Abstract
Model species often provide initial hypotheses and tools for studies of development, genetics, and molecular evolution in closely related species. Flour beetles of the genus Tribolium MacLeay (1825) are one group with potential for such comparative studies. Tribolium castaneum (Herbst 1797) is an increasingly useful developmental genetic system. The convenience with which congeneric and other species of tenebrionid flour beetles can be reared in the laboratory makes this group attractive for comparative studies on a small phylogenetic scale. Here we present the results of phylogenetic analyses of relationships among the major pest species of Tribolium based on two mitochondrial and three nuclear markers (cytochrome oxidase 1, 16S ribosomal DNA, wingless, 28S ribosomal DNA, histone H3). The utility of partitioning the dataset in a manner informed by biological structure and function is demonstrated by comparing various partitioning strategies. In parsimony and partitioned Bayesian analyses of the combined dataset, the castaneum and confusum species groups are supported as monophyletic and as each other’s closest relatives. However, a sister group relationship between this clade and Tribolium brevicornis (Leconte 1859) is not supported. Therefore, we suggest transferring brevicornis group species to the genus Aphanotus Leconte (1862). The inferred phylogeny provides an evolutionary framework for comparative studies using flour beetles.
Keywords: Tribolium, Aphanotus, Bayesian phylogenetics, data partitioning, RNA doublet modeling
Introduction
Studies of developmental evolution have increasingly focused on small phylogenetic scales. In part, this has been driven by technical advances, such as RNA interference (Brown et al., 1999) and promiscuous transposon-based transgenic methods (Berghammer et al., 1999; Pavlopoulos et al., 2004), which allow investigators to more easily study genetic functions in related non-model species. The availability of whole genome sequence data has also facilitated the isolation and study of orthologous genes from non-model species. Such comparative studies have revealed the evolutionary flexibility of developmental mechanisms at the species level (e.g. Raff et al., 1999; Kopp et al., 2000; Gompel and Carroll, 2003; Hoekstra and Nachman, 2003; Voss and Smith, 2005; Kronforst et al., 2006; Protas et al., 2006), allowing insights into the evolution of novel phenotypes (e.g. Wittkopp et al., 2002) as well as the conservation of ontogenetic functions (e.g. Abouheif and Wray, 2002).
Many of these studies have explored genera in which one species is an established developmental genetic model organism. Comparative studies of development in drosophilids (e.g. Kopp and True, 2002) and teleosts related to the zebrafish (Parichy, 2006) have benefited from the genetic models and experimental methods transferable from the model species Drosophila melanogaster and Danio rerio, respectively. In these and other groups, congeneric species often possess divergent phenotypes, which are experimentally accessible by developmental genetics, morphometrics, or other methods. A phylogenetic framework facilitates work in groups of closely related species because it identifies species at various evolutionary distances to the model species and provides information about the polarity of evolutionary changes.
Here we present a phylogenetic study of pest flour beetle species in the genus Tribolium MacLeay (1825), based on five markers from the mitochondrion and nucleus totaling 3106 bp. Tribolium castaneum (Herbst 1797) is a promising genetic model organism with a genome sequencing project currently underway (Brown et al., 2003) and amenable to sophisticated methods of genetic manipulation. The genetic and developmental methods applicable to T. castaneum provide a starting point for examination of related species. The genus Tribolium includes 36 described species (Table 1), many of which are easily cultured in the lab. A robust phylogenetic framework will inform future comparative studies employing additional members of this genus.
Table 1.
A list of known Tribolium species, the relative degree to which they are agricultural pests, and their described distributions.
| Tribolium MacLeay (1825) species | pest | known distribution |
|---|---|---|
| brevicornis group | North and South America | |
| T. brevicornis (Leconte 1859) | ++ | California |
| T. carinatum Hinton (1948) | Argentina | |
| T. gebieni Uyttenboogaart (1934) | Paraguay | |
| T. linsleyi Hinton (1948) | Mexico | |
| T. parallelus (Casey 1890) | + | western North America |
| T. setosum Triplehorn (1978) | Arizona | |
| T. uezumii Nakane (1963)a | Honshu (Japan)* | |
| confusum group | Africa | |
| T. anaphe Hinton (1948) | + | central Africa |
| T. arndti Grimm (2001) | South Africa | |
| T. beccarii Gridelli (1950)a | Aïr (Niger) | |
| T. bremeri Grimm (2001) | South Africa | |
| T. confusum Jacquelin du Val (1868) | +++ | cosmopolitan* |
| T. destructor Uyttenboogaart (1934) | +++ | cosmopolitan* |
| T. downesi Hinton (1948) | Mali, Sudan, Chad | |
| T. ferreri Grimm (2001) | Gambia | |
| T. indicum Blair (1930) | central Africa, Saudi Arabia*, Iran*, India* | |
| T. risbeci Lepesme (1943) | Senegal | |
| T. semele Hinton (1948) | Mali, Mauritania, Sudan, Chad | |
| T. semicostata (Gebien 1910) | Kenya | |
| T. sulmo Hinton (1948) | Ethiopia, Gambia, Ghana | |
| T. thusa Hinton (1948) | + | Chad, South Africa, Botswana, Namibia |
| alcine group | Madagascar | |
| T. alcine Hinton (1948) | Madagascar | |
| T. ceto Hinton (1948) | Madagascar | |
| T. quadricollis (Fairmaire 1902) | Madagascar | |
| castaneum group | South and Southeast Asia | |
| T. apiculum Neboiss (1962) | Australia | |
| T. audax Halstead (1969) | ++ | North America* |
| T. caledonicum Kaszab (1982) | Lifou (New Caledonia) | |
| T. castaneum (Herbst, 1797) | +++ | cosmopolitan* |
| T. cylindricum Hinton (1948) | Malay peninsula, Indonesia, Borneo, Philippines | |
| T. freemani Hinton (1948) | + | Kashmir, Japan*, Brazil* |
| T. madens (Charpentier 1825) | +++ | cosmopolitan* |
| T. parki Hinton (1948) | Bali, Larat (Tanimbar archipelago, Indonesia) | |
| T. politum Hinton (1948) | Doerian (Indonesia) | |
| T. waterhousei Hinton (1948) | Queensland, New South Wales | |
| myrmecophilum group | Australia | |
| T. antennatum Hinton (1948) | Queensland | |
| T. myrmecophilum Lea (1904) | Victoria |
Possibly introduced by human activity.
T. uezumii may be synonymous with T. carinatum, and T. beccarii may be a subspecies of T. downesi (Halstead, 1967).
Tribolium as an experimental organism
The red flour beetle Tribolium castaneum is an increasingly versatile model organism. Classically, this species has been used in population genetic studies (e.g. Park et al., 1964; Wade, 1976) and studies of mutagenesis (e.g. Sokoloff et al., 1963; Sulston and Anderson, 1996). In recent decades, this knowledge of Tribolium biology and genetics has enabled studies of population differentiation (DeMuth and Wade, 2007a; 2007b), comparative reverse genetic studies (Brown et al., 1994; Tomoyasu et al., 2005; Ober and Jockusch, 2006; Savard et al., 2006), and sophisticated transgenic experiments (Eckert et al., 2004; Lorenzen et al., 2007). The experimental potential of T. castaneum is likely to expand rapidly, due to sequencing and annotation of the genome now underway (Brown, et al., 2003) and the recent publication of a fine-scale physical and molecular map of the genome (Lorenzen et al., 2005).
The beetles (Coleoptera) are the most species-rich order of eukaryotes, with more than 350,000 described species occupying a wide range of ecological niches (Daly et al., 1998). Coleoptera are thought to have diverged from other holometabolous insect lineages relatively early, and to retain many primitive features of the Holometabola (Kjer, 2004; Grimaldi and Engel, 2005). Tribolium castaneum is arguably the most experimentally tractable of the Coleoptera. As such, developmental and genetic data from T. castaneum are often used in macroevolutionary comparisons to other common model insect species, especially Drosophila melanogaster (e.g. Jockusch et al., 2004; Angelini and Kaufman, 2005). For example, the larvae of T. castaneum and other coleopterans possess robust ventral appendages, and adult appendages develop from epidermal precursor cells in the larval limbs. This contrasts with the cyclorrhaphous Diptera, such as D. melanogaster, in which larvae lack appendages and adult limbs develop from internal imaginal discs. Moreover, Tribolium are economically important as destructive cosmopolitan pests of stored flour, corn, peanuts, and other dried agricultural products (Sokoloff, 1972; Throne et al., 2003).
Habits and diversity of Tribolium species
The genus Tribolium includes 36 described species (Table 1). Ten have become pests in dry, stored agricultural products (Nakakita, 1983), and some, including T. castaneum and T. confusum Jacquelin du Val (1868), are now cosmopolitan due the international shipment of infested grain and flour. Very little is known about the biology of Tribolium species that are not synanthropic or about species in their ancestral habitats. Outside of human foodstuffs, Tribolium have been found under the bark of trees (Blair, 1930; Magis, 1954), where they are thought to feed on fungi (Sokoloff, 1972), and in the nests of domesticated and wild bees (Magis, 1954; Neboiss, 1962) and ants (Lea, 1904), where adults have been observed feeding on pollen. These conditions likely represent the ancestral habitats of the genus (Sokoloff, 1972). Tenebrionids are adapted to arid environments, with features for increased water retention (Duncan, 2003). These features have presumably pre-adapted Tribolium species, as well as other tenebrionids, for the invasion of agricultural products, such as flour, where water is extremely limited.
Hinton’s (1948) synopsis of the genus Tribolium, which placed the species into five species groups (Table 1) based on geographic distributions and a few morphological characters, is the most complete taxonomic treatment thus far. The castaneum group includes 10 species distributed across southern and southeastern Asia. The myrmecophilum group, known from Australia, includes only two species with dorsoventrally flattened antennae. These two groups are characterized by antennae in which the three distal segments are enlarged into a club, while all other species of the genus have a less prominent club including the five distal segments. The confusum group contains 14 species from Africa, including the pests T. confusum and T. destructor Uyttenboogaart (1934). Three species endemic to Madagascar comprise the alcine group. Finally, the brevicornis group is found in North and South America and includes seven species that are much larger than others in the genus.
Proposed relationships among Tribolium species
Phylogenetic relationships within Tribolium remain unclear. The relationships among species of Tribolium were loosely considered by Hinton (1948), based on their geographic distributions and morphological characters such as body size, the number of enlarged antennal segments forming the club, and the form of margins on the vertex and pronotum. Hinton proposed the castaneum and myrmecophilum groups as sisters, with the basal split separating the brevicornis group from the rest of the genus. Some new species have been described in recent decades (Kaszab, 1982; Grimm, 2001). However, since their description by Hinton, nearly 60 years ago, most species have not been reported in the literature, presumably because many species are rare or endemic to remote areas. Nevertheless, in an attempt to understand the relationships and evolutionary history of this economically important group, several studies have considered the relationships among pest species.
Previous phylogenetic studies have presented conflicting phylogenetic hypotheses. A chemotaxonomic study based on the secreted organic chemical compounds produced by eight Tribolium species (those pests used in the present study) concluded that the castaneum and confusum species groups were well supported but also that T. brevicornis was sister to the castaneum group (Howard, 1987). Alternatively, isozyme data have suggested a closer relationship between T. confusum and T. brevicornis, with T. castaneum basal to this pair and to the confusum group species T. destructor (Wool, 1982). These relationships were also weakly supported when the chemotaxonomic evidence (Howard, 1987) was combined with satellite DNA data (Juan et al., 1993).
Several lines of evidence have supported monophyly of the castaneum species group. A study of satellite sequences and orientation in T. confusum, T. castaneum, Tribolium freemani Hinton (1948), and Tribolium madens (Charpentier 1825) grouped the latter three, and suggested that T. freemani was sister to T. castaneum (Ugarković et al., 1996b). Two other species of the castaneum group, T. madens and Tribolium audax Halstead (1969), are morphologically very similar and were not recognized as distinct until 1969 (Halstead, 1969). Examination of chromosome number and banding also provides support for the confusum species group. Tribolium confusum and T. destructor share a large autosomal translocation to the X chromosome relative to the karyotype of T. castaneum and other tenebrionids (Smith, 1952a).
Meštrović et al. (2006) inferred phylogenies based on cox1 and 16S rDNA sequences from eight Tribolium species (those pests used in the present study). Their results from parsimony and Bayesian analyses conflicted on the placement of T. brevicornis but supported monophyly of the castaneum and confusum species groups. This study used different outgroup species than our present study, contributing to important differences in the conclusions regarding the monophyly of Tribolium.
In the present study, the relationships of Tribolium species are clarified with DNA sequence data from two mitochondrial and three nuclear markers using several phylogenetic inference methods, including parsimony, likelihood, and Bayesian inference using various partitioning strategies. Within the limits of taxon sampling, monophyly of the castaneum and confusum species groups is strongly supported, and the combined analyses strongly support a sister group relationship between these lineages. However, Tribolium is not resolved as monophyletic in these analyses because of the placement of T. brevicornis. Inclusion of T. brevicornis in Tribolium is also rejected in individual analyses of four of five markers. By comparing random partitioning strategies to those informed by sequence structure and function, we conclude that the confidence of phylogenetic inference is significantly improved through the use of informed partitioning strategies.
Materials and Methods
Taxon sampling
Tribolium castaneum adults were obtained from Carolina Biological Supply and kept as a breeding laboratory culture. Cultures of T. freemani, T. madens, T. confusum, and T. brevicornis were obtained from the USDA Grain Marketing Research Center in Manhattan, Kansas. Additionally, cox1 and 16S rDNA sequences were obtained from GenBank for three other Tribolium pest species: T. audax (AJ438086, AJ438151), Tribolium anaphe Hinton (1948) (AJ438083, AJ438150) and T. destructor (AJ438089, AJ438147). Taxon sampling was limited to economically detrimental pest species because material from other species was unavailable. We included three additional tenebrionid outgroup species. Selection of these species was guided by hypothesized tribal relationships, since a comprehensive molecular phylogenetic analysis of the group is not yet available. Tenebrionidae includes roughly 20,000 species in 16 tribes (Daly, et al., 1998). Currently, Tribolium is placed in the tribe Triboliini (Doyen, 1985), along with Latheticus and Palorus, genera also including agricultural pests, as well as Aesymnus, Lyphia, Metulosonia, Mycotrogus, Tharsus, and Ulosonia. We included Latheticus oryzae Waterhouse (1880), a species of pest Triboliini, in this study. Until recently, Tribolium was placed in the tribe Ulomini (Horn, 1870; Sokoloff, 1972), from which we included Gnathocerus cornutus (Fabricius 1798). Finally, we also included the mealworm beetle Tenebrio molitor Linnaeus (1758) (Tenebrionini), which has been considered a close ally of the Ulomini (Kwieton, 1982) and Triboliini (Doyen, 1985). Gnathocerus cornutus and T. molitor are also common pest beetles that are easily kept in lab culture.
Greater taxonomic sampling was included in a separate analysis of additional tenebrionid 28S rDNA sequences from GenBank (AY310661; AY310668; AY310671; PBO565954; PGR565947; PCO565971; TSC565974). This marker was chosen because it provided the most extensive available sampling of tenebrionid taxa.
Isolation of DNA sequences
Genomic DNA was isolated from the larvae of laboratory beetle stocks using the NucleoSpin DNA extraction kit (Clontech). Because the genus Tribolium is globally distributed it has traditionally been assumed that its major lineages radiated in the Middle Cretaceous (Hinton, 1948). Therefore we chose five markers that span a range of evolutionary rates. Mitochondrial cytochrome oxidase subunit I (cox1) and 16S ribosomal DNA, as well as the nuclear loci wingless (wg), histone H3, and 28S ribosomal DNA were amplified using a combination of previously published and novel primers (Table 2) using Titanium Taq DNA polymerase (Clontech). In most cases, this provided an amplicon that could be sequenced directly using a dye-terminator mix with the ABI3100 capillary electrophoresis system. For some templates and primer pairs, PCR did not yield a single band. These PCR products were excised from an agarose gel and cloned into the Topo4-TA vector (Invitrogen). Multiple clones were sequenced and polymorphic positions, which were rare, were coded as missing data. Chromatogram base-calls were inspected by eye and edited as necessary in Sequencher 4.1 (Gene Codes Corporation). Sequences obtained for this study have been deposited in GenBank (accession numbers: EU048277-EU048316).
Table 2.
Primers used to amplify genomic sequence data.
| gene | amplicon | primer name | sequence | reference |
|---|---|---|---|---|
| COI | 820 bp | C1-J-2195 | TTGATTTTTTGGTCATCCAGAAGT | Simon et al., 1994 |
| TL2-N-3014 | TCCAATGCACTAATCTGCCATATTA | |||
| 16S rRNA | 512 bp | 16Sbr | CCGGTCTGAACTCAGATCACGT | Simon et al., 1994 |
| 16Sar | CGCCTGTTTAACAAAAACAT | |||
| wingless | 468 bp | wg1MP-F3 | GArTGyAArTGyCAyGGCATGTCsGG | M. Polihronakis, unpubl. |
| wg1MP-R3 | ACyICGCArCACCArTGGAAIGTGCA | |||
| 472 bp | Tsp'wg-f1 | ACnAThAArACnTGyTGGATGCGnCT | this study | |
| Tsp'wg-r1 | CrCArCACCArTGrAAnGTrCArAT | |||
| 28S rRNA | 1044 bp | D2-3665F (Bel28S) | AGAGAGAGTTCAAGAGTACGTG | Hancock et al., 1988 |
| D5-4749R (28SD4rev) | GTTACACACTCCTTAGCGGA | |||
| histone H3 | 330 bp | HexAF | ATGGCTACCAAGCAGACGGC | Ogden & Whiting, 2003 |
| HexAR | ATATCCTTGGGCATGATGGTGAC |
Sequence alignment and model selection
Alignment of orthologous sequences was done manually for most genes. For the expansion region of 28S rDNA, alignment was assisted by ClustalX (Higgins et al., 1996), using a transition-transversion weight ratio of 0.5 and gap penalties of 10 for opening and 0.2 for extension. The aligned combined dataset is available in TreeBASE (accession number SN3489). For each data partition, the best-fitting model of nucleotide evolution was determined using the Akaike information criterion as implemented in Modeltest 3.7 (Posada and Crandall, 1998). Because there was no variation in the second codon position of histone H3, the Jukes-Cantor method was arbitrarily selected to model these data when partitioned individually. For ribosomal stem regions, we also tested the usefulness of a nucleotide doublet model (Ronquist and Huelsenbeck, 2003). The evolutionary model selected for each data partition is listed in Table 3.
Table 3.
Information on sequence data and partitions.
| partition | length (bp) | parsimony informative sites | model used in MrBayes | maximum likelihood divergence from T. castaneum |
||||
|---|---|---|---|---|---|---|---|---|
| T. freemani | T. confusum | T. brevicornis | L. oryzae | |||||
| combined | 3106 | 653 | (21.0%) | GTR+I+Γ | 8.3% | 13.6% | 14.6% | 16.6% |
| COI | 813 | 234 | (28.8%) | GTR+I+Γ | 16.2% | 17.6% | 20.9% | 20.0% |
| pos. 1 | 271 | 47 | (17.3%) | GTR+Γ | 7.4% | 10.7% | 12.0% | 13.0% |
| pos. 2 | 271 | 10 | (3.7%) | GTR+Γ | 2.2% | 3.7% | 3.8% | 3.7% |
| pos. 3 | 271 | 177 | (65.3%) | HKY+Γ | 39.1% | 38.4% | 47.0% | 43.5% |
| 16S rRNA | 451 | 115 | (25.5%) | GTR+I+Γ | 13.1% | 16.6% | 15.4% | 17.2% |
| stem | 130 | 17 | (13.1%) | HKY+Γ | 5.4% | 10.8% | 6.2% | 6.9% |
| loop | 321 | 98 | (30.5%) | GTR+I+Γ | 16.3% | 19.0% | 19.5% | 21.6% |
| wingless | 468 | 121 | (25.9%) | GTR+Γ | 7.6% | 15.5% | 16.5% | 26.2% |
| pos. 1 | 156 | 11 | (7.1%) | GTR+Γ | 2.6% | 7.4% | 6.5% | 8.5% |
| pos. 2 | 156 | 6 | (3.8%) | HKY+I | 0.6% | 2.0% | 5.2% | 4.5% |
| pos. 3 | 156 | 104 | (66.7%) | HKY+Γ | 19.6% | 38.9% | 37.7% | 65.9% |
| 28S rRNA | 1044 | 97 | (9.3%) | GTR+I+Γ | 1.6% | 7.1% | 6.5% | 9.0% |
| stem | 394 | 46 | (11.7%) | GTR+Γ | 1.8% | 9.1% | 7.1% | 11.2% |
| loop | 650 | 51 | (7.8%) | GTR+I+Γ | 1.6% | 5.9% | 6.1% | 7.6% |
| histone H3 | 330 | 86 | (26.1%) | GTR+I+Γ | 4.5% | 17.6% | 21.1% | 18.2% |
| pos. 1 | 110 | 8 | (7.3%) | GTR+Γ | 0.9% | 2.7% | 5.5% | 6.0% |
| pos. 2 | 110 | 0 | JC+I | 0 | 0 | 0 | 0 | |
| pos. 3 | 110 | 78 | (70.9%) | HKY+Γ | 12.7% | 50.0% | 58.3% | 48.4% |
Data partitioning and evaluation of its effectiveness
Large differences in nucleotide divergence among markers (Table 3) suggested the need for data partitioning. Within markers, we also tested the effects of partitioning protein-coding genes by codon position. For ribosomal RNA genes, each sequence was partitioned into putative stem and loop regions, as determined by FoldAlign (Havgaard et al., 2005).
Using the same dataset, separate Bayesian analyses using different partitioning strategies or evolutionary models may be compared by Bayes factors. Bayes factor (BF) can be approximated as the ratio of harmonic mean of likelihoods in the stationary phase of the null model analysis (L0) and the analysis of a second model of interest (L1) (Brandley et al., 2005). Better fit of a model is reflected in a positive value of the ln Bayes factor. The evidence against the null hypothesis is considered to be strong when 2ln BF > 10. Evidence against the null hypothesis is weaker when 0 < 2ln BF < 10, and the null hypothesis is preferred when 2ln BF < 0 (Kass and Raftery, 1995). Several potential partitioning strategies were tested with the combined dataset: 1) no partitioning, 2) partitioning by marker, 3) partitioning by marker and by codon position or stem-loop structures within markers; and 4) random partitions (Table 4). Because of high variation in base composition at third codon positions of wg and H3, we also conducted analyses in which these partitions were excluded.
Table 4.
A summary of phylogenetic analyses performed on various data subsets. Inclusion of data from each marker is denoted by the shaded boxes at left. The partitioning strategy with markers is indicated within the box. Combination of all characters of a marker is denoted by “+”. Coding sequence partitioning based on codon positions is denoted by “1/2/3”, etc. Additional random partitioning within codon position partitions is denoted “11/22/33”. Partitioning by stem-loop structure is denoted “S/L”. Doublet modeling in stem regions is denoted “Sd/L”. Tree scores for maximum likelihood (ML) analyses represent the log likelihood of the best tree according to the model. For Bayesian inference (BI) analyses, tree scores are given as the harmonic mean of the log likelihood. Tree scores from maximum parsimony (MP) analyses are given in the number of tree steps. For Bayesian analyses topologies reflect majority consensus trees. Topologies listed for ML analyses are from 50% bootstrap consensus trees. For MP analyses the most parsimonious tree or strict consensus of equally most parsimonious trees is shown. Tree topologies are described by abbreviating species and species groups as follows: CST, castaneum species group with internal topology as in Fig. 1; CNF, confusum species group; cnf, T. confusum; ana, T. anaphe; dst, T. destructor; brv, T. brevicornis; Lth, Latheticus oryzae; Gnt, Gnathocerus cornutus; Ten, Tenebrio molitor.
| dataset |
method | tree score | 2(lnL0−lnL1) | topology | Fig. | ||||
|---|---|---|---|---|---|---|---|---|---|
| cox1 | 16S | wg | 28S | H3 | |||||
| 1/2/3 | Sd/L | 12/3 | Sd/L | 1/2/3 | BI | 12332.8 | 2799.0 | ((((CST,CNF),Lth),Gnt),brv),Ten | 1 |
| + | + | + | + | + | BI | 13348.3 | 767.8 | ((CST,CNF),(Gnt,Lth)),brv),Ten | |
| 12 random partitions of set length | BI | 13749.0 | −33.4 | ((((CST,CNF),Lth),Gnt),brv),Ten | |||||
| 5 random partitions of set length | BI | 13728.8 | 6.9 | ((CST,CNF),(Gnt,Lth)),brv),Ten | |||||
| 5 random partitions of random length | BI | 13729.9 | 4.6 | ((CST,CNF),(Gnt,Lth)),brv),Ten | |||||
| combined | BI | 13732.2 | (L0) | (CST,CNF),brv,(Gnt,Lth),Ten | |||||
| combined | ML | 13705.2 | (CST,CNF),brv,(Gnt,Lth),Ten | ||||||
| combined | MP | 2272 | (((CST,(CNF,Lth)),brv),Gnt),Ten | ||||||
| 1/2/3 | Sd/L | 12 | Sd/L | 1/2/3 | BI | 11154.2 | ((((CST,CNF),Lth),Gnt),brv),Ten | ||
| 1/2/3 | Sd/L | 12 | Sd/L | 1/2 | BI | 10260.7 | ((((CST,CNF),Lth),Gnt),brv),Ten | (1) | |
| combined (w/o wg&H3 3rd pos) | MP | 1726 | ((((CST,CNF),Lth),brv),Gnt),Ten | 2 | |||||
| 1/2 | Sd/L | 12 | Sd/L | 1/2 | BI | 7700.3 | ((((CST,CNF),Lth),Gnt),brv),Ten | ||
| combined (only 3rd positions) | BI | 4284.8 | (((CST,(cnf,Lth)),brv),Gnt),Ten | ||||||
| 1/2/3 | Sd/L | BI | 6390.4 | 1055.2 | ((((CST,Lth),CNF),Gnt),brv),Ten | ||||
| combined | BI | 6918.0 | (L0) | (((CST,Lth),CNF,Gnt),brv),Ten | |||||
| combined | ML | 7699.1 | ((Lth w/in CST),(CNF,brv),Gnt),Ten | ||||||
| combined | MP | 1321 | ((CST,Lth),Gnt),(CNF,brv),Ten | ||||||
| 12/3 | Sd/L | 1/2/3 | BI | 5902.7 | 1318.1 | (CST,cnf),brv,(Gnt,Lth),Ten | |||
| combined | BI | 6561.8 | (L0) | ((CST,cnf),brv),(Gnt,Lth),Ten | |||||
| combined | ML | 6542.1 | (CST,cnf),brv,(Gnt,Lth),Ten | ||||||
| combined | MP | 937 | (((CST,(cnf,Lth)),brv),Gnt),Ten | ||||||
| 12 | Sd/L | 1/2/3 | BI | 4724.7 | 939.1 | resolves only (CST,cnf) | |||
| combined (w/o wg pos3) | BI | 5194.3 | (L0) | ((CST,cnf),brv),(Gnt,Lth),Ten | |||||
| 12 | Sd/L | 1/2 | BI | 3765.6 | 751.0 | ((CST,cnf),brv),Gnt,Lth,Ten | |||
| combined (no pos3) | BI | 4141.1 | (L0) | ((CST,cnf),brv),(Gnt,Lth),Ten | |||||
| combined (no pos3) | MP | 390 | ((((CST,cnf),Lth),Gnt),brv),Ten | ||||||
| 11/22/33 | BI | 4193.4 | 540.0 | ((CST,CNF,Lth,Gnt),brv),Ten | |||||
| 1/2/3 | BI | 4192.2 | 542.4 | ((CST,CNF,Lth,Gnt),brv),Ten | 4A | ||||
| 12/3 | BI | 4235.7 | 455.5 | resolves only CST | |||||
| rand (3) | BI | 4472.5 | −18.0 | resolves only CST and (cnf,dst) | |||||
| + | BI | 4463.4 | (L0) | ((CST,Lth,Gnt),(cnf,dest),ana,brv),Ten | |||||
| + | ML | 4431.1 | (((CST,Lth),Gnt),CNF),brv),Ten | ||||||
| + | MP | 897 | (((((Lth w/in CST),cnf),ana),(brv,Gnt)),dst),Ten | 4F | |||||
| Sd/L | BI | 2187.5 | 213.4 | ((CST,Lth),CNF,Gnt),brv),Ten | 4B | ||||
| S/L | BI | 2275.7 | 36.8 | ((CST,Lth),CNF,Gnt),brv),Ten | |||||
| rand | BI | 2292.0 | 4.3 | ((CST,Lth),CNF,Gnt),brv),Ten | |||||
| + | BI | 2294.2 | (L0) | ((CST,Lth),CNF,Gnt),brv),Ten | |||||
| + | ML | 2265.7 | (((CST,Lth)(CNF,Gnt)),brv),Ten | ||||||
| + | MP | 421 | ((((Lth w/in CST),Gnt),CNF),brv),Ten | 4G | |||||
| 1/2/3 | BI | 1985.4 | 214.1 | (((Gnt,Lth),cnf),brv),(Ten w/in CST) | |||||
| 12/3 | BI | 1971.0 | 242.8 | (((Gnt,Lth),brv),cnf),(Ten w/in CST) | 4C | ||||
| rand | BI | 2089.4 | 6.0 | (((Gnt,Lth),cnf),brv),(Ten w/in CST) | |||||
| + | BI | 2092.4 | (L0) | (((Gnt,Lth),brv),cnf),(Ten w/in CST) | |||||
| + | ML | 2074.4 | resolves only (Gnt,Lth) and (cst,Ten) | ||||||
| + | MP | 361 | (((Lth,Gnt),brv),cnf),mad,fre,cst,Ten | 4H | |||||
| 1/2 | BI | 752.0 | 3.0 | (((CST,brv),cnf),(Gnt,Lth)),Ten | |||||
| 12 | BI | 753.5 | (L0) | (((CST,cnf),brv),(Gnt,Lth)),Ten | 4C' | ||||
| 12 | MP | 57 | resolves only CST | ||||||
| Sd/L | BI | 2614.5 | 681.9 | ((CST,cnf),brv),Lth,Gnt,Ten | 4D | ||||
| S/L | BI | 2899.0 | 112.9 | ((CST,cnf),brv),Lth,Gnt,Ten | |||||
| rand | BI | 2953.5 | 3.9 | ((CST,cnf),brv),Lth,Gnt,Ten | |||||
| + | BI | 2955.5 | (L0) | ((CST,cnf),brv),Lth,Gnt,Ten | |||||
| + | ML | 2932.4 | ((CST,cnf),brv),Lth,Gnt,Ten | ||||||
| + | MP | 312 | ((((CST,cnf),Lth),Gnt),brv),Ten | 4I | |||||
| 1/2/3 | BI | 1294.2 | 317.9 | (CST,cnf,Lth),brv,Gnt,Ten | 4E | ||||
| 12/3 | BI | 1317.4 | 271.6 | (CST,cnf,Lth),brv,Gnt,Ten | |||||
| rand | BI | 1446.2 | 13.9 | (CST,cnf),Lth),brv),Gnt),Ten | |||||
| + | BI | 1453.1 | (L0) | (CST,cnf),brv,Lth,Gnt,Ten | |||||
| + | ML | 1426.0 | ((((CST,cnf),Lth),brv),Gnt),Ten | ||||||
| + | MP | 247 | (((CST,(cnf,Lth)),Gnt),brv),Ten | 4J | |||||
| 1/2 | BI | 407.8 | 25.1 | ((CST,cnf),brv,Gnt),Lth,Ten | |||||
| 12 | BI | 420.4 | (L0) | resolves only (cst,fre,mad,cnf) | |||||
| 12 | MP | 17 | (((cst,mad),fre,(cnf,brv),Gnt),Lth),Ten | ||||||
Phylogenetic analyses
Phylogenies were inferred using a number of methods. Parsimony and maximum likelihood analyses were conducted in PAUP* 4.0beta10 (Swofford, 2003). Heuristic maximum likelihood (ML) searches included 10 random addition replicates and 100 bootstrapping iterations each with a single random addition replicate. Parsimony analyses used the branch and bound algorithm, and gaps were treated as missing data. The program TreeRot (Sorenson, 1999) was used for the calculation of partitioned decay indices (Baker and DeSalle, 1997). In ML searches, values of parameters for rate matrices, site heterogeneity and invariance were estimated by Modeltest on a neighbor-joining tree. MrBayes 3.1 was used for Bayesian phylogenetic inference (Ronquist and Huelsenbeck, 2003). In most analyses, four Markov chains were allowed to sample parameters and tree topologies every 1000 generations for 1,100,000 generations. Inspecting logged parameter values by eye with Tracer (Rambaut and Drummond, 2004) demonstrated that values typically stabilized after 3000 to 4000 generations, and the first 100,000 were discarded as a conservative burn-in. For each Bayesian analysis, Tracer was also used to verify that autocorrelation between samples was limited enough to produce effective sample sizes greater than 100 for each parameter. In highly partitioned analyses making use of doublet RNA models, it became necessary to run analyses longer (2.5 × 107 generations) to ensure adequate effective sample sizes for doublet parameters. In all analyses, Tenebrio molitor was used to root trees.
Results and Discussion
Sequence data were obtained from two mitochondrial genes and three nuclear loci from five Tribolium species and three other tenebrionids. These data were supplemented with orthologous mitochondrial sequences of three additional Tribolium species from GenBank. Thus, the combined dataset includes 11 taxa and 3106 aligned nucleotide positions. Selected models and corrected divergence levels between T. castaneum and several other species are listed in Table 3 for each data partition. Between species groups, coding sequences appear to be saturated at third codon positions.
As illustrated by nucleotide divergence (Table 3), the evolutionary rates of individual markers and partitions vary greatly, suggesting that analyses would benefit from a method of data partitioning. MrBayes 3.1 currently implements data partitioning in a Bayesian phylogenetic framework (Ronquist and Huelsenbeck, 2003). Bayesian methods also have the advantage of sampling many trees through a multi-chain Monte Carlo (MCMC) process, and the posterior distribution of topologies and model parameters provides estimations of support for particular branches and parameter values. By testing various partitioning strategies, we determined that a highly partitioned, combined analysis was preferred over unpartitioned analysis (Δ 2ln BF = 2799.0; Table 4; Fig. 1) and over partitioning by gene alone (Δ 2ln BF = 2031.1). After discussing the phylogenies inferred using the results of combined analyses, we will address methodological questions related to individual markers and data partitioning.
Figure 1.
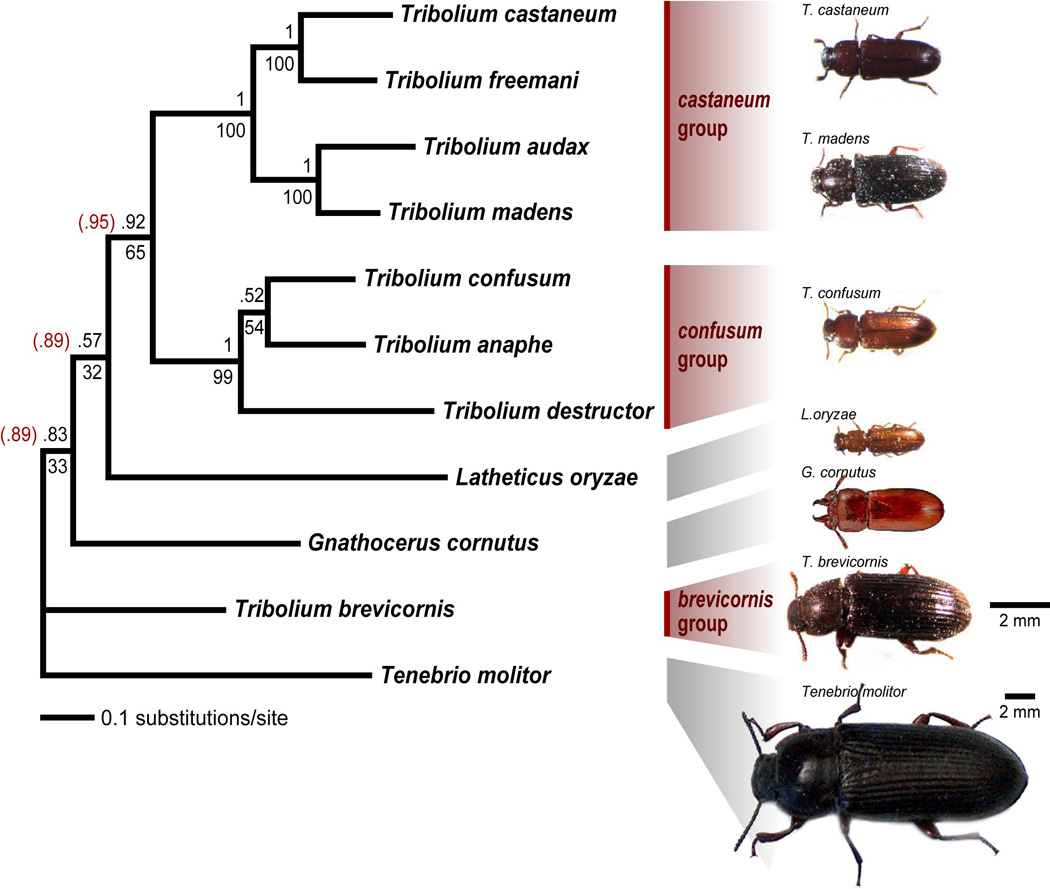
Consensus phylogram from the complete combined partitioned Bayesian analysis. Bayesian posterior probabilities (above the branch) and bootstrap values > 50% in the combined ML analysis are shown (below the branch). Posterior probabilities given in parentheses result from exclusion of partitions in which base composition is not homogeneous across taxa (third codon positions of wingless and histone H3). Adults of selected species are shown to scale, with the exception of Tenebrio molitor.
Monophyly is strongly supported for the castaneum and confusum species groups
The consensus phylogram from the partitioned Bayesian analysis using the complete, combined dataset is presented in Figure 1. The castaneum and confusum species groups are recovered as monophyletic in all the sampled trees and also in all ML bootstrap replicates. Within the castaneum group, relationships are resolved, with T. castaneum + T. freemani sister to T. madens + T. audax. A close relationship between T. castaneum and T. freemani has been suggested by their ability to produce hybrid offspring (Nakakita et al., 1981; Wade and Johnson, 1994b). Tribolium madens and T. audax are morphologically quite similar, and a close relationship has been suggested by Halstead (1969). Relationships among the three confusum group species are less certain. The consensus topology unites T. confusum and T. anaphe to the exclusion of T. destructor but this relationship is weakly supported (Bayesian posterior support: 0.52; ML bootstrapping: 54%), with the other two possible arrangements receiving roughly equal support in the Bayesian posterior trees. All combined Bayesian and ML analyses provide moderate to high support for a close relationship between the castaneum and confusum species groups relative to other species in this study. The monophyly of species groups was also recovered using parsimony (Fig. 2). Bootstrap support and decay indices for the relationships within the castaneum species group are stronger than within the confusum group (Fig. 2). The most parsimonious tree in combined analyses groups T. anaphe and T. destructor, but the decay index for this branch is low and comes exclusively from cox1 (Fig. 2).
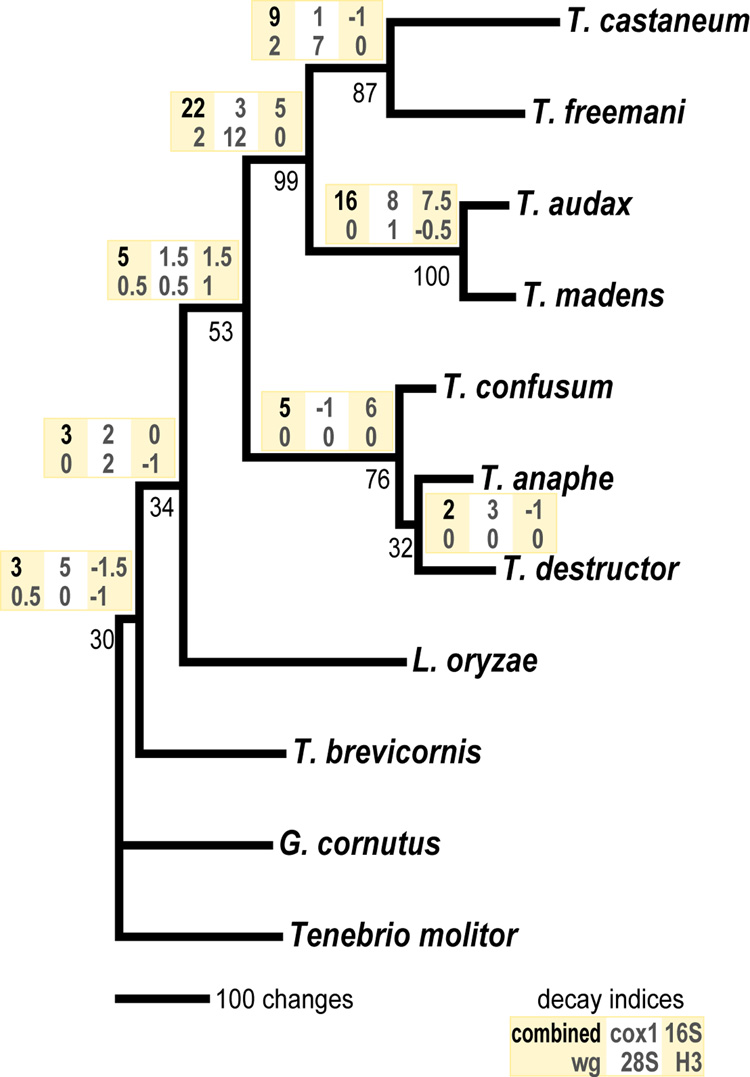
Figure 2.
Most parsimonious tree based on combined analysis of cox1, 16S rDNA, wg, 28S rDNA, and histone H3. This analysis excluded codon third positions from wg and histone H3 based on failure of the base homogeneity test (P ≪ 0.01). Decay indices based on the combined dataset and each partition are given above each branch. Bootstrap values are given below branches.
These results support the relationships suggested by Hinton (1948), and contradict the phylogenetic hypotheses of later studies based on chemotaxonomy (Howard, 1987), isozyme cladistics (Wool, 1982), and satellite DNAs (Juan, et al., 1993). The mitochondrial phylogeny of Meštrović et al. (2006) also strongly supports the monophyly of the castaneum and confusum species groups, although using parsimony and Bayesian methods they obtain conflicting species relationships within the confusum group.
The genus Tribolium is not monophyletic
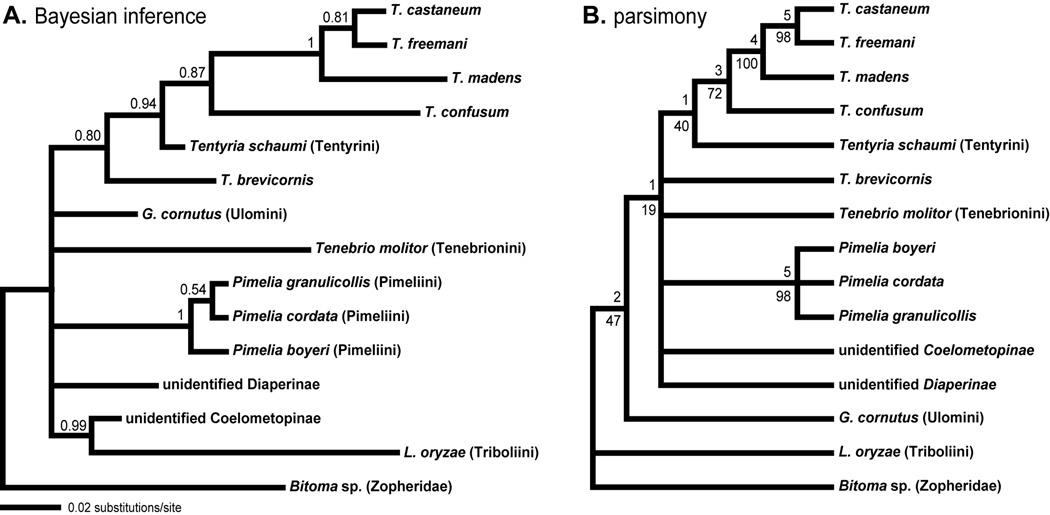
Multiple analyses suggest that Tribolium is not monophyletic (Table 4; Fig. 1, 2, 3). In the combined Bayesian analysis both L. oryzae and G. cornutus are inferred to be more closely related to the castaneum + confusum species groups than is T. brevicornis. Although neither of these relationships is strongly supported (<0.83 posterior probability), monophyly of Tribolium had a very low posterior probability (0.052). Maximum likelihood analysis in PAUP is equivocal on the relations of L. oryzae, G. cornutus, T. brevicornis, and other Tribolium species (which are grouped as in Fig. 1). The ML tree unifies Tribolium (−lnL = 13705.2; Table 4); however, the second best tree (−lnL = 13705.4) places both L. oryzae and G. cornutus as more closely related to the castaneum + confusum groups, as in the Bayesian analysis. Only five of the ten best ML trees (−lnL < 13709.5) support the monophyly of Tribolium. Moreover, monophyly is supported in only 37% of bootstrap trees. Parsimony also places L. oryzae as sister to the clade of castaneum + confusum species groups (Fig. 2). Although the decay index is low for this branch, support comes from multiple markers.
Figure 3.
Trees produced from analyses of tenebrionid 28S rDNA sequences. (A) Majority consensus phylogram from Bayesian inference. Posterior probabilities are given above each branch. (B) Consensus cladogram of six equally most parsimonious trees. Decay indices are shown above branches. Bootstrap values are given below each branch. In both trees, taxa are poorly resolved at the subfamilial and tribal levels. Notably Tribolium is rendered polyphyletic in these analyses.
Other evidence is consistent with the hypothesis that the brevicornis group is not sister to the other Tribolium species. For more than 80 years, brevicornis group species were regarded as a distinct genus, Aphanotus (Leconte, 1862; Casey, 1890), until they were subsumed into Tribolium by Hinton (1948). Despite this merger, exceptions for brevicornis group morphology are made in the Tribolium diagnosis with respect to the form of the prosternal and pronotal apices. The brevicornis group species were regarded as primitive in these characters relative to other Tribolium species (Hinton, 1948). Hinton also conceded that the brevicornis group might merit recognition at the genus level because of similarity between other Tribolium groups and the genus Lyphia. While other Tribolium species are native to the tropical Old World, brevicornis group species are distributed in the New World and occur in more temperate climates (Table 1). Furthermore, a recent analysis of satellite DNA sequences from T. brevicornis has identified satellites in this species that are unrelated to others in the genus Tribolium (Mravinac et al., 2005).
In an effort to further examine the relationship of T. brevicornis to other species of Tribolium, we conducted analyses using sequence data from additional tenebrionids. A previous mitochondrial phylogeny of Tribolium has suggested monophyly of the genus, but this is likely due to the fact that fewer and different outgroup (non-Tribolium) species were included (Meštrović, et al., 2006). Here we sought to include as broad a sampling of tenebrionids as possible. We used 28S rDNA sequences available from GenBank, yielding a total of 15 tenebrionid species representing four tenebrionid tribes and three subfamilies, rooted with a sequence from the allied family Zopheridae. The relationships between T. castaneum, T. freemani, T. madens, and T. confusum are consistent in these analyses (Fig. 3) and the multigene analyses (Fig. 1, 2; Table 4). Most basal relationships could not be resolved. However, the monophyly of Tribolium was again recovered with very low probability (0.029) from the Bayesian posterior sample. Instead, Tentyria schaumi Kraatz (1865) (Tentyrini) is supported as the closest relative of the castaneum + confusum species groups (Fig. 3A; 0.94 posterior probability). Tribolium brevicornis appears as sister to this larger clade with modest support (0.80). Parsimony also groups Tentyria schaumi as sister to the castaneum + confusum species groups (Fig. 3B), although this branch does not receive strong support (40% bootstrap support; decay index of 1). While it is clear that greater taxon sampling is needed to definitively assign relationships among Tenebrionidae, these results reinforce the conclusion that T. brevicornis is not the closest relative of other Tribolium species groups.
Transfer of brevicornis group species from Tribolium MacLeay (1825) to Aphanotus Leconte (1862)
Considering the results of this study and the morphological observations of Hinton (1948), it is appropriate that brevicornis group species be removed from Tribolium. The genus Aphanotus Leconte (1862) previously contained T. brevicornis and T. parallelus (Casey 1890) before being subsumed by Hinton (1948) into Tribolium. This taxon is currently available, containing no species. Therefore, all brevicornis group species are here transferred to Aphanotus in order to reflect their independent evolutionary history. Thus, the genus Aphanotus contains seven known species: Aphanotus brevicornis (Leconte, 1859), Aphanotus carinatum (Hinton, 1948) n. com., Aphanotus gebieni (Uyttenboogaart, 1934) n. com., Aphanotus linsleyi (Hinton, 1948) n. com., Aphanotus parallelus (Casey, 1890), Aphanotus setosum (Triplehorn, 1978) n. com., and Aphanotus uezumii (Nakane, 1963) n. com. The descriptions of Aphanotus given by Leconte (1859; 1862) are very brief. Therefore, Hinton’s (1948) description of the brevicornis species group remains the best description of the genus Aphanotus. This change does not affect the use of Hinton’s key for the identification of species.
Problems with dating divergences among Tribolium
The dates of lineage splits separating T. castaneum from its relatives remain obscure. It was Hinton’s supposition that Tribolium species groups were very old due to their wide geographic distributions. However, the fact that T. castaneum and T. freemani as well as T. madens and T. audax are capable of hybridization may suggest more recent origins. The topologies of all analyses in this study were not clock-like, preventing a reliable estimate of divergence times from either a fixed or relaxed clock model. Branch lengths are most consistent among the castaneum and confusum group species where a clock might be applicable. Using various mitochondrial molecular clock estimates (Venanzetti et al., 1993; Farrell, 2001), divergence times for T. castaneum and T. freemani range from 11.6 to 47.0 million years ago (Mya), and for T. castaneum and T. confusum between 13.9 and 60.7 Mya. While fossil data on various beetle families could be used to constrain a relaxed clock analysis, the extreme rate variation among lineages would still make such an undertaking problematic. Therefore, a reliable estimate of divergence dates within Tribolium will ultimately require calibration information specific to this lineage.
Conflict among individual markers and combined analyses
The five markers used in this study present a moderate degree of conflict (Fig. 4; Table 4). Conflict among characters or multiple loci is a frequent problem which has been considered extensively (e.g. Olmstead and Sweere, 1994; Mason-Gamer and Kellogg, 1996; Nixon and Carpenter, 1996; Gatesy et al., 1999; Kjer et al., 2001; Funk and Omland, 2003). Partitions including cox1, 16S, and first and second codon positions of histone H3 contribute negative decay indices for at least one branch in the combined parsimony analysis (Fig. 2). In the Bayesian analyses of this study, nuclear and mitochondrial markers differ mostly in their placement of L. oryzae, with mitochondrial markers weakly supporting this species as sister to the castaneum group (0.57), and nuclear markers supporting a clade containing L. oryzae and G. cornutus (0.67 posterior probability; 0.95 probability of castaneum + confusum groups).
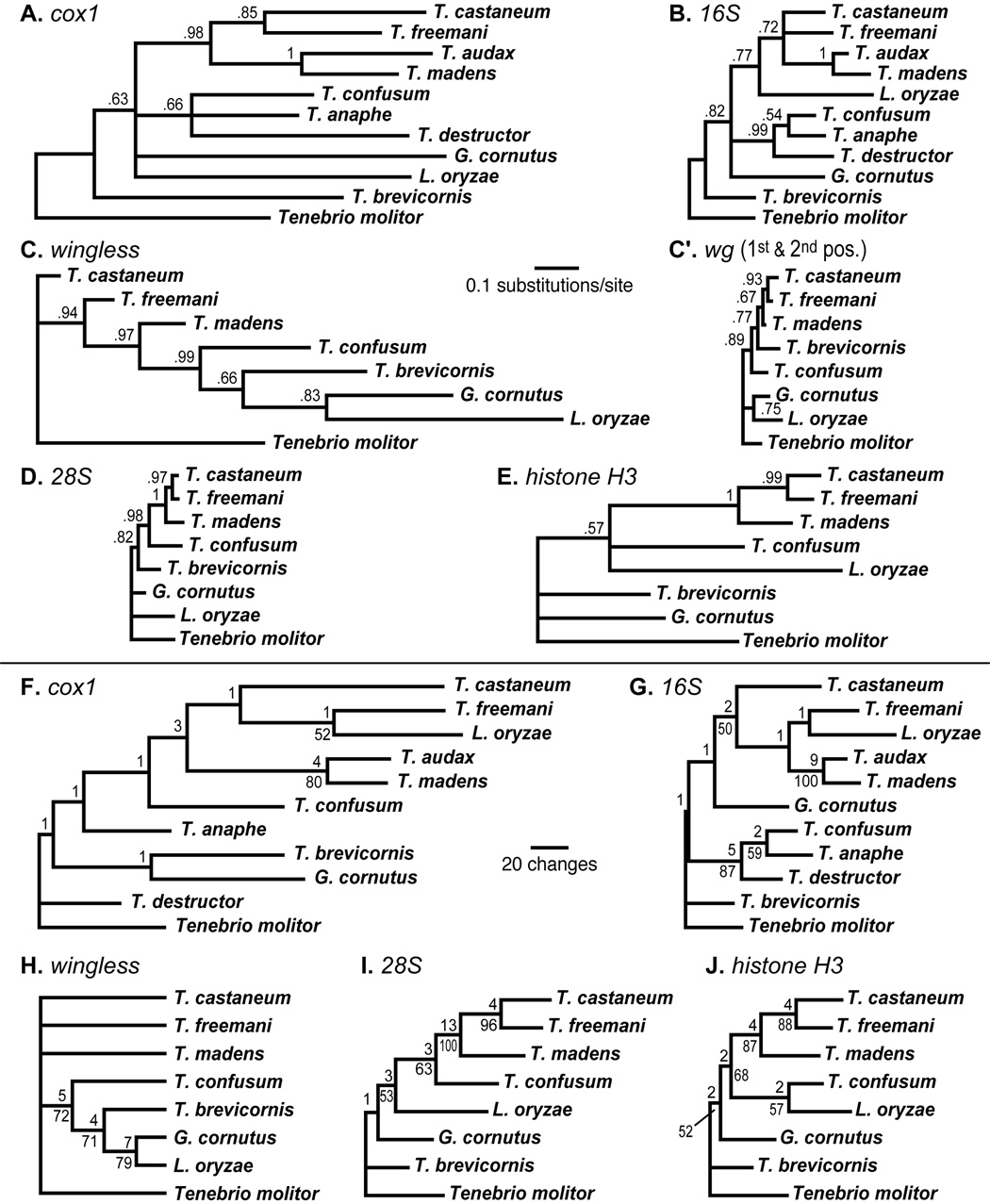
Figure 4.
Trees from single marker analyses. The scale of branch lengths is comparable among Bayesian (A–E) and parsimony (F–J) analyses. In Bayesian consensus phylograms posterior probabilities are given above each branch. A single most parsimonious tree was obtained for most markers (F,G,I,J). However analysis of wg yielded two equally parsimonious trees. The consensus cladogram from this analysis is shown (H); note that branch lengths are arbitrary in this tree. Decay indices for parsimony trees is given above each branch, and bootstrap values > 50% are shown below branches.
The monophyly of Tribolium is recovered only in analyses of 28S rDNA alone (Fig. 4D, 0.82 Bayesian posterior probability; Fig. 4I, 63% bootstrap support, decay index of 3). However, in the expanded 28S analyses, Tribolium is rendered paraphyletic by Tentyria schaumi (Fig. 3). Partitioned decay indices reveal that most support for the topology in combined parsimony analysis comes from cox1 and somewhat from 28S (Fig. 2). Thus, while single marker analyses are not sufficient to resolve relationships among Tribolium species groups, together they provide strong support for the main conclusions of this study: that the castaneum and confusum species groups are monophyletic and together form a clade, and that T. brevicornis is not included with other Tribolium species.
In Bayesian analyses, most individual markers suggest the monophyly of Tribolium species groups (Fig. 4A–E). The only exception is wingless. When trees are rooted with Tenebrio molitor, the castaneum species group is rendered paraphyletic by a clade containing all other species, and the pattern of branch lengths suggests that the tree is improperly rooted (Fig. 4C). If the tree is instead rooted with G. cornutus or L. oryzae, then the castaneum group is instead rendered paraphyletic by the inclusion of Tenebrio molitor. However, this placement of Tenebrio molitor appears with posterior support of less than 0.0002 in all combined Bayesian analyses. One possible explanation of this surprising result is a shared base composition bias. Tenebrio molitor and T. castaneum share high G+C content in third codon positions of the wingless gene (85.5% and 75.3%, respectively) relative to other species in this study (31.0% for L. oryzae to 65.6% for T. brevicornis). Excluding third codon positions reduces support for Tenebrio molitor + T. castaneum to a posterior probability of 0.051 (Fig. 4C'). Similarly, base composition differs at third codon positions of histone H3, where G+C content is low in L. oryzae (17.2%) and T. confusum (27.3%), relative to other taxa (37.3% for T. freemani to 66.4% for Tenebrio molitor). With the exclusion of H3 third codon positions, monophyly of the castaneum + confusum species groups is supported in Bayesian analysis (Table 4). Therefore, it is possible that selection on G+C content or codon bias has led to nucleotide convergence in synonymous nuclear sites of some species. Exclusion of third codon positions of both wg and H3 from the combined five-marker Bayesian analysis, does not change the consensus topology (Table 4), however, support is increased for basal branches of the tree (indicated parenthetically in Fig. 1). Inclusion of the two partitions with heterogeneous base composition alters phylogenetic relationships in parsimony grouping (Table 4).
Data partitioning
A major advantage of phylogenetic studies incorporating relatively few taxa is that it becomes feasible to explore a variety of analytical methods. Here, we have examined the merit of a wide range of data partitioning strategies in combined Bayesian analyses. As implemented in MrBayes, separate data partitions allow individual models of nucleotide evolution, and parameters, such as rate matrices, base frequencies, and rate heterogeneity, to be estimated independently for each partition.
Data are often partitioned in a manner informed by biological structure and function, for example by gene or by codon position within protein-coding sequences. However, if a dataset contains characters with an effectively multimodal distribution of model parameters, any partitioning strategy, including random partitioning, has the potential to isolate these conflicting characters and improve performance. Therefore we compared the effects of biologically informed partitioning strategies to those of several different strategies for randomly assigning nucleotide characters to partitions. For single genes, either two (ribosomal DNA) or three (protein-coding) random partitions were created. Because the biologically informed, partitioned analyses resulted in 5 (by gene) or 12 (by codon position and stem-loop structure) partitions, combined analyses were performed with 5 or 12 partitions to which characters where randomly assigned. In these analyses, the length of partitions was set equal to their biologically based counterparts. An additional five-partition analysis was conducted in which the length of each partition was also randomly determined.
In 6 of 8 analyses, random partitions improved model fit relative to unpartitioned data, but the improvements were small, especially compared to the improvement from biologically-informed partitioning (Table 4). This improved fit is likely due to the random partitions capturing some of the otherwise un-modeled complexity in the dataset. However, in two cases, the full dataset with 12 partitions and cox1, random partitioning was highly detrimental. The length of partitions did not have a strong effect, but the Bayes factor became significantly more negative in the more highly partitioned analysis. Furthermore, increased partitioning may not be effective when the data contains little or no additional structure. For example, when partitions consisting of first, second, and third codon positions from cox1 were each further randomly partitioned in two, the resulting analysis was not favored over partitioning by codon position alone (2ln BF = −2.4). These results suggest that random partitioning is not an effective method to reliably improve model fit. Instead, comparison to partitions based on sequence structure and function emphasize the importance of biological relevance to partitioning.
Given the limited utility of random partitioning, to what extent is biologically informed partitioning useful? Partitioning of the combined dataset by marker or by structure within markers resulted in significantly improved fit relative to unpartitioned analysis (2ln BF = 767.8 and 2782.1, respectively). In analyses of single markers, partitioning by codon position or stem-loop structure was highly favored (2ln BF between 36.8 and 542.4). However, increased partitioning was not always favored. For each of the three protein-coding markers used in this study, we tested partitioning third codon positions and each position individually compared to an unpartitioned analysis. For cox1 and histone H3, partitioning each codon individually was a significantly better fit than combining first and second positions (2ln BF = 46.3 and 86.9, respectively). However, for wingless partitioning each codon position did not produce as good a fit as when first and second positions were combined (2ln BF = −28.7). Therefore, it is not universally true that greater biologically-based partitioning is consistently beneficial to analyses (Brandley, et al., 2005).
RNA doublet modeling significantly improves the performance of rDNA partitions
In addition to testing partitions within protein-coding markers, the effect of partitioning ribosomal DNA sequences based on their predicted RNA secondary structure was also examined. The complement of nucleotides in predicted RNA stem structures was used to specify nucleotide doublet pairs in MrBayes. Doublet models account for the fact that mutation in one nucleotide of a pair increases the likelihood of complementary substitution at the apposing site (Schoniger and Von Haeseler, 1994), effectively reducing branch lengths.
Partitioning of predicted stem and loop regions significantly improved the performance of analyses for 16S (2ln BF = 36.8) and 28S rDNA (2ln BF = 112.9). However, a drastic gain in fit was obtained by applying the doublet model to stem regions. The difference in 2ln BF was 176.6 for 16S and 569.0 for 28S. Therefore, stem-loop partitioning and stem doublet modeling appear to be beneficial to parameter fit in combined Bayesian analyses. While it seems likely that partitions based on very accurate stem-loop predictions, such as those of nuclease-protection assays or x-ray crystallography would perform still better, a relatively simple prediction method such as FoldAlign is highly effective.
Conclusions
Through the use of multiple evolutionary models in data partitions based on sequence structure and function in combined Bayesian analyses, we have inferred a well-supported phylogeny for pest species of Tribolium (Fig. 1). This is also supported by combined parsimony analysis (Fig. 2). The developmental genetic model species T. castaneum appears to be highly nested within Tribolium. Thus, other species of the genus provide a range of phylogenetic distances for comparison to T. castaneum. The monophyly of the castaneum and confusum species groups is recovered with high support. These groups also appear to be each other’s closest relatives. Significantly, the genus Tribolium as previously recognized is not recovered as monophyletic. Tribolium brevicornis is more distantly related to Tribolium sensu stricto than are representatives of other tenebrionid genera in analyses of the combined dataset and four of five single markers. Based on this and other evidence, brevicornis group species are removed to the available genus Aphanotus. Thus, derived characters shared by Aphanotus and Tribolium likely provide examples of convergence. These conclusions should provide an evolutionary framework for comparative studies of Tribolium and other flour beetles, utilizing the genetic and genomic tools of T. castaneum in related, phenotypically distinct species.
Acknowledgements
Beetle stocks were kindly provided by the labs of Susan J. Brown and Robin Denell. Jonathan Q. Richmond and Paul O. Lewis generously provided guidance regarding phylogenetic methods and software. Thanks also to Brigid O’Donnell and Maxi Polihronikas for sharing useful PCR primers. The image of Gnathocerus cornutus used in Figure 1 was kindly provided by James Dunford. We are grateful for comments on the manuscript from the Jockusch lab group and two anonymous reviewers. This work was supported through an NIH NRSA Kirschstein postdoctoral fellowship to DRA (5F32GM074365-02) and research funds from the University of Connecticut to ELJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Abouheif E, Wray GA. Evolution of the gene network underlying wing polyphenism in ants. Science. 2002;297:249–252. doi: 10.1126/science.1071468. [DOI] [PubMed] [Google Scholar]
- Angelini DR, Kaufman TC. Insect appendages and comparative ontogenetics. Dev. Biol. 2005;286:57–77. doi: 10.1016/j.ydbio.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Baker RH, DeSalle R. Multiple sources of character information and the phylogeny of Hawaiian Drosophilids. Syst. Biol. 1997;46:654–673. doi: 10.1093/sysbio/46.4.654. [DOI] [PubMed] [Google Scholar]
- Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- Blair KG. The Indian species of Palorus, Muls. (Coleoptera : Tenebrionidae) and some associated beetles. Indian Forest Rec. 1930;14:1–20. (133–152) illus. [Google Scholar]
- Brandley MC, Schmitz A, Reeder TW. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst. Biol. 2005;54:373–390. doi: 10.1080/10635150590946808. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Denell RE, Beeman RW. Beetling around the genome. Gen. Res. 2003;82:155–161. doi: 10.1017/s0016672303006451. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Hilgenfeld RB, Denell RE. The beetle Tribolium castaneum has a fushi tarazu homolog expressed in stripes during segmentation. Proc. Natl. Acad. Sci. USA. 1994;91:12922–12926. doi: 10.1073/pnas.91.26.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, Mahaffey JP, Lorenzen MD, Denell RE, Mahaffey JW. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol. Dev. 1999;1:11–15. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- Casey TL. Coleopterological notices, part II. An. New York Acad. Sci. 1890;5:307–504. [Google Scholar]
- Charpentier T. Horae Entomologicae, adjectis tabulis novem coloratis. Wratislaviae. 1825;16:261. [Google Scholar]
- Daly HV, Doyen JT, Purcell AHI. Oxford: Oxford University Press; 1998. Introduction to Insect Biology and Diversity. [Google Scholar]
- DeMuth JP, Wade MJ. Population differentiation in the beetle Tribolium castaneum. I. Genetic architecture. Evolution. 2007a;61:494–509. doi: 10.1111/j.1558-5646.2007.00048.x. [DOI] [PubMed] [Google Scholar]
- DeMuth JP, Wade MJ. Population differentiation in the beetle Tribolium castaneum. II. Haldane's Rule and incipient speciation. Evolution. 2007b;61:694–699. doi: 10.1111/j.1558-5646.2007.00049.x. [DOI] [PubMed] [Google Scholar]
- Doyen JT. Reconstruction of the tribes Ulomini and Triboliini for North and Central America (Tenebrionidae: Coleoptera) Proc. Entomol. Soc. Wash. 1985;87:512–524. [Google Scholar]
- Duncan FD. The role of the subelytral cavity in respiration in a tenebrionid beetle, Onymacris multistriata (Tenebrionidae: Adesmiini) J. Insect Physiol. 2003;49:339–346. doi: 10.1016/s0022-1910(03)00018-0. [DOI] [PubMed] [Google Scholar]
- Eckert C, Aranda M, Wolff C, Tautz D. Separable stripe enhancer elements for the pair-rule gene hairy in the beetle Tribolium. EMBO Rep. 2004;5:638–642. doi: 10.1038/sj.embor.7400148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairmaire L. Matériaux pour la faune coléoptérique de la région Malgache. An. Soc. Entomol. France. 1902;71:325–388. [Google Scholar]
- Farrell BD. Evolutionary assembly of the milkweed fauna: cytochrome oxidase I and the age of Tetraopes beetles. Mol. Phyl. Evol. 2001;18:467–478. doi: 10.1006/mpev.2000.0888. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Omland KE. Species-Level Paraphyly and Polyphyly: Frequency, Causes, and Consequences, with Insights from Animal mitochondrial DNA. An. Rev. Ecol. Evol. Syst. 2003;34:397–423. [Google Scholar]
- Gatesy J, O'Grady P, Baker R. Corroboration among data sets in simultaneous analysis: Hidden support for phylogenetic relationships among higher level artiodactyl taxa. Cladistics. 1999;15:271–313. doi: 10.1111/j.1096-0031.1999.tb00268.x. [DOI] [PubMed] [Google Scholar]
- Gebien H. Tenebrionidae. In: Junk W, Schenkling S, editors. Tenebrionidae. Berlin: W. Junk; 1910. pp. 1–354. [Google Scholar]
- Gompel N, Carroll SB. Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature. 2003;424:931–935. doi: 10.1038/nature01787. [DOI] [PubMed] [Google Scholar]
- Gridelli E. Contribution à l'étude de l'Aïr (Col. Tenebrionidæ) Mém. Inst. Français d'Afrique Noire. 1950;10:153–180. [Google Scholar]
- Grimaldi D, Engel MS. New York: Cambridge University Press; 2005. Evolution of the Insects. [Google Scholar]
- Grimm R. Faunistik und Taxonomie einiger Arten der Gattung Tribolium MacLay, 1825, mit Beschreibung von drei neuen Arten aus Africa (Coleoptera, Tenebrionidae) Entomofauna. 2001;22:393–404. [Google Scholar]
- Halstead DGH. Notes on the systematics and distribution of some Tribolium species (Coleoptera: Tenebrionidae) J. Stored Prod. Res. 1967;3:269–272. [Google Scholar]
- Halstead DGH. A new species of Tribolium from North America previously confused with Tribolium madens (Charp) (Coleoptera: Tenebrionidae) J. Stored Prod. Res. 1969;4:295–304. [Google Scholar]
- Hancock JM, Tautz D, Dover GA. Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster. Mol. Biol. Evol. 1988;5:393–414. doi: 10.1093/oxfordjournals.molbev.a040501. [DOI] [PubMed] [Google Scholar]
- Havgaard JH, Lyngso RB, Gorodkin J. The FOLDALIGN web server for pairwise structural RNA alignment and mutual motif search. Nuc. Acids Res. 2005;33:W650–W653. doi: 10.1093/nar/gki473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst JFW. Berlin: Der Käfer; 1797. p. 346. [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Meth. Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hinton HE. A synopsis of the genus Tribolium Macleay, with some remarks on the evolution of its species-groups (Coleoptera: Tenebrionidae) Bull. Entomol. Res. 1948;39:13–55. doi: 10.1017/s0007485300024287. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Nachman MW. Different genes underlie adaptive melanism in different populations of rock pocket mice. Mol. Ecol. 2003;12:1185–1194. doi: 10.1046/j.1365-294x.2003.01788.x. [DOI] [PubMed] [Google Scholar]
- Horn GH. Revision of the Tenebrionidæ of America, North of Mexico. Trans. Amer. Phil. Soc., New Ser. 1870;14:253–404. [Google Scholar]
- Howard RW. Chemosystematic studies of the Triboliini (Coleoptera: Tenebrionidae): Phylogenic inferences from the defensive chemicals of eight Tribolium spp., Palorus ratzeburgi (Wissmann), and Latheticus oryzae Waterhouse. An. Entomol. Soc. Amer. 1987;80:398–405. [Google Scholar]
- Jacquelin du Val PNC, Fairmaire L. Manuel entomologique: Genera des coléoptères d'Europe. Paris: 1868. [Google Scholar]
- Jockusch EL, Williams TA, Nagy LM. The evolution of patterning of serially homologous appendages in insects. Dev. Genes Evol. 2004;214:324–338. doi: 10.1007/s00427-004-0412-6. [DOI] [PubMed] [Google Scholar]
- Juan C, Vazquez P, Rubio JM, Petitpierre E, Hewitt GM. Presence of highly repetitive DNA sequences in Tribolium flour-beetles. Heredity. 1993;70:1–8. doi: 10.1038/hdy.1993.1. [DOI] [PubMed] [Google Scholar]
- Kass RE, Raftery AE. Bayes Factors. J Amer. Stat. Assoc. 1995;90:773–795. [Google Scholar]
- Kaszab Z. Die Tenebrioniden Neukaledoniens und der Loyauté-Inseln. Fol. Entomol. Hungarica. 1982;43:1–294. [Google Scholar]
- Kaszab Z. Insects of Saudi Arabia. Coleoptera: Fam. Tenebrionidae. Part 2. Fauna Saudi Arabia. 1982;4:124–243. [Google Scholar]
- Kjer KM. Aligned 18S and insect phylogeny. Syst. Biol. 2004;53:506–514. doi: 10.1080/10635150490445922. [DOI] [PubMed] [Google Scholar]
- Kjer KM, Blahnik RJ, Holzenthal RW. Phylogeny of Trichoptera (Caddisflies): Characterization of Signal and Noise Within Multiple Datasets. Syst. Biol. 2001;50:781–816. doi: 10.1080/106351501753462812. [DOI] [PubMed] [Google Scholar]
- Kopp A, Duncan I, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Kopp A, True JR. Evolution of male sexual characters in the oriental Drosophila melanogaster species group. Evol. Dev. 2002;4:278–291. doi: 10.1046/j.1525-142x.2002.02017.x. [DOI] [PubMed] [Google Scholar]
- Kronforst MR, Young LG, Kapan DD, McNeely C, O'Neill RJ, Gilbert LE. Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc. Natl. Acad. Sci. USA. 2006;103:6575–6580. doi: 10.1073/pnas.0509685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwieton E. Revue critique des systèmes récents de la famille des Tenebrionidae (Col.) Acta Musei Natl. Prague. 1982;38:79–100. [Google Scholar]
- Lea AM. On Nepharis and other ants' nest beetles taken by Mr. J.C. Goudie at Birchip. Proc. Royal Soc. Victoria. 1904;17:371–385. [Google Scholar]
- Leconte JL. Catalogue of the Coleoptera of Fort Tejon, California. Proc. Acad. Nat. Sci. Philadelphia. 1859;11:69–90. [Google Scholar]
- Leconte JL. Classification of the Coleoptera of North America prepared for the Smithsonian Institution. Smithsonian Misc. Col. 1862;233:209–286. [Google Scholar]
- Lepesme P. Un Tribolium inédit du Sénégal (Col. Tenebrionidae) Rev. Français Entomol. 1943;10:45–46. [Google Scholar]
- Lorenzen MD, Doyungan Z, Savard J, Snow KJ, Crumly LR, Shippy TD, Stuart JJ, Brown SJ, Beeman RW. Genetic linkage maps of the red flour beetle, Tribolium castaneum, based on bacterial artificial chromosomes and expressed sequence tags. Genetics. 2005;170:741–747. doi: 10.1534/genetics.104.032227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen MD, Kimzey T, Shippy TD, Brown SJ, Denell RE, Beeman RW. piggyBac-based insertional mutagenesis in Tribolium castaneum using donor/helper hybrids. Insect Mol. Biol. 2007;16:265–275. doi: 10.1111/j.1365-2583.2007.00727.x. [DOI] [PubMed] [Google Scholar]
- Magis N. Aperçu de l'histoire naturelle des complexes d'espèces du genre Tribolium (McLeay, 1825) (Coleoptera: Tenebrionidae) Bull. Inst. Royal Sci. Nat. Belgique. 1954;30:1–10. [Google Scholar]
- Mason-Gamer RJ, Kellogg EA. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae) Syst. Biol. 1996;45:522–543. [Google Scholar]
- Meštrović N, Mravinac B, Plohl M, Ugarković Đ, Bruvo-Mađarić B. Preliminary phylogeny of Tribolium beetles (Coleoptera: Tenebrionidae) resolved by combined analysis of mitochondrial genes. Euro. J. Entomol. 2006;103:709–715. [Google Scholar]
- Mravinac B, Ugarković Đ, Franjević D, Plohl M. Long Inversely Oriented Subunits Form a Complex Monomer of Tribolium brevicornis Satellite DNA. J. Mol. Evol. 2005;60:513–525. doi: 10.1007/s00239-004-0236-z. [DOI] [PubMed] [Google Scholar]
- Nakakita H. Rediscovery of Tribolium freemani Hinton: a stored product insect unexpected to entomologists for past 100 years. Japan Ag. Res. Q. 1983;16:239–245. [Google Scholar]
- Nakakita H, Imura O, Winks RG. Hybridization between Tribolium freemani Hinton and Tribolium castaneum (Herbst), and some preliminary studies of the biology of Tribolium freemani (Coleoptera: Tenebrionidae) Appl. Entomol. Zool. 1981;16:209–215. [Google Scholar]
- Nakane T. New or little-known Coleoptera from Japan and its adjacent regions. XIX. Heteroma. Family Tenebrionidae. Fragmenta Coleopterologica. 1963;7:26–30. [Google Scholar]
- Neboiss A. Notes on the distribution and descriptions of new species. Mem. Natl. Mus. Victoria. 1962;25:243–258. [Google Scholar]
- Nixon KC, Carpenter JM. On Simultaneous Analysis. Cladistics. 1996;12:221–241. doi: 10.1111/j.1096-0031.1996.tb00010.x. [DOI] [PubMed] [Google Scholar]
- Ober KA, Jockusch EL. The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev. Biol. 2006;294:391–405. doi: 10.1016/j.ydbio.2006.02.053. [DOI] [PubMed] [Google Scholar]
- Ogden TH, Whiting MF. The problem with the Paleoptera problem: sense and sensitivity. Cladistics. 2003;19:432–442. doi: 10.1111/j.1096-0031.2003.tb00313.x. [DOI] [PubMed] [Google Scholar]
- Olmstead RG, Sweere JA. Combining Data in Phylogenetic Systematics: An Empirical Approach Using Three Molecular Data Sets in the Solanaceae. Syst. Biol. 1994;43:467–481. [Google Scholar]
- Parichy DM. Evolution of Danio pigment pattern development. Heredity. 2006;97:200–210. doi: 10.1038/sj.hdy.6800867. [DOI] [PubMed] [Google Scholar]
- Park T, Leslie P, Mertz D. Genetic strains and competition in populations of Tribolium. Phys. Zool. 1964;37:97–162. [Google Scholar]
- Pavlopoulos A, Berghammer AJ, Averof M, Klingler M. Efficient transformation of the beetle Tribolium castaneum using the Minos transposable element: quantitative and qualitative analysis of genomic integration events. Genetics. 2004;167:737–746. doi: 10.1534/genetics.103.023085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinf. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Gen. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Raff EC, Popodi EM, Sly BJ, Turner FR, Villinski JT, Raff RA. A novel ontogenetic pathway in hybrid embryos between species with different modes of development. Dev. 1999;126:1937–1945. doi: 10.1242/dev.126.9.1937. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. computer program. Oxford, UK: 2004. Tracer: MCMC Trace Analysis Tool. ver 1.2.1. http://evolve.zoo.ox.ac.uk/software.html. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinf. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Savard J, Marques-Souza H, Aranda M, Tautz D. A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell. 2006;126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- Schoniger M, Von Haeseler A. A Stochastic Model for the Evolution of Autocorrelated DNA Sequences. Mol. Phyl. Evol. 1994;3:240–247. doi: 10.1006/mpev.1994.1026. [DOI] [PubMed] [Google Scholar]
- Simon C, Rati FF, Eckenbach AB, Respi BC, Iu HL, Look PF. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. An. Entomol. Soc. Amer. 1994;87:651–701. [Google Scholar]
- Smith SG. The evolution of heterochromatin in the genus Tribolium (Tenebrionidae: Coleoptera) Chromosoma. 1952a;4:585–610. doi: 10.1007/BF00325793. [DOI] [PubMed] [Google Scholar]
- Sokoloff A. The Biology of Tribolium. Vol 1. Oxford: Clarendon Press; 1972. [Google Scholar]
- Sokoloff A, Dawson PS, Englert DC. Linkage studies in Tribolium castaneum Herbst. VIII. Short antenna, a dominant marker for the seventh linkage group. Can. J. Gen. Cytol. 1963;5:299–306. [Google Scholar]
- Sorenson MD. computer program. Boston, MA: Boston University; 1999. TreeRot. ver 2c. [Google Scholar]
- Sulston IA, Anderson KV. Embryonic patterning mutants in Tribolium castaneum. Dev. 1996;122:805–814. doi: 10.1242/dev.122.3.805. [DOI] [PubMed] [Google Scholar]
- Swofford DL. computer program. Sunderland, Massachusetts: Sinauer Associates; 2003. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods). ver 4.0beta10. http://paup.csit.fsu.edu/ [Google Scholar]
- Throne JE, Hallman GJ, Johnson JA, Follett PA. Post-harvest entomology research in the United States Department of Agriculture-Agricultural Research Service. Pest Man. Sci. 2003;59:619–628. doi: 10.1002/ps.690. [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y, Wheeler SR, Denell RE. Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature. 2005;433:643–647. doi: 10.1038/nature03272. [DOI] [PubMed] [Google Scholar]
- Triplehorn CA. A new species of Tribolium from Arizona (Coleoptera: Tenebrionidae) Coleop. Bull. 1978;32:73–75. [Google Scholar]
- Ugarković Đ, Podnar M, Plohl M. Satellite DNA of the red flour beetle Tribolium castaneum—comparative study of satellites from the genus Tribolium. Mol. Biol. Evol. 1996b;13:1059–1066. doi: 10.1093/oxfordjournals.molbev.a025668. [DOI] [PubMed] [Google Scholar]
- Uyttenboogaart DL. Revision des Genus Tribolium (Col. Ten.) Entomol. Blätt. 1934;30:20–31. [Google Scholar]
- Venanzetti F, Cesaroni D, Mariottini P, Sbordoni V. Molecular phylogenies in Dolichopoda cave crickets and mtDNA rate calibration. Mol. Phyl. Evol. 1993;2:275–280. doi: 10.1006/mpev.1993.1026. [DOI] [PubMed] [Google Scholar]
- Voss SR, Smith JJ. Evolution of salamander life cycles: a major-effect quantitative trait locus contributes to discrete and continuous variation for metamorphic timing. Genetics. 2005;170:275–281. doi: 10.1534/genetics.104.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ. Group selections among laboratory populations of Tribolium. Proc. Natl. Acad. Sci. USA. 1976;73:4604–4607. doi: 10.1073/pnas.73.12.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ, Johnson NA. Reproductive isolation between two species of flour beetles, Tribolium castaneum and T. freemani: variation within and among geographical populations of T. castaneum. Heredity. 1994;72:155–162. doi: 10.1038/hdy.1994.22. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila Yellow and Ebony proteins in the development and evolution of pigment patterns. Dev. 2002;129:1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- Wool D. Critical examination of postulated cladistic relationships among species of flour beetles (Genus Tribolium, Tenebrionidae, Coleoptera) Biochem. Gen. 1982;20:333–349. doi: 10.1007/BF00484428. [DOI] [PubMed] [Google Scholar]