Abstract
The Escherichia coli umuDC operon is induced in response to replication-blocking DNA lesions as part of the SOS response. UmuD protein then undergoes an RecA-facilitated self-cleavage reaction that removes its N-terminal 24 residues to yield UmuD′. UmuD′, UmuC, RecA, and some form of the E. coli replicative DNA polymerase, DNA polymerase III holoenzyme, function in translesion synthesis, the potentially mutagenic process of replication over otherwise blocking lesions. Furthermore, it has been proposed that, before cleavage, UmuD together with UmuC acts as a DNA damage checkpoint system that regulates the rate of DNA synthesis in response to DNA damage, thereby allowing time for accurate repair to take place. Here we provide direct evidence that both uncleaved UmuD and UmuD′ interact physically with the catalytic, proofreading, and processivity subunits of the E. coli replicative polymerase. Consistent with our model proposing that uncleaved UmuD and UmuD′ promote different events, UmuD and UmuD′ interact differently with DNA polymerase III: whereas uncleaved UmuD interacts more strongly with β than it does with α, UmuD′ interacts more strongly with α than it does with β. We propose that the protein–protein interactions we have characterized are part of a higher-order regulatory system of replication fork management that controls when the umuDC gene products can gain access to the replication fork.
The Escherichia coli SOS response is the paradigm for how a cell responds to DNA damage (1). Although most of the repair and damage tolerance pathways induced as part of the SOS response are error-free, one component of the response is umuDC-dependent translesion synthesis (TLS), which is responsible for most of the mutagenesis induced by UV radiation and many chemicals (2, 3). TLS requires the UmuD′ protein (a posttranslationally modified form of the umuD gene product), UmuC, and RecA. Collectively, these proteins are thought to function in concert with the replicative polymerase, DNA polymerase III (pol III) holoenzyme, to enable replication over lesions in damaged DNA that otherwise would be strongly blocking (1, 2).
After the umuDC operon is induced in response to DNA damage, the UmuD protein undergoes an RecA-facilitated self-cleavage reaction that removes its N-terminal 24 residues to yield UmuD′ (4, 5), an event that activates it for TLS (6). Uncleaved UmuD is not only inactive in TLS, but is an inhibitor of this process as well (7). Recently, we have proposed that uncleaved UmuD acting together with UmuC plays a positive role in helping cells survive DNA damage by acting as a prokaryotic DNA damage cell-cycle checkpoint system that regulates DNA synthesis after DNA damage, thereby allowing time for accurate repair to take place (8). Thus, RecA-facilitated cleavage of UmuD to UmuD′ appears to function as a molecular switch that regulates the release of the checkpoint control while simultaneously helping to restart stalled replisomes by TLS.
Echols and colleagues (9) were the first to show that the addition of UmuD′, UmuC, and RecA, the three proteins genetically shown to be required for SOS mutagenesis (1), to pol III holoenzyme resulted in TLS. This finding since has been reproduced by others (10, 11). UmuC is a member of a large family of related proteins referred to as the UmuC superfamily (1–3, 12), which can be divided into four subfamilies defined by E. coli umuC, E. coli dinB, Saccharomyces cerevisiae REV1, and S. cerevisiae and human RAD30 (13, 14). Importantly, key representatives of the UmuC superfamily, including E. coli DinB (pol IV) (15), S. cerevisiae Rev1 (16), S. cerevisiae Rad30 (pol η) (17), and human Rad30 homolog, XP-V (18), all have been shown to exhibit a DNA polymerase activity. Consistent with this, UmuD′2C recently has been shown to contain an intrinsic, error-prone DNA polymerase activity (pol V) that, in combination with pol III, permits efficient replication past a synthetic abasic site in vitro (19). How this catalytic activity of UmuD′2C complex is coordinated with the action of E. coli’s replicative polymerase, an 18-polypeptide protein machine (20), is not yet understood.
In this paper we describe our efforts to determine whether the UmuD and/or UmuD′ proteins interact with E. coli pol III. Our results represent the first biochemical evidence that these two forms of the umuD gene product can interact with specific components of E. coli’s replicative DNA polymerase and suggest that the interactions are part of a higher-order regulatory system of replication fork management that serves to regulate access of the umuDC gene products to the replication fork.
Materials and Methods
Bacterial Strains and Plasmid DNAs.
Constructs for overproduction of pol III subunits were gifts from C. S. McHenry, University of Colorado, Health Sciences Center, and are described in the legend to Fig. 5. BL21(DE3) (Novagen) was used for overproduction of UmuD, UmuD′, and kinase-tagged derivatives. The latter two derivatives were constructed by PCR amplification of the wild-type umuD or recombinantly engineered UmuD′ gene as described (21) by using a synthetic DNA primer coding for four additional amino acids (RASV) subsequent to the native C terminus of UmuD. After amplification, the product was ligated to NdeI- and EcoRI-digested pET5a (22). pSU18DC is a pACYC184 derivative (23) containing the umuDC operon in a PstI fragment.
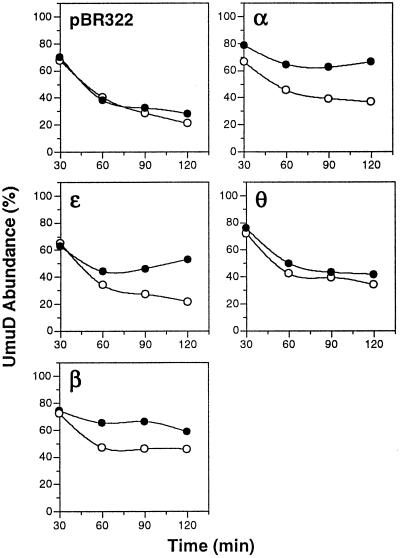
Figure 5.
Overexpression of α, ɛ, or β inhibits RecA*-facilitated self-cleavage of UmuD to UmuD′ in vivo. The kinetics of self-cleavage of UmuD to UmuD′ in E. coli AB1157 bearing either pBR322 or a pBR322 derivative directing overproduction of α (pDNAE), ɛ (pHN1), θ (pHN101), or β (pJRC210), in addition to a compatible plasmid containing the umuDC operon (pSU18DC), were measured very carefully. Cultures were grown in supplemented M9 medium at 37°C as described (37). When cultures reached early-log phase, they were split in half and isopropyl β-d-thiogalactoside (20 μM) was added to one half of each culture (filled circles) to induce expression of the indicated pol III subunit 30 min before induction of expression and subsequent self-cleavage of UmuD by UV irradiation (20 J/m2). At the indicated times, aliquots were removed for immunoblotting for detection of UmuD and UmuD′ by chemiluminescence, as described (37). The absolute abundance of uncleaved UmuD and UmuD′ were determined by scanning densitometry of appropriately exposed films relative to UmuD and UmuD′ standard curves by using the molecular analyst software package (Bio-Rad). The abundance of UmuD is presented as the percent abundance of uncleaved UmuD divided by the sum of the abundance of UmuD and UmuD′. Before 30 min post-UV irradiation, levels of uncleaved UmuD and UmuD′ were insufficient for accurate quantitation by this method. Open circles represent the abundance of UmuD in the absence of added isopropyl Β-d-thiogalactoside, and filled circles represent the abundance of UmuD in the presence of added isopropyl β-d-thiogalactoside.
Proteins and Reagents.
αΔN1 (24), UmuD, UmuD′, and kinase-tagged derivatives (22) were purified as described in the indicated references. αΔN1 contains a His6 and a biotin tag at its N terminus, neither of which affect its activity (24). Purified α, α– ɛ complex, pol III core, and β, as well as the corresponding polyclonal antisera, were gifts from C. S. McHenry. Purified DnaB helicase was a gift from J. M. Kaguni, Michigan State University. Affinity-purified polyclonal antisera specific to UmuD and UmuD′ has been described (7). Other reagents were bovine heart muscle cAMP-dependent protein kinase (catalytic subunit), BSA (Fraction V; Sigma); [γ-32P]ATP (DuPont/NEN); Affi-Gel A-15 (Bio-Rad); and Western Light protein detection kit (Tropix, Bedford, MA).
Affinity Chromatography.
Highly purified UmuD, UmuD′, and BSA were coupled to Affi-Gel A-15 according to the manufacturer’s recommendations (≈5–7 mg protein/ml for Fig. 3 and ≈1 mg/ml for Fig. 4). Chromatography was done at 4°C by using buffer A (10 mM sodium phosphate, pH 6.8/75 mM NaCl/15% glycerol/0.5 mM EDTA/1 mM DTT). After applying samples, columns were washed with five column volumes of buffer A. Bound proteins were then eluted stepwise with five column volumes of buffer A containing 250 mM followed by 1 M NaCl.
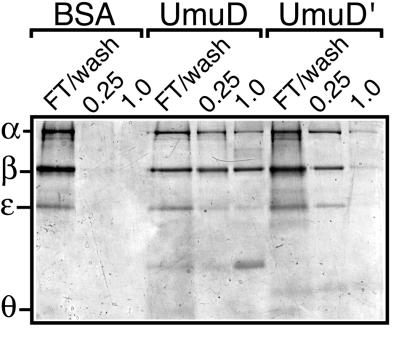
Figure 3.
UmuD and UmuD′ affinity chromatography with purified pol III core and β. Purified pol III core (11 μg) supplemented with an equivalent molar amount of β (2 μg) was applied to each of the three affinity columns as described in Materials and Methods. The flow-through and wash fractions were combined for each column. Bound proteins then were eluted stepwise with buffer A containing 0.25 M followed by 1 M NaCl. After TCA precipitation, aliquots of each sample were fractionated by SDS/PAGE and then stained with Coomassie brilliant blue R-250. The faint bands visible in the UmuD and UmuD′ column fractions migrating between ɛ and θ are UmuD and UmuD′, respectively, which presumably leached from the columns.
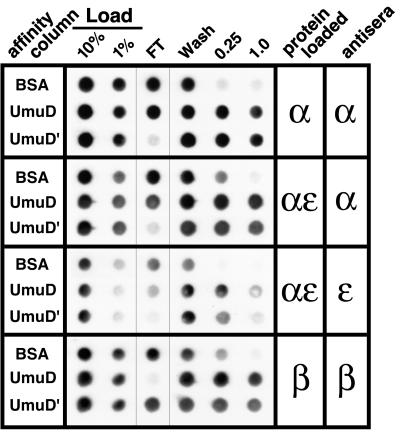
Figure 4.
UmuD and UmuD′ affinity chromatography of individual pol III subunits. αNΔ1 (10 μg), α–ɛ complex (20 μg), or β (10 μg) was applied to each column as described in Materials and Methods. For this experiment, the flow-through and wash fractions for each column were kept separate. Bound proteins then were eluted stepwise with buffer A containing 0.25 M followed by 1 M NaCl. Aliquots (20%) of each fraction, as well as 1% and 10% of each load, were applied to poly(vinylidene difluoride) membrane by using a dot-blot manifold (Bio-Rad). Fractions containing the respective polymerase subunits were identified by probing the membranes with polyclonal antiserum specific to either α, ɛ, or β followed by chemiluminescence detection, as described (37). The identity of each affinity column is indicated to the left, and the protein loaded onto each column and antisera used for immunodetection are indicated to the right.
Results
Radiolabeled UmuD and UmuD′ Interact Physically with Components of pol III.
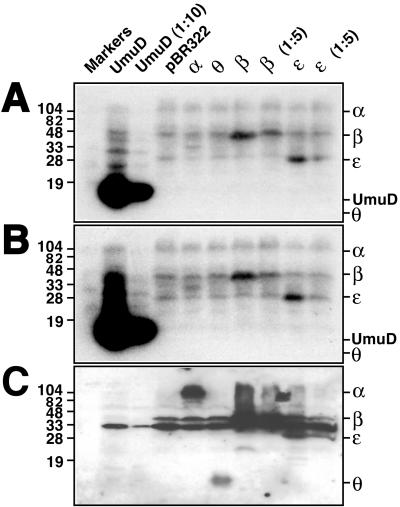
Our initial attempts to demonstrate a direct, physical interaction between the umuDC gene products and pol III by coimmunoprecipitation or by gel filtration were unsuccessful. Therefore, we searched for such interactions by the very sensitive technique of far Western blotting (25, 26). We prepared derivatives of uncleaved UmuD and UmuD′ that carry a C-terminal 4-aa sequence that permits the proteins to be radiolabeled with [32P]phosphate (26). In a preliminary experiment, we used these 32P-labeled UmuD and UmuD′ derivatives to probe a whole-cell extract of a non-SOS-induced E. coli strain that had been separated by SDS/PAGE and transferred to a membrane. The results we observed were strikingly specific. For both UmuD and UmuD′, the only clear bands of hybridization we observed occurred at positions of polypeptides of approximately 28 and 42 kDa (data not shown). Intriguingly, these correspond to the molecular masses of the ɛ and β subunits of pol III, respectively (20).
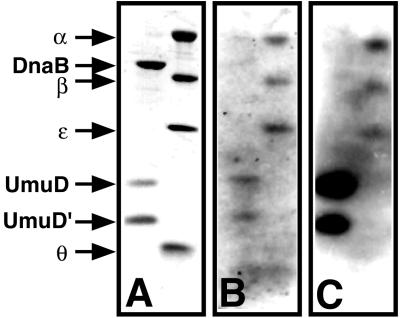
We then used these 32P-labeled UmuD and UmuD′ derivatives to probe whole-cell extracts of a non-SOS-induced lexA+ E. coli strain (AB1157) that harbored either pBR322 or a pBR322 derivative that directed overproduction of the α, ɛ, θ, or β subunit of pol III (Fig. 1, lanes 4–10). Because UmuD and UmuD′ exist as homo- and heterodimers in solution (4, 7), we used a whole-cell extract of a different E. coli strain [BL21(DE3)] that overproduces UmuD protein as a positive control (Fig. 1). As expected, radiolabeled UmuD and UmuD′ interacted with UmuD protein adsorbed to the membrane. Again, polypeptides of approximately 28 and 42 kDa interacted specifically with UmuD and UmuD′. The possibility that the 28- and 42-kDa polypeptides corresponded to the ɛ and β subunits of pol III was supported by the observation that the extracts enriched in ɛ or β, respectively, contained equivalent interacting species but of an apparently greater abundance (Fig. 1, lanes 7–10). In contrast, UmuD and UmuD′ did not interact with θ by this method (Fig. 1). A >100-kDa diffuse species also in the non-SOS-induced AB1157 extract did not appear more intense when the extract was enriched with α, but an ≈35-kDa species specific to the α-enriched extract did interact with both UmuD and UmuD′ (Fig. 1, lane 5). Although this species did not cross-react with our anti-pol III antibodies (Fig. 1C), it remains possible that it is a proteolytically truncated form of α that arose during the preparation of the extract and lacks the epitopes recognized by the antiserum preparation.
Figure 1.
Far Western blot of crude E. coli extracts. Crude cell extracts (≈0.1 OD600 units) were fractionated by SDS/PAGE and then transferred to poly(vinylidene difluoride) membrane in triplicate. Membranes then were probed with either 32P-labeled UmuD (A) or UmuD′ (B). Radiolabeling of the UmuD and UmuD′ kinase-tagged derivatives and subsequent far Western blotting were carried out as described (26). To identify the positions of α, β, ɛ, and θ, one membrane (C) was processed as an immunoblot by using polyclonal anti-pol III antiserum. The ≈30-kDa species visible in lanes 2–10 corresponds to a protein that cross-reacts with the antibody preparation and is not related to pol III. The UmuD, β, and ɛ crude cell extracts were diluted as indicated (1:5 or 1:10) before electrophoresis.
Further evidence that the 28- and 42-kDa polypeptides correspond to the ɛ and β subunits came from immunoblot analysis that indicated that all of the AB1157 extracts, but not the BL21(DE3) extract (compare lanes 2–3 and 4–10 of Fig. 1C), contained β at levels easily detectable with our anti-pol III antibodies. Longer exposures of the same immunoblot (data not shown) indicated that a similar result was true for ɛ as well. Our failure to detect ɛ and β in the BL21(DE3) extract either in immunoblot or far Western experiments is presumably because a lesser amount of that extract was loaded on the gel. UmuD and UmuD′ also interacted very strongly with partially purified UmuC protein, which is known to form complexes with both UmuD and UmuD′ (10, 27), whereas we observed no additional interactions when we probed an extract of an AB1157 derivative that overproduced DnaB, the replicative helicase (data not shown).
To confirm that the interacting 28- and 42-kDa polypeptides present in the whole-cell extracts were, in fact, subunits of pol III, we repeated the experiment but this time by using highly purified β and pol III core, which consists of a stable complex containing the α, ɛ, and θ subunits in a 1:1:1 stoichiometry. UmuD and UmuD′ were used as positive controls, and DnaB helicase served as a negative control to confirm the specificity of the interactions. Consistent with our inferences from the experiments using whole-cell extracts (Fig. 1), both UmuD and UmuD′ interacted with UmuD, UmuD′, ɛ, and β, but not θ or DnaB helicase (Fig. 2). However, in contrast to our observations using whole-cell extracts, both UmuD and UmuD′ also were able to interact with α. Crude extract contains insufficient levels of α to detect an interaction with UmuD and UmuD′, probably because of proteolytic instability of α in the extract, but we were able to observe an interaction between UmuD or UmuD′ and α by using a more concentrated cell extract enriched with α (data not shown). That UmuD, UmuD′, α, ɛ, β, and DnaB helicase are all acidic proteins (calculated pI ca. 4.3–5.6), whereas θ is basic (calculated pI ca. 9.2), suggests that the interactions we observed in these experiments did not result from nonspecific ionic interactions but, rather, from specific interactions between the pairs of proteins.
Figure 2.
Far Western blot of purified pol III subunits. UmuD, UmuD′, β, DnaB helicase (0.5 μg each), and pol III core (3 μg) were fractionated by SDS/PAGE and then transferred to poly(vinylidene difluoride) membrane in triplicate. Membranes then were either stained with Coomassie brilliant blue R-250 (A) or probed with 32P-labeled UmuD (B) or UmuD′ (C) as described in the legend to Fig. 1. The position of each protein is indicated to the left.
Uncleaved UmuD and UmuD′ Interact with Subunits of pol III in Reciprocal Fashions.
Because of the high local concentration of the immobilized ligand, affinity chromatography can be a very effective method for characterizing weak protein–protein interactions (28). We therefore utilized affinity chromatography to test whether the interactions we had observed using crude, denatured extracts could be detected by using purified proteins in their native conformation. Highly purified BSA, UmuD, and UmuD′ were covalently coupled to Affi-Gel A-15. Because all three proteins are similarly acidic (calculated pI ca. 4.3–5.3), the BSA column serves as a control to distinguish between ionic and specific interactions. We then investigated whether UmuD or UmuD′ was able to interact with purified pol III core supplemented with an equivalent molar amount of β. α and β interact only weakly in solution (29, 30), making it unlikely that significant amounts of either pol III core or β would be retained by UmuD or UmuD′ by any type of interaction other than a direct one. Aliquots of each flow-through and wash, 0.25 M elution, and 1 M NaCl elution were TCA-precipitated and then fractionated by SDS/PAGE. Coomassie staining indicated that although neither pol III core nor β was retained by the BSA column, both were efficiently retained by the UmuD and UmuD′ columns (Fig. 3), thereby clearly demonstrating that both UmuD and UmuD′ interact physically and directly with pol III.
Careful comparison of the elution profiles of pol III core relative to β for the UmuD and UmuD′ columns suggested that UmuD and UmuD′ have different affinities for pol III core and β; UmuD appeared to bind β more strongly than pol III core whereas UmuD′ appeared to bind pol III core more strongly than β (Fig. 3). To investigate this possibility, we used UmuD and UmuD′ columns similar to those described above to carry out affinity chromatography of highly purified α, α–ɛ complex, and β. We chose α–ɛ complex instead of ɛ because ɛ must be resolubilized from an insoluble form after overproduction (31), whereas the α–ɛ complex is readily purified in a soluble form (32). Finally, to permit a better evaluation of the relative affinities of UmuD and UmuD′ for the individual pol III subunits, we collected the flow-through and wash fractions separately. In our earlier experiment (Fig. 3), the flow-through and 75 mM NaCl wash fractions were combined.
Both the UmuD and UmuD′ columns were effective in retaining α, α–ɛ complex, and β whereas the BSA column was not (Fig. 4). However, comparison of the flow-through and wash fractions for α, α–ɛ complex, and β confirmed that UmuD and UmuD′ differ with respect to their affinities for particular subunits of pol III. In the case of the UmuD column, β was virtually absent from the flow-through, indicating that it was completely retained on the column. In contrast, α and α–ɛ complex were not completely retained on the column. Quantitative analysis of the data presented in Fig. 4 indicated that whereas <1% of the β loaded onto the UmuD column was found in the flow-through, approximately 5% of the α and α–ɛ complex was present in the flow-through. A reciprocal result was obtained with the UmuD′ column. Whereas <1% of the α and α–ɛ complex loaded was detected in the flow-through, nearly 10% of the β was present in the flow-through. Taken together, these observations suggest that UmuD interacts more strongly with β than does UmuD′ and that UmuD′ interacts more strongly with α and α–ɛ complex than does UmuD. Although interactions of UmuD and UmuD′ with ɛ were detected by far Western analysis (Figs. 1 and 2), quantitative analysis of the data shown in Fig. 4 indicated that both UmuD and UmuD′ interacted similarly with α and α–ɛ complex.
Overexpression of Each of the Three pol III Subunits That Interact with UmuD in Vitro Inhibits RecA-Facilitated Cleavage of UmuD to UmuD′ in Vivo.
To complement the experiments described above, we searched for evidence that the interactions between UmuD and the α, ɛ, and β subunits of pol III occurred in vivo as well as in vitro. We reasoned that, if we overproduced the various pol III subunits, interaction between the relevant pol III subunit and UmuD might retard RecA-facilitated cleavage of UmuD to yield UmuD′. This might occur through their ability to compete or interfere with the interactions between UmuD and RecA nucleoprotein filaments that are necessary for UmuD cleavage to occur (33, 34). Therefore, we carried out an analysis in which we very carefully quantitated whether the overexpression of certain pol III subunits affected the kinetics of UmuD cleavage in vivo. In these experiments, we used lexA+ E. coli derivatives carrying plasmids that overproduced either the α, ɛ, θ, or β subunit of pol III upon the addition of isopropyl β-d-thiogalactoside (IPTG) to the culture medium. To facilitate immunodetection of UmuD and UmuD′, we also introduced into these strains a compatible plasmid containing the umuDC operon under control of its native, LexA-regulated promoter (6, 35, 36). Thirty minutes before UV irradiation, IPTG was added to initiate the overexpression of the respective pol III subunits. As shown in Fig. 5, overproduction of the θ subunit of pol III for 30 min before UV irradiation had no discernible effect on the kinetics of UmuD cleavage; the rate of UmuD cleavage in these cells was indistinguishable from that observed with identically treated cells that did not overproduce any pol III subunit (Fig. 5). In contrast, overproduction of the α, ɛ, or β subunit of pol III for 30 min before UV irradiation caused a distinct slowing of the rate of UmuD cleavage (Fig. 5). That the kinetics of umuD expression were essentially indistinguishable among the various strains (UmuD was detectable within 10–15 min after UV irradiation for all five strains; data not shown) indicated that overproduction of either α, ɛ, or β did not result in a general inhibition of SOS induction but, rather, produced a specific effect on UmuD cleavage. Although indirect, these observations are consistent with the hypothesis that interactions between UmuD and the α, ɛ, and β subunits of pol III occur in vivo as well as in vitro.
Discussion
In this report, we present biochemical evidence that uncleaved UmuD and UmuD′ are capable of a highly specific, direct physical interaction with the catalytic (α) and processivity (β) subunits of the E. coli replicative polymerase, DNA polymerase III holoenzyme. Furthermore, we present evidence based on far western experiments that both uncleaved UmuD and UmuD′ interact physically with the proofreading (ɛ) subunit of pol III. Our results represent the direct demonstration of a physical interaction between the catalytic subunit of any replicative DNA polymerase and proteins not required for normal DNA synthesis.
Analysis of the UmuD/UmuD′–pol III interactions by affinity chromatography indicated that UmuD and UmuD′ have reciprocal affinities for α and β: UmuD favored β relative to α or α–ɛ complex whereas UmuD′ favored α and α–ɛ complex relative to β. Our conclusion that UmuD has a higher affinity for β than does UmuD′ has also been supported additionally by recent cross-linking experiments (M.D.S. and G.C.W., unpublished results). Taken together, our observations suggest that direct physical interactions between UmuD and UmuD′ and specific pol III subunits play a role in determining the different biological functions exerted by the umuDC gene products.
Based on our observation that UmuD has higher affinity for β than does UmuD′, we suggest that the UmuD–β interaction is important for the checkpoint-like role we have proposed for UmuD and UmuC (8). In eukaryotes, regulation of the G1-to-S transition is governed in part by p21 (38). In addition to its ability to block progression through the cell cycle by inhibiting cyclin–CDK complexes, p21 is also able to inhibit DNA replication directly by tightly binding to proliferating cell nuclear antigen (39–41), the eukaryotic counterpart to the E. coli pol III β processivity clamp, thereby precluding binding of polymerase δ (40, 41). That UmuD interacts with other subunits besides β suggests that it could inhibit DNA replication by a different mechanism from p21. Based on the crystal structure, the central hole of the β dimer is sufficiently large to accept double–strand DNA (42). Modeling studies suggest that interactions between the β dimer and double-strand DNA could remain largely nonspecific if mediated by water molecules (42). However, the nonspecific nature of these interactions was predicted to be highly dependent on the orientation of β to the DNA; only a perpendicular arrangement would limit possible specific interactions between β and the major and/or minor grooves (42). Consequently, if uncleaved UmuD were to contact both α and β simultaneously, it is conceivable that its presence could alter the orientation of β relative to α, perturbing the perpendicular arrangement of β and the DNA. Such a perturbation might lead to specific interactions between β and the DNA, thus stopping DNA synthesis by pol III. Alternatively, a UmuD–β–α ternary complex might affect translocation of the lagging-strand polymerase, which must occur upon completion of each Okazaki fragment, or about once every second (43), thereby uncoupling leading- and lagging-strand synthesis, leading to replisome collapse.
In the model we proposed (8), cleavage of UmuD to UmuD′ acts as a molecular switch that regulates the release of the checkpoint control while enabling TLS. Based on our results discussed above, we propose that RecA-mediated cleavage of UmuD serves to convert the umuD gene product into a form (UmuD′) that has a diminished affinity for β yet an increased affinity for α. Furthermore, we suggest that both UmuD and UmuD′ interact with more than one component of pol III simultaneously, but in reciprocally distinct fashions. Presently, it is unknown whether UmuC also interacts directly with components of pol III. However, UmuD, UmuD′, and UmuC are known to interact with RecA protein (44, 45). Interestingly, UmuD interacts with RecA differently than does UmuD′: whereas UmuD′ acting together with UmuC inhibits RecA-mediated homologous recombination, UmuD together with UmuC does not (36, 46). Taken together, these findings suggest that UmuD cleavage changes how the two forms of the umuD gene product interact with pol III and RecA protein, thereby helping to reposition the UmuD′2C complex within the replisome relative to where the UmuD2C complex had resided. We suggest that the net result of this repositioning is the release of the checkpoint and the onset of TLS.
Based on genetic evidence, UmuD′, UmuC and RecA protein have long been thought to modulate some form of pol III to result in TLS (1). However, the recent finding that UmuD′2C complex contains an intrinsic error-prone DNA polymerase activity (19) led Tang et al. to suggest that UmuD′2C (pol V) might displace pol III core stalled at a lesion and, in conjunction with the pol III processivity apparatus, copy over the lesion. Once past the lesion, Tang et al. speculate that UmuD′2C might dissociate from the primer–template terminus, allowing pol III to complete replication of the genome. We suggest that instead of displacing pol III core in the vicinity of the lesion, UmuD′2C might reside within the replisome, as discussed above. Then, upon encountering a replication-stalling lesion, the replisome might back up, exposing the 3′ primer terminus. The interacting UmuD′2C complex then might synthesize 1 or 2 nt past the lesion in a template-dependent but error-prone fashion (19) before returning the primer–template terminus to the replicative polymerase for continued processive, high-fidelity synthesis. The apparent copurification of α with UmuD′2C (10) and the stimulation of bypass in vitro by β and γ complex (10, 19), both of which suggest that UmuD′2C interacts with components of pol III during TLS, are consistent with this suggestion. An attractive aspect of this model is that the replicative polymerase would regulate when UmuD′2C could gain access to the primer terminus, thereby providing an additional level of regulation upon error-prone repair. Viewed in such a way, the alternative umuDC-encoded polymerase, pol V, would be serving as an accessory protein of the replicative DNA polymerase.
Additional evidence that a UmuD′–α interaction is important for TLS was provided by the finding that a temperature-sensitive mutant of α (DnaE1026) was stabilized by UmuD′2C in vitro (19). Furthermore, comparison of the replication activity of UmuD′2C purified from a strain lacking the gene encoding pol II and containing a temperature-sensitive pol III dnaE allele (19) with that of UmuD′2C purified from a strain containing the wild-type allele for both pol II and dnaE (10) indicates that the former possessed a far weaker polymerase activity than the latter. This would be consistent with, in the wild-type case (10), the UmuD′2C preparation being contaminated with small, yet significant, amounts of α (47). Further evidence that physical interactions involving UmuD′2C and pol III are instrumental in enabling TLS is the finding that low levels of UmuD′2C in combination with low levels of α were significantly more active for lesion bypass than were high levels of UmuD′2C in the absence of α (19).
That UmuD′2C is endowed with a polymerase activity helps to demystify the molecular mechanism of TLS in E. coli. However, this finding does not on its own rule out a possible noncatalytic role of the umuDC gene products in TLS. It should be stressed that noncatalytic and catalytic roles for the umuDC gene products in TLS are not necessarily mutually exclusive (47). It is known that a mismatch in the first 3 to 4 bp from the 3′ primer terminus can cause a polymerase to stall (48), suggesting that translocation is tightly coupled to accurate base pairing. For example, Bacillus stearothermophilus DNA polymerase makes extensive interactions over the first 4 bp from the 3′ primer terminus that are independent of the DNA sequence but dependent on the structure of the minor groove (49). It is conceivable that the presence of a lesion within the first 4 bp from the 3′ primer terminus could lead to replisome stalling and subsequent processing of the primer end by the exonuclease domain (48). Consequently, if pol III reacquires the 3′ primer terminus from UmuD′2C one or two bases past the lesion, direct interaction of UmuD′2C with the catalytic and proofreading subunits of pol III might help to make the replisome less sensitive to possible stalling because of an atypical, lesion-containing base pair in the vicinity of the primer–template terminus.
Our results suggest that a higher-order regulatory system that one might term “replication fork management” is an emergent property that is evident only when the action of the known DNA replication machinery is considered in the context of the entire cell. Such a replication fork management system would be based on a series of protein–protein and protein–DNA interactions and would determine which events occur, and in what order, when a DNA lesion is encountered. If the bacterial DNA replication machinery is indeed a stationary factory that pulls the DNA through (50), then the consequence of a complete dissociation of the replicative polymerase from the fork at a lesion would be even greater than previously thought and, hence, the greater the need would be for such a management system. Members of the UmuC subfamily are presently the only proteins of the UmuC superfamily known to have specific partner proteins (the umuD gene product and homologs) that control the nature of their interactions with the replication fork (1). Is the action of other members of the superfamily similarly coordinated with other cellular components involved in DNA replication, either directly or via interactions with as yet unidentified partner proteins? For example, it is conceivable that the S. cerevisiae Rev1 protein (16) might interact with DNA polymerase ζ, an error-prone polymerase encoded by the REV3 and REV7 genes (51), and/or the replicative DNA polymerase. Similarly, it is possible that human pol η, the XP-V (Xeroderma pigmentosum variant) gene product (18, 52, 53), which can replicate over a thymine–thymine cis-syn cyclobutane dimer (18), has its action coordinated with, or directed by, the human replicative DNA polymerase machinery.
Acknowledgments
This work was supported by Public Health Service Grant CA21615 to G.C.W. from the National Cancer Institute. M.D.S. was supported by a fellowship (5 F32 CA79161-01) from the National Cancer Institute. We thank Charles McHenry, Brad Glover, and Cherie Mueller for generously supplying strains, proteins, and antisera and for their many helpful discussions. We thank Jon Kaguni and Kevin Carr for DnaB helicase. We appreciate the help of Ruchi Mathur and Andrew Wright with the far western experiments. We also thank members of our lab, in particular, Brad Smith and Brett Pellock, for their comments on the manuscript.
Abbreviations
- pol III and pol II
DNA polymerase III and II, respectively
- TLS
translesion synthesis
References
- 1.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Smith B T, Walker G C. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodgate R, Levine A S. Cancer Surv. 1996;28:117–140. [PubMed] [Google Scholar]
- 4.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinagawa H, Iwasaki H, Kato T, Nakata A. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nohmi T, Battista J R, Dodson L A, Walker G C. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battista J R, Ohta T, Nohmi T, Sun W, Walker G C. Proc Natl Acad Sci USA. 1990;87:7190–7194. doi: 10.1073/pnas.87.18.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Opperman T, Murli S, Smith B T, Walker G C. Proc Natl Acad Sci USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman M F, Echols H. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O’Donnell M, Goodman M F. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reuven N B, Tomer G, Livneh Z. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 12.Kulaeva O I, Koonin E V, McDonald J P, Randall S K, Rabinovich N, Connaughton J F, Levine A S, Woodgate R. Mutat Res. 1996;357:245–253. doi: 10.1016/0027-5107(96)00164-9. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg E C, Gerlach V L. Cell. 1999;98:413–416. doi: 10.1016/s0092-8674(00)81970-4. [DOI] [PubMed] [Google Scholar]
- 14.Woodgate R. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 15.Wagner J, Gruz P, Kim S-R, Yamada M, Matsui K, Fuchs R P P, Nohmi T. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 16.Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 17.Johnson R E, Prakash S, Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 18.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang M, Xuan S, Frank E G, O’Donnell M, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelman Z, O’Donnell M. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 21.Lee M H, Ohta T, Walker G C. J Bacteriol. 1994;176:4825–4837. doi: 10.1128/jb.176.16.4825-4837.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferentz A E, Opperman T, Walker G C, Wagner G. Nat Struct Biol. 1997;4:979–983. doi: 10.1038/nsb1297-979. [DOI] [PubMed] [Google Scholar]
- 23.Bartolome B, Jubete Y, Martinez E, de la Cruz F. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 24.Kim D R, McHenry C S. J Biol Chem. 1996;271:20690–20698. doi: 10.1074/jbc.271.34.20690. [DOI] [PubMed] [Google Scholar]
- 25.Blanar M A, Rutter W J. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- 26.Hale C A, de Boer P A. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 27.Woodgate R, Rajagopalan M, Lu C, Echols H. Proc Natl Acad Sci USA. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Formosa T, Barry J, Alberts B M, Greenblatt J. Methods Enzymol. 1991;208:24–45. doi: 10.1016/0076-6879(91)08005-3. [DOI] [PubMed] [Google Scholar]
- 29.Kim D R, McHenry C S. J Biol Chem. 1996;271:20699–20704. doi: 10.1074/jbc.271.34.20699. [DOI] [PubMed] [Google Scholar]
- 30.Stukenberg P T, Studwell-Vaughan P S, O’Donnell M. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 31.Scheuermann R H, Echols H. Proc Natl Acad Sci USA. 1984;81:7747–7751. doi: 10.1073/pnas.81.24.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D R, McHenry C S. J Biol Chem. 1996;271:20681–20689. doi: 10.1074/jbc.271.34.20681. [DOI] [PubMed] [Google Scholar]
- 33.Craig N L, Roberts J W. J Biol Chem. 1981;256:8039–8044. [PubMed] [Google Scholar]
- 34.Sassanfar M, Roberts J W. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 35.McDonald J P, Frank E G, Levine A S, Woodgate R. Proc Natl Acad Sci USA. 1998;95:1478–1483. doi: 10.1073/pnas.95.4.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sommer S, Bailone A, Devoret R. Mol Microbiol. 1993;10:963–971. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- 37.Ohta T, Sutton M D, Guzzo A, Cole S, Ferentz A E, Walker G C. J Bacteriol. 1999;181:177–185. doi: 10.1128/jb.181.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 39.Gulbis J M, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 40.Flores-Rozas H, Kelman Z, Dean F B, Pan Z Q, Harper J W, Elledge S J, O’Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waga S, Hannon G J, Beach D, Stillman B. Nature (London) 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 42.Kong X P, Onrust R, O’Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 43.Stukenberg P T, Turner J, O’Donnell M. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 44.Frank E G, Hauser J, Levine A S, Woodgate R. Proc Natl Acad Sci USA. 1993;90:8169–8173. doi: 10.1073/pnas.90.17.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freitag N, McEntee K. Proc Natl Acad Sci USA. 1989;86:8363–8367. doi: 10.1073/pnas.86.21.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehrauer W M, Bruck I, Woodgate R, Goodman M F, Kowalczykowski S C. J Biol Chem. 1998;273:32384–32387. doi: 10.1074/jbc.273.49.32384. [DOI] [PubMed] [Google Scholar]
- 47.Walker G C. Proc Natl Acad Sci USA. 1998;95:10348–10350. doi: 10.1073/pnas.95.18.10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson K A. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 49.Kiefer J R, Mao C, Braman J C, Beese L S. Nature (London) 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 50.Lemon K P, Grossman A D. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 51.Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 52.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 53.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]