Google Scholar]
- 27.Hunte, C., E. Screpanti, M. Venturi, A. Rimon, E. Padan, and H. Michel. 2005. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 435:1197–1202. [DOI] [PubMed] [Google Scholar]
- 28.Yamashita, A., S. K. Singh, T. Kawate, Y. Jin, and E. Gouaux. 2005. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 437:215–223. [DOI] [PubMed] [Google Scholar]
- 29.Yernool, D., O. Boudker, Y. Jin, and E. Gouaux. 2004. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 431:811–818. [DOI] [PubMed] [Google Scholar]
- 30.Abramson, J., H. R. Kaback, and S. Iwata. 2004. Structural comparison of lactose permease and the glycerol-3-phosphate antiporter: members of the major facilitator superfamily. Curr. Opin. Struct. Biol. 14:413–419. [DOI] [PubMed] [Google Scholar]
- 31.Hirai, T., and S. Subramaniam. 2003. Structural insights into the mechanism of proton pumping by bacteriorhodopsin. FEBS Lett. 545:2–8. [DOI] [PubMed] [Google Scholar]
- 32.Botfield, M. C., and T. H. Wilson. 1988. Mutations that simultaneously alter both sugar and cation specificity in the melibiose carrier of Escherichia coli. J. Biol. Chem. 263:12909–12915. [PubMed] [Google Scholar]
- 33.Pourcher, T., S. Leclercq, G. Brandolin, and G. Leblanc. 1995. Melibiose permease of Escherichia coli: large scale purification and evidence that H+, Na+, and Li+ sugar symport is catalyzed by a single polypeptide. Biochemistry. 34:4412–4420. [DOI] [PubMed] [Google Scholar]
- 34.Rigaud, J. L., M. T. Paternostre, and A. Bluzat. 1988. Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 2. Incorporation of the light-driven proton pump bacteriorhodopsin. Biochemistry. 27:2677–2688. [DOI] [PubMed] [Google Scholar]
- 35.Goormaghtigh, E., V. Raussens, and J. M. Ruysschaert. 1999. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim. Biophys. Acta. 1422:105–185. [DOI] [PubMed] [Google Scholar]
- 36.le Coutre, J., L. R. Narasimhan, C. K. Patel, and H. R. Kaback. 1997. The lipid bilayer determines helical tilt angle and function in lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 94:10167–10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Axelsen, P. H., B. K. Kaufman, R. N. McElhaney, and R. N. Lewis. 1995. The infrared dichroism of transmembrane helical polypeptides. Biophys. J. 69:2770–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venyaminov, S., and N. N. Kalnin. 1990. Quantitative IR spectrophotometry of peptide compounds in water (H2O) solutions. I. Spectral parameters of amino acid residue absorption bands. Biopolymers. 30:1243–1257. [DOI] [PubMed] [Google Scholar]
- 39.Chirgadze, Y. N., O. V. Fedorov, and N. P. Trushina. 1975. Estimation of amino acid residue side-chain absorption in the infrared spectra of protein solutions in heavy water. Biopolymers. 14:679–694. [DOI] [PubMed] [Google Scholar]
- 40.Goormaghtigh, E., V. Cabiaux, and J. M. Ruysschaert. 1994. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. I. Assignments and model compounds. Subcell. Biochem. 23:329–362. [DOI] [PubMed] [Google Scholar]
- 41.Marsh, D., M. Muller, and F. J. Schmitt. 2000. Orientation of the infrared transition moments for an α-helix. Biophys. J. 78:2499–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsh, D., and T. Pali. 2001. Infrared dichroism from the x-ray structure of bacteriorhodopsin. Biophys. J. 80:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reisdorf, W. C., Jr., and S. Krimm. 1995. Infrared dichroism of amide I and amide II modes of αI- and αII-helix segments in membrane proteins. Biophys. J. 69:271–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubner, W., and H. H. Mantsch. 1991. Orientation of specifically 13C=O labeled phosphatidylcholine multilayers from polarized attenuated total reflection FT-IR spectroscopy. Biophys. J. 59:1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Bihan, T., and M. Pezolet. 1998. Study of the structure and phase behavior of dipalmitoylphosphatidylcholine by infrared spectroscopy: characterization of the pretransition and subtransition. Chem. Phys. Lipids. 94:13–33. [Google Scholar]
- 46.Hauser, H., and G. Poupart. 1992. Lipid structure. In The Structure of Biological Membranes. CRC Press, Boca Raton, FL. Chapter 1.
- 47.Kauppinen, J. K., D. J. Moffatt, H. H. Mantsch, and D. G. Cameron. 1981. Fourier self-deconvolution: a method for resolving intrinsically overlapped bands. Appl. Spectrosc. 35:271–276. [Google Scholar]
- 48.Lorenz-Fonfria, V. A., J. Villaverde, and E. Padros. 2002. Fourier deconvolution in non-self-deconvolving conditions. Effective narrowing, signal-to-noise degradation, and curve fitting. Appl. Spectrosc. 56:232–242. [Google Scholar]
- 49.Lórenz-Fonfria, V. A., and E. Padros. 2004. Curve-fitting of Fourier manipulated spectra comprising apodization, smoothing, derivation and deconvolution. Spectrochim. Acta A Mol. Biomol. Spectrosc. 60:2703–2710. [DOI] [PubMed] [Google Scholar]
- 50.Lorenz-Fonfría, V. A., and E. Padrós. 2004. Curve-fitting overlapped bands: quantification and improvement of curve-fitting robustness in the presence of errors in the model and in the data. Analyst. 129:1243–1250. [DOI] [PubMed] [Google Scholar]
- 51.Reference deleted in proof.
- 52.Goormaghtigh, E., V. Cabiaux, and J. M. Ruysschaert. 1994. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. II. Experimental aspects, side chain structure, and H/D exchange. Subcell. Biochem. 23:363–403. [DOI] [PubMed] [Google Scholar]
- 53.de Jongh, H. H., E. Goormaghtigh, and J. M. Ruysschaert. 1997. Monitoring structural stability of trypsin inhibitor at the submolecular level by amide-proton exchange using Fourier transform infrared spectroscopy: a test case for more general application. Biochemistry. 36:13593–13602. [DOI] [PubMed] [Google Scholar]
- 54.Fabian, H., and D. Naumann. 2004. Methods to study protein folding by stopped-flow FT-IR. Methods. 34:28–40. [DOI] [PubMed] [Google Scholar]
- 55.Arrondo, J. L., and F. M. Goñi. 1999. Structure and dynamics of membrane proteins as studied by infrared spectroscopy. Prog. Biophys. Mol. Biol. 72:367–405. [DOI] [PubMed] [Google Scholar]
- 56.Jackson, M., and H. H. Mantsch. 1995. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 30:95–120. [DOI] [PubMed] [Google Scholar]
- 57.Nevskaya, N. A., and Y. N. Chirgadze. 1976. Infrared spectra and resonance interactions of amide-I and II vibration of α-helix. Biopolymers. 15:637–648. [DOI] [PubMed] [Google Scholar]
- 58.Torii, H., and M. Tasumi. 1992. Application of the three-dimensional doorway-state theory to analyses of the amide-I infrared bands of globular proteins. J. Chem. Phys. 97:92–98. [Google Scholar]
- 59.Chirgadze, Y. N., E. V. Brazhnikov, and N. A. Nevskaya. 1976. Intramolecular distortion of the α-helical structure of polypeptides. J. Mol. Biol. 102:781–792. [DOI] [PubMed] [Google Scholar]
- 60.Krimm, S., and J. Bandekar. 1986. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 3:181–364. [DOI] [PubMed] [Google Scholar]
- 61.Pourcher, T., M. L. Zani, and G. Leblanc. 1993. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. I. Effect on Na+-dependent transport and binding properties. J. Biol. Chem. 268:3209–3215. [PubMed] [Google Scholar]
- 62.Boudker, O., R. M. Ryan, D. Yernool, K. Shimamoto, and E. Gouaux. 2007. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 445:387–393. [DOI] [PubMed] [Google Scholar]
- 63.Ganea, C., T. Pourcher, G. Leblanc, and K. Fendler. 2001. Evidence for intraprotein charge transfer during the transport activity of the melibiose permease from Escherichia coli. Biochemistry. 40:13744–13752. [DOI] [PubMed] [Google Scholar]