Summary
The yeast Vps27/Hse1 complex and homologous mammalian Hrs/STAM complex deliver ubiquitinated transmembrane proteins to the ESCRT endosomal sorting pathway. The Vps27/Hse1 complex directly binds to ubiquitinated transmembrane proteins and recruits both ubiquitin ligases and deubiquitinating enzymes. We have solved the crystal structure of the core responsible for the assembly of the Vps27/Hse1 complex at 3.0Å resolution. The structure consists of two intertwined GAT domains, each consisting of two helices from one subunit and one from the other. The two GAT domains are connected by an antiparallel coiled-coil to form a 90 Å-long barbell-like structure. This structure places the domains of Vps27 and Hse1 that recruit ubiquitinated cargo and deubiquitinating enzymes close to each other. Coarse-grained Monte Carlo simulations of the Vps27/Hse1 complex on a membrane show how the complex binds cooperatively to lipids and ubiquitinated membrane proteins and acts as a scaffold for ubiquitination reactions.
Introduction
Protein ubiquitination is a widespread, multifunctional regulatory mechanism. Ubiquitin is conjugated to proteins via an isopeptide bond between the C-terminus of ubiquitin and Lys residues in the ubiquitinated protein. This reaction is carried out by an ubiquitin activating enzyme (E1), an ubiquitin conjugating enzyme (E2), and an ubiquitin protein ligase (E3) (Hershko et al., 2000; Hochstrasser, 2000; Pickart, 2001; Weissman, 2001). Ubiquitination is a major regulator of endocytosis and vesicular trafficking (Hicke, 2001; Raiborg et al., 2003). Ubiquitinated proteins are targeted to and regulate the vesicular trafficking machinery via interactions between the ubiquitin moiety and proteins that contain ubiquitin binding domains (Harper and Schulman, 2006; Hicke et al., 2005; Hurley et al., 2006).
The ESCRT protein network targets ubiquitinated transmembrane proteins for degradation in the lysosome or yeast vacuole (Babst, 2005; Bowers and Stevens, 2005; Hurley and Emr, 2006; Slagsvold et al., 2006). These proteins were discovered in yeast, where defects in their genes lead to an enlarged cargo-rich compartment adjacent to the vacuole (Bowers and Stevens, 2005). This phenotype is referred to as a class E vacuolar protein sorting (VPS) defect. Yeast class E VPS genes encode the subunits of four hetero-oligomeric protein complexes: the Vps27/Hse1 complex (Bilodeau et al., 2003; Bowers and Stevens, 2005; Piper et al., 1995), and ESCRT-I, II, and III (Babst, 2005; Bowers and Stevens, 2005; Hurley and Emr, 2006; Slagsvold et al., 2006). The ESCRT network is conserved from yeast to human and sorts ubiquitinated transmembrane proteins into small vesicles that bud into the lumen of endosomes, thus forming multivesicular bodies (MVBs) (Gruenberg and Stenmark, 2004; Piper and Luzio, 2001). In mammalian cells, the ESCRT network directs the lysosomal degradation of signaling molecules such as the EGF receptor (Clague and Urbe, 2001; Haglund et al., 2003; Katzmann et al., 2002; Slagsvold et al., 2006). Further, this network is hijacked by viruses such as HIV, which use a process topologically equivalent to MVB formation to bud from cells (Demirov and Freed, 2004; Morita and Sundquist, 2004).
Vps27/Hse1 is a multifunctional complex required for MVB sorting of ubiquitinated cargo molecules, as well as the efficient recycling of late Golgi proteins including the carboxypeptidase Y (CPY) sorting receptor, Vps10 (Bilodeau et al., 2002; Bilodeau et al., 2003; Piper et al., 1995). Human Vps27 is known as Hrs (Hepatocyte growth factor receptor substrate), and Hse1 has two human orthologs, STAM1 and STAM2 (Signal transducing adaptor molecule) (Komada and Kitamura, 2005) (Fig. 1A). The Vps27/Hse1 and Hrs/STAM complexes sort cargo proteins from early endosomes to the ESCRT-I complex (Bilodeau et al., 2003; Katzmann et al., 2003) via clathrin coated domains (Lloyd et al., 2002; Raiborg et al., 2002). The Vps27/Hse1 complex is targeted to early endosomes via the FYVE domains of Vps27 or Hrs (Raiborg et al., 2001), which bind to phosphatidylinositol 3-phosphate (PI(3)P). The Vps27/Hse1 complex recruits clathrin via a short peptide motif near the C-termini of Vps27 and Hrs (Raiborg et al., 2002), and both proteins contain P(S/T)XP motifs that recruit ESCRT-I (Bilodeau et al., 2003; Katzmann et al., 2003; Lu et al., 2003).
Figure 1. Modular Organization of Vps27 and Hse1 and Related Proteins, and Alignment of GAT Domains.

(A) Modular organization of Vps27, Hse1, and other GAT-domain containing proteins. Domain name abbreviations are as follows: VHS, Vps27/Hrs/STAM; UIM, ubiquitin-interacting motif; SH3, Src homology-3; GAT, GGA and TOM; GGA, Golgi-localized, gamma-ear containing, ADP-ribosylation-factor-binding protein; TOM, target of Myb; FYVE, Fab1/YOTP/Vac1/EEA1; CB, clathrin-binding; DUIM, double UIM; NGAT, the N-terminal region preceding GAT domain, responsible for binding to Arf1-GTP; GAE, γ-adaptin ear. A helical region of Hrs is a putative, but unproven, GAT domain. (B) GAT domains were aligned based on three-dimensional structural superposition where available (Vps27, Hse1, GGA1, GGA3, and Tom1), and otherwise by sequence homology with the most similar protein of known structure. Colored dots or triangles indicate residues of Vps27 (blue) and Hse1 (orange) that participate in the heterodimer interface. Residues shown in triangles were mutated in this study. The letter “A” above the alignment denotes residues of Vps27 mutated to Ala (Bilodeau et al., 2003). The major site 1 ubiquitin-binding motif of GGA1, GGA3, and Tom1 as discussed in the text is outlined in black.
The Vps27/Hse1 and Hrs/STAM complexes are scaffolds for binding of ubiquitinated cargo proteins and coordinating ubiquitination and deubiquitination reactions that regulate sorting. The yeast complex recruits ubiquitinated cargo via two tandem UIMs (Ubiquitin Interacting Motif) in Vps27 (Bilodeau et al., 2002; Shih et al., 2002; Swanson et al., 2003) and one in Hse1. Hse1 and STAM isoforms recruit the deubiquitinating enzymes (DUBs) UBPY (Kaneko et al., 2003; Kato et al., 2000), AMSH (McCullough et al., 2006), and, in yeast, Ubp7 (Ren et al., 2007) through their SH3 domains. The Hse1 SH3 domain also recruits the adaptor protein, Hua1, which in turn recruits a complex of the ubiquitin ligase Rsp5, the DUB Ubp2, and the regulatory protein Rup1 (Ren et al., 2007). There is a second mechanism in which the C-terminus of Hse1 binds the ubiquitin ligase Rsp5 directly (Bowers et al., 2004; Ren et al., 2007). These mechanisms regulate different cargo to different extents. CPY sorting is slowed, but not blocked, when the complex is disrupted by the loss of Hse1 (Bilodeau et al., 2002). In contrast, cargo such as carboxypeptidase S (Cps1) and the mating factor receptor Ste3 are profoundly dependent on the integrity of the complex, and their sorting is largely blocked by loss of Hse1 (Bilodeau et al., 2002; Ren et al., 2007).
While much is known about individual domains of the Vps27/Hse1 complex, little is known about how the subunits of the complex associate with one another. A predicted coiled-coil region in the Vps27/Hse1 (Bilodeau et al., 2003) and Hrs/STAM (Mizuno et al., 2004) complexes is necessary for complex formation, and an adjacent region of STAM1 dubbed the “STAM specific motif” (SSM) has also been implicated (Mizuno et al., 2004). To characterize the core around which the complex assembles, we carried out sequence similarity searches in the region of the predicted coiled-coil using PSI-Blast (Altschul et al., 1997). To our surprise, these regions of Vps27, Hse1, and STAM all showed statistically significant similarity (E values of ∼ 10-9) to the GAT (GGAs and TOM) domains of the GGA and TOM trafficking adaptor proteins (Fig. 1, A and B). GAT domains are monomeric three-helix bundles (Collins et al., 2003; Shiba et al., 2003; Suer et al., 2003; Zhu et al., 2003) that bind to ubiquitin (Mattera et al., 2004; Puertollano and Bonifacino, 2004; Scott et al., 2004; Shiba et al., 2004). We sought to test the prediction that Vps27 and Hse1 contain GAT domains and to understand how a GAT domain-based core could assemble an oligomeric complex by determining the crystal structure of what we refer to as the Vps27/Hse1 core complex. The structure shows that the core complex consists of two intertwined GAT domains, each consisting of two helices from one subunit, and one from the other. The two GAT domains are connected by a two-stranded coiled-coil. Residues in the interface between the subunits are shown to be essential for the normal sorting functions of these proteins. Finally, the role of the core in organizing the cooperative interactions of other domains of the Vps27/Hse1 complex was explored using coarse-grained Monte Carlo simulations.
Results
The Vps27/Hse1 Core Complex
Based on secondary structure predictions and the putative homology to known GAT domains, a protein construct comprising residues 345-440 of Vps27 fused to an N-terminal hexahistidine tag was co-expressed in Escherichia coli with a second untagged construct comprising residues 275-375 of Hse1. The Vps27 and Hse1 fragments co-eluted from the chelating column and co-migrated on size exclusion chromatography (Fig. 2A). All of the material consisted of the binary complex, with no observable population of free monomeric subunits. Since these portions of Vps27 and Hse1 are competent to form a highly stable binary complex, and there is no indication that other domains of these proteins are involved in complex formation, we refer to this complex as the “Vps27/Hse1 core complex” throughout.
Figure 2. Hydrodynamic Properties of the Vps27/Hse1 Core Complex.

(A) Gel filtration analysis of the recombinant Vps27/Hse1 core showing co-migration at an apparent mass of 29 kDa. This is slightly higher than the calculated mass of 23.2 kDa for a 1:1 complex, and is therefore consistent with an elongated 1:1 complex, but is not consistent with any oligomer with a greater number of subunits. (B) Sedimentation equilibrium profiles at 4.0°C plotted as a distribution of the absorbance at 280 nm vs. r at equilibrium. Data were collected at 13 (orange), 16 (yellow), 19 (green), 22 (cyan), 25 (blue) and 28 (brown) krpm at a loading A280 of 0.75. The solid lines show the best-fit global analysis (carried out for the three loading concentrations) in terms of a single ideal solute, with the corresponding residuals shown in the panels above the plot. (C) Sedimentation equilibrium profile at 4.0°C and 22 krpm plotted in terms of lnA280 vs. r2. The data shown correspond to a loading A280 of 0.25. The solid line indicates the plot expected for a monodisperse 1:1 Vps27:Hse1 complex. (D) Sedimentation equilibrium profile of the Vps27 core region in isolation at 4.0°C and 22 krpm plotted in terms of lnA280 vs. r2. The data shown correspond to a loading A280 of 0.60. The solid line indicates the plot expected for a monomeric Vps27, indicating the presence of higher oligomers.
To determine the oligomeric state of the Vps27/Hse1 core complex in solution, the Vps27/Hse1 complex was analyzed by analytical ultracentrifugation. The global data analysis was consistent with a single ideal solute having a molecular mass of 23.2 ± 0.4 kDa. This compares well to the calculated molecular mass of 23,703 Da for a 1:1 complex of the Vps27 and Hse1 core fragments. The experimental stoichiometry of 1:1 heterodimers is n = 0.98 ± 0.02 (Fig. 2B,C, Fig. S1). The core region of Hse1 expressed alone was too unstable to characterize by analytical ultracentrifugation. The isolated core region of Vps27 was found to be relatively stable, however. The isolated core region of Vps27 was characterized by sedimentation equilibrium experiments (Fig. 2D). The global data analysis in terms of a single ideal solute resulted in poor data fits, but analysis in terms of two non-interacting solutes, returned excellent fits consistent with the presence of monomeric Vps27 and a higher molecular mass species (Fig. 2D, Fig. S2). The best fit showed that the isolated Vps27 core sample contained 94% monomer and 6% aggregate on a molar basis.
Structure of the Vps27/Hse1 Core Complex
The crystal structure of the Vps27/Hse1 core complex was determined at 3.0 Å resolution by a two-wavelength multiwavelength anomalous dispersion experiment at the Se edge (Fig. 3A). The overall structure is barbell-like, with dimensions of roughly 90 × 20 × 20 Å (Fig. 3B, C). The structures of the two subunits are very similar to each other, with an r.m.s.d. of 1.8 Å for the overlay of 67 Cα atoms. The two subunits are intimately intertwined such that 2104 Å2 of solvent accessible surface area is buried per subunit (Fig. 3D, E). The N-termini of the subunits are close together (17 Å apart) at the middle of the barbell. In contrast, the C-termini are at opposite ends of the barbell, 87 Å away from each other.
Figure 3. Crystal Structure of the Vps27/Hse1 Complex.
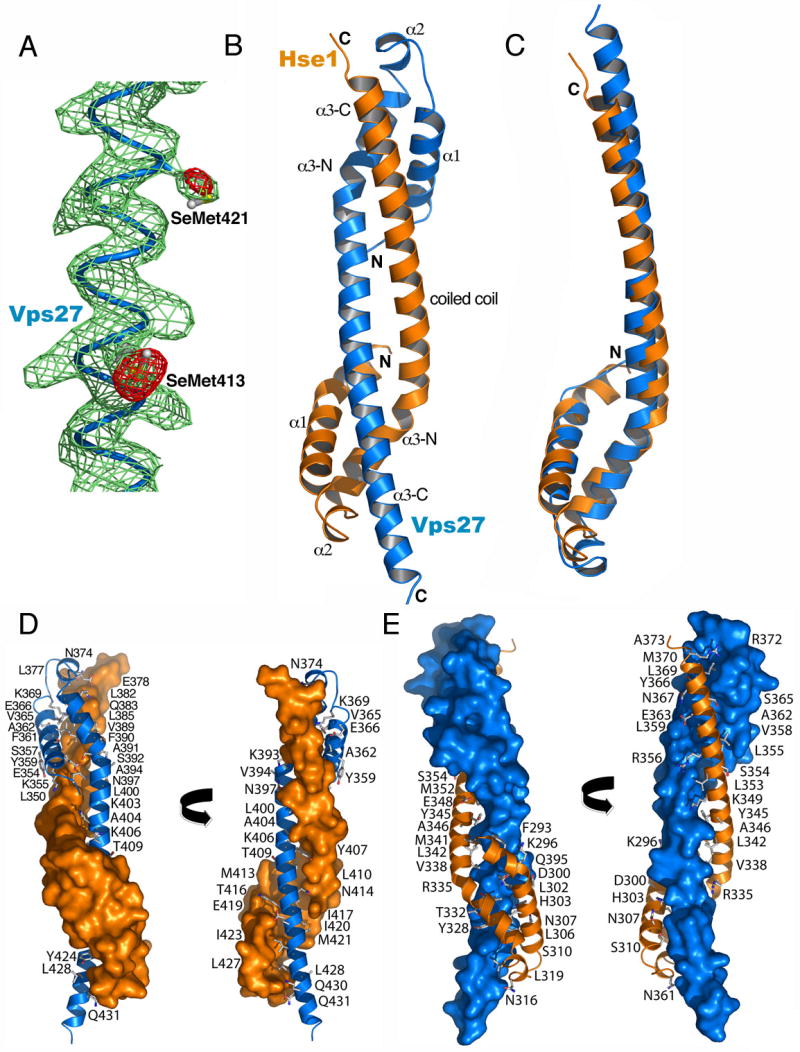
(A) Density-modified MAD Fourier synthesis (green) contoured at 1.0 σ and Se anomalous difference Fourier (red) contoured at 4.0 σ superimposed on the refined structure, with SeMet residues highlighted. (B) Overall structure of the core heterodimer, with Vps27 in blue and Hse1 in orange. (C) Superposition of the core portions of the Vps27 and Hse1 monomers. (D) Surface of Hse1 showing interactions with labeled residues of Vps27. (E) Surface of Vps27 showing interactions with labeled residues of Hse1.
Each subunit consists of three α-helices. At 89 and 72 Å long, respectively, the α3 helices are the longest in the structure. The α3 helices have three distinct structural roles. The N- and C-terminal portions of α3 contribute to the formation of three-helix bundles at each end of the barbell. One bundle (HHV) consists of α1 (residues 288-312) and α3-N (322-341) of Hse1, together with α3-C of Vps27 (410-438). The second bundle (VVH) consists of α1 (residues 351-371) and α3-N (381-399) of Vps27, together with α3-C of Hse1 (354-372). The narrow center of the barbell consists of a two-stranded coiled-coil formed by the central portion of each α3 helix. The coiled-coil region spans residues 396-414 of Vps27 and 338-356 of Hse1. Several residues of the coiled-coil are thus also part of the helical bundles.
Domain-Swapped GAT Domains in Vps27 and Hse1
The two three-helix bundles closely resemble the structures of GAT domains (Fig. 4A, B). A search of the structural database with Dali (Holm and Sander, 1995) using the HHV bundle as the probe structure identified the GAT domain of Tom1 (Akutsu et al., 2005) as the top-scoring match, with a Z-score of 7.7 (Fig. 4C). The Tom1-GAT domain overlays the HHV bundle with an r.m.s.d. of 2.6 Å over 73 Cα positions. The VVH bundle overlays with an r.m.s.d. of 2.2 Å over 47 Cα positions. These r.m.s.d. values compare to values of 1.6 to 1.8 Å over 86-90 Cα positions when comparing the structures of GAT domains of GGA1, GGA3 and Tom1. Given the significant sequence and structural similarity, it seems appropriate to designate the Vps27/Hse1 three-helix bundles as new members of the GAT domain family (Fig. 1, A and B). The α3-N segment corresponds to α2 of the GAT domain, and α3-C of the opposing subunit corresponds to α3 of the GAT domain. The major difference between the GAT domains of Vps27/Hse1 compared to GGAs and Tom1 is that the former are heterodimeric whereas the latter are monomeric. Further, the Vps27/Hse1 GAT domains contain a very short helix, α2 (Fig. 4, A and B), which has no counterpart in the GGA GAT structures.
Figure 4. GAT Domains in Vps27 and Hse1.

(A) The HHV helical bundle. (B) The VVH helical bundle. (C) The GAT domain of Tom1 shown in the same orientation as in parts A and B. (D) Superposition of the HHV and VVH bundles and the Tom1 GAT domain. (E) Model for the closed monomeric conformation of the Vps27 GAT domain, generated by superimposing the Vps27 structure on the Tom1 GAT domain monomer.
Mutational Analysis of the Heterodimer Interface
To confirm the physiological importance of the subunit contacts observed in the Vps27/Hse1 core complex crystal structure, point mutations were introduced into the heterodimer interface. Vps27 residues Leu-410, Ile-417, and Ile-420 are deeply buried in the dimer interface (Fig. 3E) and were selected for mutagenesis to the charged residue Asp. The mutations were introduced into a full-length HA-tagged Vps27 construct to generate the mutants, Vps27-I417D, Vps27-I420D, and Vps27-L410D. For comparison, a UIM mutation (Vps27-A266Q) that was predicted not to abolish core complex assembly was also generated. These point mutants, as well as wild-type (WT) HA-tagged Vps27 (Vps27-WT), were co-expressed with wild-type Hse1-myc in vps27Δ hse1Δ yeast cells. To determine the expression levels of these proteins, whole cell lysates from these strains were subjected to SDS-PAGE and immunoblot analysis with antibodies to the HA and myc epitopes. No significant difference in expression level was detected among Vps27-WT, core complex mutants or the Vps27-A266Q mutant (Fig. 5A, lanes 1-5). Hse1-myc levels in each strain were also equivalent. The lysates were then subjected to immunoprecipitation with anti-HA antibody and analyzed by immunoblotting with anti-HA and anti-myc antibodies. As expected, Hse1-myc protein co-immunoprecipitated with both Vps27-WT and the UIM mutant, Vps27-A266Q (Fig. 5A; lanes 6 and 7); however, this interaction was abolished by all three core complex mutations (Fig, 5A; lanes 8-10).
Figure 5. The Vps27/Hse1 Interface is Required for Sorting.

(A) Expression levels and co-immunoprecipitation of HA-tagged Vps27 and myc-tagged Hse1 proteins. Left panels. Whole cell lysates from hse1Δ vps27Δ cells co-transformed with Hse1-myc and Vps27-HA constructs were lysed and subjected to SDS-PAGE and immunoblot analysis with anti-HA and anti-myc antibodies. Right panels. Rabbit Anti-HA immunoprecipitation from the above lysates, followed by SDS-PAGE and immunoblotting with mouse anti-HA and mouse anti-myc antibodies. (B-I) Fluorescence microscopy images of GFP chimeras of Ste3 and Cps1 in vps27Δ cells expressing Vps27-WT or the indicated core complex mutants. (J) CPY maturation in hse1Δ vps27Δ cells expressing various constructs. Cells were metabolically labeled with 35S-metionine for 10 min (pulse), chased for 15 min in complete medium, and endogenous CPY was immunoprecipitated with anti-CPY antibody. The 15 min point samples were analyzed by SDS-PAGE followed by fluorography. (K) CPY colony blot assay on hse1Δ vps27Δ cells co-expressing Hse1-WT and various Vps27 constructs. Colonies from each strain were spotted onto selective medium and overlayed with nitrocellulose. Secreted CPY was detected by immunoblotting the nitrocellulose with an anti-CPY antibody.
We next sought to determine the functional consequence of core complex mutations. MVB sorting was tested using GFP chimeras of the biosynthetic cargo protein, Cps1 (GFP-Cps1) and the plasma membrane receptor, Ste3 (Ste3-GFP) expressed in vps27Δ cells. Both GFP-Cps1 and Ste3-GFP accumulated in the class E compartment in vps27Δ cells expressing only empty vector (Fig. 5B-C). In addition, GFP-Cps1 labeled the limiting membrane of the vacuole (Fig. 5B). Transformation of the vps27Δ strain with Vps27-WT restored transport of both cargo proteins into the lumen of the vacuole (Fig. 5D-E). In contrast, neither of the core complex mutants, Vps27-L410D or Vps27-I420D, was capable of restoring this transport (Figure 5F-I).
To assess the sorting of the soluble vacuolar hydrolase, CPY, the proteolytic maturation of the Golgi precursor (p2) form to the vacuolar mature (m) form was examined by pulse-chase analysis of hse1Δvps27Δ cells transformed with various Vps27 and Hse1 constructs (Fig. 5J). Following a 15 min chase period, CPY species were isolated by immunoprecipitation and analyzed by SDS-PAGE (Fig. 5J). hse1Δvps27Δ cells co-expressing Vps27-WT and Hse1-WT exhibited normal CPY maturation, with the majority of the hydrolase migrating as mature CPY (Fig. 5J, lane 1). Significant differences were observed in CPY maturation in hse1Δ vps27Δ cells expressing either Vps27-WT alone or Hse1-WT alone (Figure 5J, lanes 2 and 3). Specifically, cells expressing Vps27-WT alone (Fig. 5J, lane 2) exhibited a partial CPY maturation defect, as compared to cells expressing Hse1-WT alone (Fig. 5J, lane 3), in which virtually all the CPY migrated as the p2 form. These findings are consistent with past results that Vps27 is essential for CPY maturation, while loss of Hse1 results in only a modest reduction in CPY processing (Bilodeau et al., 2002). Interestingly, none of the core complex mutants (Vps27-I420D and Vps27-L410D shown) fully complemented the loss of Vps27 in hse1Δ vps27Δ cells expressing Hse1-WT alone (Fig. 5J, lanes 5 and 6), while the Vps27-A266Q behaved identically to Vps27-WT (Fig. 5J, lane 4). Similar results were obtained when the effects of the Vps27-A266Q and core complex mutations were analyzed in a CPY secretion colony blot assay (Fig. 5K). In this assay, secreted CPY was undetectable in hse1Δ vps27Δ cells expressing Vps27-WT, and barely visible in hse1Δ vps27Δ cells expressing Vps27-A266Q (Fig. 5K; 1 and 2); however, significant amounts of CPY secretion were detected in hse1Δ vps27Δ cells expressing either Vps27-I420D or Vps27-L410D (Fig. 5K; 3 and 4). These data are also consistent with previous studies showing no significant role for the UIM of Vps27 in CPY maturation (Bilodeau et al., 2002). These results thus show that point mutants in Vps27 that abrogate binding to Hse1 affect CPY sorting to roughly the same extent as the deletion of the Hse1 gene. Taken together with the results showing that these mutants block sorting of Cps1 and Ste3, these data demonstrate that the Vps27 interface residues observed in the crystal structure are important for the cellular functions of the Vps27/Hse1 complex.
Monte Carlo Simulation Analysis of the Properties of the Vps27/Hse1 Complex
The Vps27/Hse1 core is the last folded domain of the Vps27/Hse1 complex to have its three-dimensional structure solved. The complex consists of several folded domains linked by unstructured segments (Fig. 6). This structural information allowed us to conduct a theoretical analysis of the organization, dynamics, and interactions of this complex with the model ubiquitinated transmembrane cargo, Cps1 (Ub-Cps1), using a coarse-grained Monte Carlo (MC) approach. The UIMs of Vps27 and Hse1 were modeled as interacting with ubiquitin moieties on Lys-8 of the Cps1 cytosolic tail (Katzmann et al., 2001). The distance distributions of all three Ub-Cps1 molecules to a non-interacting domain used as a reference point are found to be equivalent over the course of the simulation, indicating the system is well-sampled and thoroughly equilibrated (Fig. 7A)
Figure 6. A Unified Model for the Interaction of Vps27/Hse1 with Membranes and Ubiquitinated Cargo.
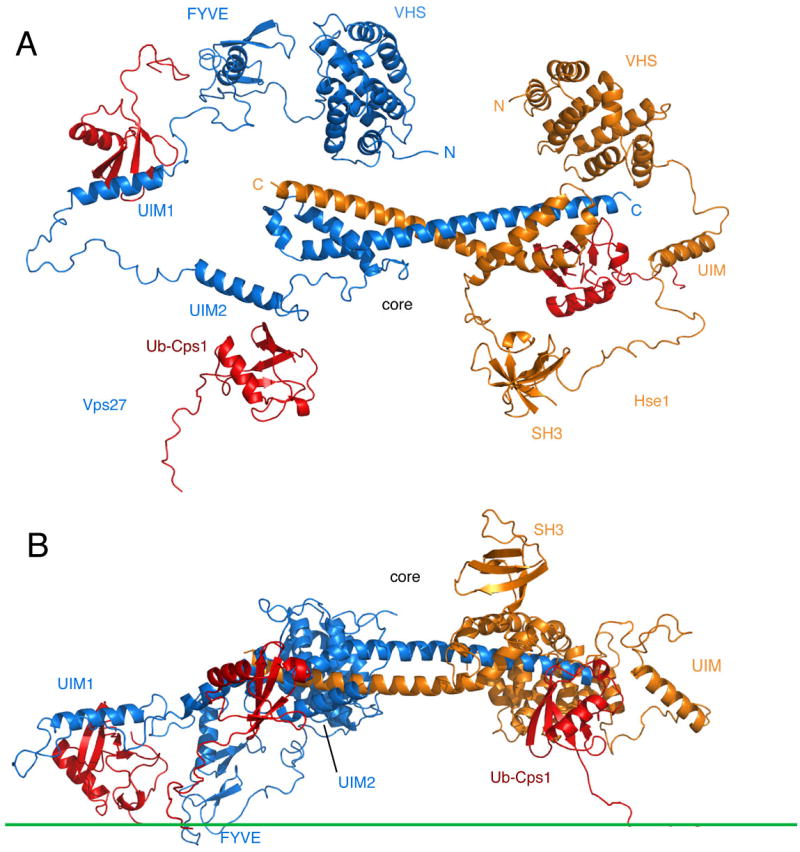
The figure shows a single snapshot from the MC simulation. (A) View looking directly down towards the membrane. (B) View normal to the plane of the membrane (membrane surface indicated by the green line). Vps27 is shown in blue, Hse1 in orange, and ubiquitinated cargo in red.
Figure 7. Dynamics of the Vps27/Hse1 complex.
(A) Distributions of the distances between the SH3 domain and three Ub-Cps1molecules. (B) Time series of the distance between the SH3 domain and the Hse1 UIM. (C) Distributions of the distances between the SH3 domain and three UIMs. (D) Average of Rg as a function of the overall system size. (E) Distributions of Rg for different system sizes. (F) Distributions of the distances between three UIMs and the membrane surface. (G) Fraction of Ub-Cps1 bound to the different UIMs (in isolation, on Vps27 alone, and on the full Hse1/Vps27 complex) as a function of the two-dimensional Ub-Cps1 concentration in the membrane.
The MC simulations showed this complex to be flexible and dynamic, capable of binding multiple ubiquitin moieties situated at different distances from the membrane. Even with the overall topology maintained, the Vps27/Hse1 complex can undergo large conformational changes, as indicated by the time series and distributions of distances between non-specifically interacting domains (Fig. 7B, C). Along the MC simulation trajectories, the Vps27/Hse1 core retains an extended and open configuration with a radius of gyration Rg between ∼40 and ∼60 Å (Fig. 7D, E). A globular protein of the same molecular weight as the simulated portion of Vps27/Hse1 would be expected to be much more compact, with Rg = ∼ 28 Å.
Interactions of the Vps27 FYVE domain with PI(3)P and the two Vps27 UIM domains with Ub-Cps1 keep this contiguous portion of the Vps27 complex near the membrane (Fig. 7F). Unlike the two Vps27 UIM domains, the single Hse1 UIM does not directly adjoin a membrane-binding domain in the primary sequence. Therefore, on average, the Hse1 UIM is found farther from the membrane (Fig. 7F). To explore the relative contributions of the different UIMs to overall binding on the membrane, the fraction of Ub-Cps1 molecules bound to the three different UIMs in isolation, in the full Vps27/Hse1 complex, and in Vps27 alone were calculated as a function of concentration (Fig. 7G). The affinities of the Vps27 UIMs in the context of the full complex are comparable to those of the isolated UIMs. In contrast, the affinity of the Hse1 UIM for Ub-Cps1 is substantially reduced in the complex. Several factors contribute to the difference. Within the complex, the Ub-Cps1/UIM binding is limited by steric restrictions and internally competitive interactions that are not present for the isolated UIM. This negative steric contribution to binding is balanced by cooperative interactions with the membrane and membrane-bound Ub-Cps1 molecules. This positive cooperative contribution is stronger for Vps27 than for Hse1 because the FYVE and two UIMs of Vps27 are close together. Therefore, the Vps27 UIM domains appear to have much stronger interactions with Ub-Cps1 than the Hse1 UIM.
Discussion
The structure of the Vps27/Hse1 core fills the last major gap in our understanding of the organization of this complex, and led us to several unexpected observations. First, the region involved in forming the core is more extensive than anticipated and extends beyond the predicted coiled-coil regions in both the N- and C-terminal directions. Second, the core comprises two GAT domains that had not previously been identified in these proteins. Third, the core assembles by the interchange of the homologous C-terminal halves of the α3 helices from each GAT domain. In addition, the resolution of this structure has allowed us to model its function as a scaffold for ubiquitin-binding and ubiquitination-deubiquitination reactions at the endosomal membrane.
The interchange of homologous α3 C-terminal halves from Vps27 and Hse1 is reminiscent of the mechanism of “domain swapping”. As originally defined, domain swapping refers to the oligomerization of identical protomers by interchange of identical regions of subunits (Liu and Eisenberg, 2002). The assembly of the Vps27/Hse1 core seems to us structurally and functionally equivalent to domain swapping in every respect other than the sequence identity of the exchanged regions.
In order to judge whether the domain-swapped complex represents the bona fide assembly mechanism for Vps27 and Hse1 in vivo, we analyzed the structure in the light of mutational analysis in the literature, and carried out additional mutational studies of complex formation. Residues 416-418 of Vps27 (sequence KIS) were critical for complex formation with Hse1 (Bilodeau et al., 2003). Vps27 Ile-417 within this sequence is deeply buried and almost completely surrounded by hydrophobic residues from Hse1 (Fig. 3), consistent with a critical role in function. We mutated Vps27 residues Leu-410, Ile-417, and Ile-420 individually to Asp, and found that these mutations prevented formation of the complex with Hse1. For each cargo tested, Cps1, Ste3 and CPY, these mutations resulted in a loss of function mirroring that seen in the deletion of Hse1. This establishes that the protein:protein interface observed in the structure is responsible for the assembly of the Vps27/Hse1 complex in yeast.
Despite the fact that Vps27 and Hse1 are subunits of a tightly assembled heterodimer, deletion of the gene encoding each subunit results in a quantitatively different defect in cargo sorting. Deletion of VPS27 causes a much stronger CPY missorting phenotype than deletion of HSE1 (Bilodeau et al., 2002) (see also Fig. 5). The missorting of Cps1 and Ste3 is also more severe in VPS27- than HSE1-deletion mutants (Bilodeau et al., 2002). HSE1 disruption phenotypes are more manifest in certain genetic backgrounds, like that of the SF8389D yeast strain used in these studies (Bilodeau et al., 2002). Our observations shed light on the probable cause for these phenotypic differences. Although the Vps27 core domain prefers to assemble as a heterodimer with the Hse1 core domain, it is nonetheless stable as a monomer when expressed in the absence of the Hse1 core domain. This is likely due to its ability to form an intramolecular GAT fold. In contrast, the Hse1 core domain expressed in isolation tends to aggregate and be degraded. Deletion of the VPS27 gene may thus lead to loss of both the Vps27 and Hse1 proteins, whereas deletion of the HSE1 gene would still leave enough Vps27 protein to sustain a modicum of function. In addition, Vps27 contains the main determinant of attachment of the complex to membranes, the FYVE domain, such that monomeric Hse1 is likely incapable of efficient recruitment to endosomes. Conversely, monomeric Vps27 could bind to membranes independently of Hse1, thus bringing its ubiquitin-, ESCRT-I- and clathrin-binding activities to bear on MVB sorting. Finally, the MC simulations show that the two UIM domains of Vps27 are closer to the membrane and exhibit more cooperativity than the single UIM domain of Hse1.
The human Hrs/STAM complex has been intensively studied, but the structural basis for its assembly remains unknown. The significant sequence homology between STAM, Hse1, and Vps27 allows us to predict that the core region of STAM will adopt the same structural fold as Vps27 and Hse1. The so-called SSM, which is needed for Hrs/STAM complex formation (Mizuno et al., 2004), corresponds to the C-terminal half of helix α1 and a few residues immediately following α1. Several of the conserved residues in the SSM correspond to key hydrophobic anchor residues in the subunit interface. The sequence of the core region of Hrs diverges from those of Vps27, Hse1, and STAM. However, the region of Hrs corresponding to the Vps27 GAT domain is predicted to be α-helical. Further, the example of Vps27 and Hse1 suggests that the STAM GAT domain requires a complementary GAT domain in Hrs with which to associate.
The unexpected observation of GAT domains in Vps27 and Hse1 highlights the parallel roles of these proteins with other GAT-domain-containing trafficking adaptors, the GGAs and Tom1 and Tom1-like proteins. The GGAs are modular proteins that contain a receptor-binding VHS domain, an Arf-binding helical hairpin domain, a ubiquitin-, Rabex-5, and ESCRT-I-binding GAT domain, an unstructured region containing autoinhibitory and clathrin-binding domains, and a GAE domain that binds to various accessory proteins (Bonifacino, 2004). Tom1 and its relatives Tom1L1 and Tom1L2 have a similar modular structure to the GGAs (Fig. 1A), and bind to ubiquitin via their GAT domains (Katoh et al., 2004; Yamakami et al., 2003) and to ESCRT-I (Puertollano, 2005). Collectively, the GGAs, Tom1 and the Tom1-like proteins, and the Vps27/Hse1 and Hrs/STAM complexes are a class of endosomal clathrin-binding proteins that sort ubiquitinated cargo proteins into the ESCRT pathway (Raiborg et al., 2006). These similarities highlight the GAT domain proteins collectively (Fig. 1A) as a family of proteins that sort ubiquitinated cargo into the ESCRT system.
The GAT domains of GGA1, GGA3, and Tom1 bind ubiquitin (Katoh et al., 2004; Puertollano and Bonifacino, 2004; Scott et al., 2004; Shiba et al., 2004) with affinities ranging from 180-410 μM (Akutsu et al., 2005; Kawasaki et al., 2005; Prag et al., 2005). However, no ubiquitin binding to the Vps27/Hse1 core was detected by isothermal titration calorimetry or surface plasmon resonance at concentrations of up to 8.0 mM and 2.0 mM, respectively (data not shown). Known ubiquitin-binding GAT domains contain two ubiquitin binding sites. Ubiquitin binds to the GGA and Tom1 GAT domains at site 1 on helices α1 and α2, and site 2 on helices α2 and α3. These sites are incompletely conserved in Vps27 and Hse1 (Fig. 1B). Unlike Vps27 and Hse1, the GGAs and Tom1 do not contain ubiquitin-binding UIM motifs. If the family of GAT-domain containing adaptors evolved from a common ancestor, the GAT domains have served multiple purposes in ubiquitin binding, dimerization, and other functions. By the same token, different ubiquitin-binding, GAT domain-containing proteins acquired different mechanisms for binding ubiquitinated cargo, some binding through the GAT domain, and others via their UIMs.
The structures and the ubiquitin and membrane affinities of individual domains from the Vps27/Hse1 and Hrs/STAM complexes are known (Diraviyam et al., 2003; Fisher et al., 2003; Hirano et al., 2006; Kaneko et al., 2003; Mao et al., 2000; Misra and Hurley, 1999; Stahelin et al., 2002; Swanson et al., 2003). Despite a wealth of information on individual domains, it has not been possible to integrate this knowledge into a unified model of the Vps27/Hse1 complex. The structure determination of the Vps27/Hse1 core provides the missing link that allows the integration of the domain information for the first time. The simulations show that cooperativity between the Vps27 and FYVE and UIM domains in membrane and ubiquitinated membrane protein binding offsets steric constraints imposed in the complex. The simulations portray the complex as open and dynamic. Vps27/Hse1 traffics a variety of ubiquitinated cargo. The molecular weight, the size of the cytosolic domain, and the location of the ubiquitination sites on these cargoes vary widely. An open, dynamic complex such as Vps27/Hse1 can adapt to these differences in cargo in ways that a rigid complex could not. Finally, the simulations show that the Hse1 SH3 domain, which targets DUBs that potentially deubiquitinate cargo, samples conformational space that frequently approaches within 20 Å of ubiquitinated cargo. This suggests that the action of the Hse1 UIM and SH3 domains coordinates ubiquitinated cargo binding and deubiquitination reactions.
A model has been proposed for Hrs, based on a 16 Å resolution cryo-EM structure of an Hrs hexamer determined in the absence of STAM (Pullan et al., 2006). In the Hrs model, three sets of membrane and ubiquitin binding domains are located 175 Å apart from each other at two sets of end caps. It is difficult to compare these models given the substantial differences in the Hrs and Vps27 core sequences and the presence of a 1:1 heterodimer in one structure vs. a homohexamer in the other.
We have visualized the core of the Vps27/Hse1 complex, which uses an elegant variation on the GAT domain, not to bind ubiquitin directly, but instead to coordinate ubiquitinated cargo binding, and ubiquitination and deubiquitination reactions. The structure of the Vps27/Hse1 complex shows how the complex can spatially confine ubiquitinated cargo and coordinate the action of DUBs (e.g., Ubp7) against a tightly localized subpopulation of substrate. Such a mechanism could help account for the biological specificity of this deubiquitinating enzyme, given the large number of potential substrates in the cell. Most importantly, the structure of the Vps27/Hse1 core complex has provided us with a unifying framework for understanding the integrated action of the many modular domains of these two proteins.
Experimental Procedures
Protein Expression, Purification and Crystallization
DNA sequences encoding Vps27 and Hse1 genes were amplified by PCR from Saccharomyces cerevisiae genomic DNA. DNA coding for residues 345-440 of Vps27 was subcloned in frame with a hexahistidine tag followed by a TEV protease cleavage sequence into the pST39 (Tan, 2001) expression vector. DNA coding for residues 275-375 of Hse1 was subsequently cloned into the same vector without a tag to yield a polycistronic expression vector for the Vps27/Hse1 core complex (pHisVps27/Hse1). The Hse1 GAT domain with similar boundaries was cloned into pHIS-Parallel.2 in-frame to an N-terminal His-tag and a TEV recognition site. Site directed mutagenesis of pHisVps27/Hse1 was performed using the QuikChange kit (Stratagene) with appropriate DNA primers, and the entire sequences were verified by DNA sequencing. The individual GAT domains of Vps27 or Hse1 were overexpressed in Escherichia coli BL21 (λDE3) Rosetta cells (Novagen) overnight at 20 °C with 1 mM IPTG. Pellets were resuspended in 0.5 M NaCl, 10% glycerol and 50 mM Tris-HCl pH8.0 supplemented with protease inhibitors. Cells were disrupted by lysozyme treatment followed by sonication and centrifugation. The proteins were then purified on a Ni-NTA agarose column. The His-tags were removed by cleavage with TEV protease and the samples were passed through a second Ni-NTA column, followed by gel-filtration. The Vps27/Hse1 core complex was overexpressed and purified in the same manner. The selenomethionyl form of the complex was expressed in the methionine auxotroph Escherichia coli strain, B834 (λDE3) grown in defined media that contained selenomethionine, and purified as described above.
The native and selenomethionyl forms of the Vps27/Hse1 core complex were concentrated to 18 mg/ml and crystallized in 1.1-1.4 M ammonium sulfate, 0.1 M Tris-HCl pH 8.85 in hanging drops at a 1:1 ratio of protein to precipitant. Crystals were improved by micro-seeding into similar conditions with 1.25 M ammonium sulfate. Individual crystals were cryoprotected with Paratone-N (Hampton Research) and were frozen in liquid nitrogen.
Structure Determination
A two wavelength MAD data set was collected from a single selenomethionyl crystal at the Advanced Photon Source (APS) synchrotron, Argonne National Laboratory at 95 K. Energies for the MAD experiment were chosen at the inflection point and remote at 12,658 and 12,750 eV respectively. Data were analyzed with DENZO and Scalepack (HKL Research). Eight selenium atoms were located by a direct methods search using the BnP interface (Weeks et al., 2002), and phases were calculated by Phasit. Density modification of the initial maps was performed using RESOLVE (Terwilliger, 2000), DM and Solomon of the CCP4 suite programs (CCP4, 1994). The resulting maps were used to build atomic models in O (Jones et al., 1991) and Coot (Emsley and Cowtan, 2004). The model was then refined using CNS (Brunger et al., 1998) (Table 1) at 3.0 Å against the inflection point dataset.
Table 1. Crystallographic data processing, phasing and refinement statistic.
| Collection Energies | Inflection | Remote |
|---|---|---|
| Data processing | ||
| Wavelength (Å) | 0.97948 | 0.97242 |
| Space group | P3221 | P3221 |
| Cell dimensions | ||
| a, b, c (Å) | 62.4, 62.4, 94.5 | 62.4, 62.4, 94.6 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 3.0 | 3.2 |
| I / σI | 18.1 (2.5) | 12.7 (0.4) |
| Redundancy | 4.1 (2.3) | 3.7 (1.2) |
| Completeness (%) | 97.5 (88.1) | 82.5 (17.3) |
| No. of reflections | 4415 | 3798 |
| Rsym | 0.052 (0.336) | 0.059 (0.744) |
| Refinement | ||
| Resolution (Å) | 3.0 | |
| Rwork / Rfree | 0.205 / 0.278 | |
| No. of protein atoms | 1420 | |
| No. of water molecules | 41 | |
| B factors | ||
| Average of main chains | 60.5 | |
| Average of side chains | 70.5 | |
| Water molecules | 60.6 | |
| Geometric R.M.S deviation | ||
| Bond length (Å) | 0.007 | |
| Bond angles (°) | 1.2 |
Gel Filtration
In vitro analysis of the oligomeric state of purified recombinant GAT domains from the individual proteins and from the complex was performed on a Superdex 200 HR 30/10 column (Amersham Pharmacia Biotech) in 150 mM NaCl, 50 mM Tris-HCl pH 8.0.
Sedimentation Equilibrium
Sedimentation equilibrium experiments were conducted at 4.0°C on a Beckman Optima XL-A analytical ultracentrifuge. Samples in 50 mM Tris (pH 8.0) were loaded at concentrations corresponding to measured A280 values of 0.25, 0.50 and 0.75. Data were acquired at 13, 16, 19, 22, 25 and 28 krpm as an average of 4 absorbance measurements at 280 nm and a radial spacing of 0.001 cm. Equilibrium was achieved within 48 h. Data were analyzed globally in SEDPHAT 4.3 (Schuck, 2003), (http://www.analyticalultracentrifugation.com/sedphat/sedphat.htm) using solution densities (ρ) and protein partial specific volumes (v) calculated in SEDNTERP (Philo, J. http://www.jphilo.mailway.com/) as described in the text. Excellent fits were observed in all cases. For the sample consisting of the Vps27 core region alone, the mass of the smallest species was fixed to that expected for the monomer to return a value of 48 kDa for the oligomeric species (n = 4.3). This higher mass species contributes approximately 20% of the total absorbance; therefore a solution having an absorbance at 280 nm of 1.0 will contain 130 μM of the monomer and 9 μM of the aggregate, demonstrating that the Vps27 monomer is the major species.
Co-immunoprecipitation
Twenty OD600 of cells grown to midlog phase were lysed in TBS-T (10mM Tris, 140mM NaCl, 0.1% Tween 20, pH 7.5) supplemented with protease inhibitor cocktail (Roche, Indianapolis, IN) using glass bead lysis. Lysates were centrifuged at 14,000 rpm at 4°C, and supernatants were pre-cleared by incubation for 60 min at 4°C with 30μl of Protein-A Sepharose beads (Amersham Pharmacia Biotech, Piscataway, NJ) and centrifugation at 8,000 g for 5 min. The pre-cleared lysates were incubated for 2 h at 4°C with 30 μl Protein A-Sepharose beads bound to rabbit polyclonal anti-HA antibody (Covance, Princeton, NJ). The beads were then washed four times with TBS-T and subjected to SDS-PAGE and immunoblotting analysis with either mouse monoclonal anti-HA or mouse monoclonal anti-myc (9E10) (Covance, Princeton, NJ).
Fluorescence Microscopy
Cells were grown to midlog phase in selective media and viewed on an Olympus IX-70 fluorescence microscope (excitation, 560 nm; dichroic mirror at 595 nm; emission, 630 nm). Images were captured with an IMAGO charge-coupled device camera controlled by TILLvisION software (TILL Photonics, Eugene, OR) and processed using Adobe Photoshop 5.0 (Adobe Systems, Mountain View, CA).
CPY Maturation Assay
Yeast hse1Δ vps27Δ transformants were metabolically labeled with the 35S Express reagent (Perkin Elmer Life Sciences) for 10 min (pulse) and chased for 30 min (Bonifacino and Dell'Angelica, 1998). Immunoprecipitations using mouse-anti-CPY (Molecular Probes, Eugene, OR) were performed overnight at 4°C, and the immunoprecipitates were analyzed by SDS-PAGE and fluorography.
CPY Secretion Assay
The CPY secretion colony blot assay was performed as described (Mullins and Bonifacino, 2001). Strains were grown to 1 to 1.5 units of OD600 /ml at 30°C and concentrated to 0.2 units of OD600 /ml. Five μl of each strain were spotted on selective medium. Plates were incubated overnight at 30°C to allow cell growth, overlayed with nitrocellulose and incubated 12 to 14 h. Membranes were rinsed with distilled H2O, and analyzed by immunoblotting with anti-CPY antibodies (Molecular Probes, Eugene, OR).
Simulations
To obtain a starting model, the Vps27 FYVE domain (Misra and Hurley, 1999) was docked to the membrane (Diraviyam et al., 2003). The Vps27 UIM1:ubiquitin complex and the Vps27 tandem UIM1-UIM2 constructs (Swanson et al., 2003) were used to model the ubiquitin complex with both Vps27 UIMs and the Hse1 UIM:ubiquitin complex. The Hse1 SH3 domain was modeled from the STAM2 SH3 domain (Kaneko et al., 2003). The VHS domains of Vps27 and Hse1 were modeled from the Hrs VHS domain (Mao et al., 2000). The core was positioned such that linkers to the nearest domains would be able to reach them. The predicted unstructured (Rost and Liu, 2003) C-terminal regions of Vps27/Hse1 were omitted.
Long-range electrostatic interactions were included at the Debye-Hückel level. The relative interaction strengths have previously been calibrated to reproduce measured protein second virial coefficients. Flexible linkers were represented as polymers at the amino acid level, and the folded protein domains were treated as rigid bodies. The resulting model has been validated by predictions of a series of protein-complex structures and binding affinities (Kim and Hummer, in preparation). Homology modeling was carried out manually in O (Jones et al., 1991) or semi-manually in Coot (Emsley and Cowtan, 2004).
Protein:membrane interactions were represented by a combination of residue-dependent short-range interactions (Miyazawa and Jernigan, 1996) and a Gouy-Chapmann-type electrostatic potential between the flat membrane and the amino acids (Kim and Hummer, in preparation). To control molecular concentrations, the proteins were confined into semi-spheres of different sizes above the membrane plane. The FYVE domain was anchored to the membrane by a harmonic potential between the known PI(3)P binding site (Misra and Hurley, 1999) and the membrane surface. The Ub-Cps1 molecules were modeled as a covalent link between Gly-76 of ubiquitin and Lys-8 of Cps1. Cps1 residues were modeled explicitly through residue 18, which was anchored to the membrane surface by a strict distance constraint.
An equilibrium ensemble of complex structures was obtained by performing replica-exchange Monte Carlo (MC) simulations, with 20 replicas covering the temperature range from 0.8T to 1.7T, where T is room temperature. For each system, a total of 20000-30000 complex structures were obtained, which amounted to about 107-108 MC steps. To test for convergence, results from three independent runs were compared.
To gauge whether the simulations provided a realistic estimate of affinity, the affinities of the isolated UIMs for ubiquitin were calculated. We obtained a calculated value of Kd = ∼800 μM for Vps27 UIM1, as compared to the experimental value of Kd = ∼300 μM (Fisher et al., 2003). The calculated Kd of Vps27 UIM1 was lower than that of Vps27 UIM2, also in agreement with experiment (Fisher et al., 2003; Swanson et al., 2003). The simulations predicted the structure of the Vps27 UIM1-ubiquitin complex to within 3 Å r.m.s.d. (Cα) of the solution structure (Swanson et al., 2003). We found that the overall topology of the Vps27/Hse1 multi-protein complex inferred from known domain interactions was roughly maintained (Fig. 7), despite substantial conformational dynamics. Ubiquitin binding events were thoroughly equilibrated. These observations suggest that the simulations provide a well-sampled and quantitatively reasonable reflection of the system.
Supplementary Material
Acknowledgments
We thank X. (Snow) Ren and W. Smith for assistance with biochemical experiments, R. Piper for strains and plasmids, J. Chrzas, J. Gonczy, M. Graham, and all of the staff of SER-CAT beamline ID-22 for user support, and Y. Ye and A. Hickman for comments on the manuscript. Use of the APS was supported by the U. S. DOE, Basic Energy Sciences, Office of Science, under Contract No.W-31-109-Eng-38. This research was supported by NIH intramural support, NIDDK (J.H.H. and G.H.), NICHD (J.S.B.) and IATAP (J.H.H. and J.S.B.).
Footnotes
COORDINATES
Crystallographic coordinates have been deposited in the Protein Data Bank with accession code XXXX.
References
- Akutsu M, Kawasaki M, Katoh Y, Shiba T, Yamaguchi Y, Kato R, Kato K, Nakayama K, Wakatsuki S. Structural basis for recognition of ubiquitinated cargo by Tom1-GAT domain. FEBS Lett. 2005;579:5385–5391. doi: 10.1016/j.febslet.2005.08.076. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. A Protein's Final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p-Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol. 2003;163:237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS. The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Dell'Angelica EC. Protein labeling and immunoprecipitation. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM, editors. Current Protocols in Cell Biology. New York, NY: John Wiley and Sons, Inc.; 1998. [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochimica Et Biophysica Acta-Molecular Cell Research. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D-Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr A. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Urbe S. The interface of receptor trafficking and signalling. J Cell Sci. 2001;114:3075–3081. doi: 10.1242/jcs.114.17.3075. [DOI] [PubMed] [Google Scholar]
- Collins BM, Watson PJ, Owen DJ. The structure of the GGA1-GAT domain reveals the molecular basis for ARF binding and membrane association of GGAs. Dev Cell. 2003;4:321–332. doi: 10.1016/s1534-5807(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Freed EO. Retrovirus budding. Virus Research. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Diraviyam K, Stahelin RV, Cho W, Murray D. Computer modeling of the membrane interaction of FYVE domains. J Mol Biol. 2003;328:721–736. doi: 10.1016/s0022-2836(03)00325-5. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D-Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–28984. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nature Structural & Molecular Biology. 2006;13:272–277. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali - a Network Tool for Protein-Structure Comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomolec Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron- Density Maps and the Location of Errors in These Models. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Kumasaka T, Ganbe T, Sato T, Miyazawa K, Kitamura N, Tanaka N. Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain. J Biol Chem. 2003;278:48162–48168. doi: 10.1074/jbc.M306677200. [DOI] [PubMed] [Google Scholar]
- Kato M, Miyazawa K, Kitamura N. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J Biol Chem. 2000;275:37481–37487. doi: 10.1074/jbc.M007251200. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Shiba Y, Mitsuhashi H, Yanagida Y, Takatsu H, Nakayama K. Tollip and Tom1 form a complex and recruit ubiquitin-conjugated proteins onto early endosomes. J Biol Chem. 2004;279:24435–24443. doi: 10.1074/jbc.M400059200. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki M, Shiba T, Shiba Y, Yamaguchi Y, Matsugaki N, Igarashi N, Suzuki M, Kato R, Kato K, Nakayama K, Wakatsuki S. Molecular mechanism of ubiquitin recognition by GGA3 GAT domain. Genes To Cells. 2005;10:639–654. doi: 10.1111/j.1365-2443.2005.00865.x. [DOI] [PubMed] [Google Scholar]
- Komada M, Kitamura N. The Hrs/STAM complex in the downregulation of receptor tyrosine kinases. Journal of Biochemistry. 2005;137:1–8. doi: 10.1093/jb/mvi001. [DOI] [PubMed] [Google Scholar]
- Liu Y, Eisenberg D. 3D domain swapping: As domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, Bellen HJ. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–269. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- Lu Q, Hope LWQ, Brasch M, Reinhard C, Cohen SN. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci U S A. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YX, Nickitenko A, Duan XQ, Lloyd TE, Wu MN, Bellen H, Quiocho FA. Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell. 2000;100:447–456. doi: 10.1016/s0092-8674(00)80680-7. [DOI] [PubMed] [Google Scholar]
- Mattera R, Puertollano R, Smith WJ, Bonifacino JS. The trihelical bundle subdomain of the GGA proteins interacts with multiple partners through overlapping but distinct sites. J Biol Chem. 2004;279:31409–31418. doi: 10.1074/jbc.M402183200. [DOI] [PubMed] [Google Scholar]
- McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate- specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Miyazawa K, Jernigan RL. Residue-residue potentials with a favorable pair term and an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- Mizuno E, Kawahata K, Okamoto A, Kitamura N, Komada M. Association with Hrs is required for the early endosomal localization, stability, and function of STAM. Journal of Biochemistry. 2004;135:385–396. doi: 10.1093/jb/mvh046. [DOI] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Mullins C, Bonifacino JS. Structural requirements for function of Yeast GGAs in Vacuolar Protein Sorting, Alpha-Factor Maturation, and Interactions with Clathrin. Mol Cell Biol. 2001;21:7981–7994. doi: 10.1128/MCB.21.23.7981-7994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. Vps27 Controls Vacuolar And Endocytic Traffic Through A Prevacuolar Compartment In Saccharomyces-Cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Luzio JP. Late endosomes: Sorting and partitioning in multivesicular bodies. Traffic. 2001;2:612–621. doi: 10.1034/j.1600-0854.2001.20904.x. [DOI] [PubMed] [Google Scholar]
- Prag G, Lee SH, Mattera R, Arighi CN, Beach BM, Bonifacino JS, Hurley JH. Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, gamma-ear-containing, ADP-ribosylation-factor-binding proteins. Proc Natl Acad Sci U S A. 2005;102:2334–2339. doi: 10.1073/pnas.0500118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R. Interactions of Tom1L1 with the multivesicular body sorting machinery. J Biol Chem. 2005;280:9258–9264. doi: 10.1074/jbc.M412481200. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- Pullan L, Mullapudi S, Huang Z, Baldwin PR, Chin C, Sun W, Tsujimoto S, Kolodziej SJ, Stoops JK, Lee JC, et al. The endosome-associated protein Hrs is hexameric and controls cargo sorting as a “master molecule”. Structure. 2006;14:661–671. doi: 10.1016/j.str.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D'Arrigo A, Stang E, Stenmark H. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Wesche J, Malerod L, Stenmark H. Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J Cell Sci. 2006;119:2414–2424. doi: 10.1242/jcs.02978. [DOI] [PubMed] [Google Scholar]
- Ren J, Kee Y, Huibergtse JM, Piper RC. Hse1, a Component of the Yeast Hrs-STAM Ubiquitin Sorting Complex, Associates with Ubiquitin Peptidases and a Ligase to Control Sorting Efficiency into Multivesicular Bodies. Mol Biol Cell. 2007;18:324–335. doi: 10.1091/mbc.E06-06-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Liu JF. The PredictProtein server. Nucleic Acids Res. 2003;31:3300–3304. doi: 10.1093/nar/gkg508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Analytical Biochemistry. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Scott PM, Bilodeau PS, Zhdankina O, Winistorfer SC, Hauglund MJ, Allaman MM, Kearney WR, Robertson AD, Boman AL, Piper RC. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat Cell Biol. 2004;6:252–259. doi: 10.1038/ncb1107. [DOI] [PubMed] [Google Scholar]
- Shiba T, Kawasaki M, Takatsu H, Nogi T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Nakayama K, Wakatsuki S. Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat Struct Biol. 2003;10:386–393. doi: 10.1038/nsb920. [DOI] [PubMed] [Google Scholar]
- Shiba Y, Katoh Y, Shiba T, Yoshino K, Takatsu H, Kobayashi H, Shin HW, Wakatsuki S, Nakayama K. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J Biol Chem. 2004;279:7105–7111. doi: 10.1074/jbc.M311702200. [DOI] [PubMed] [Google Scholar]
- Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends In Cell Biology. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- Suer S, Misra S, Saidi LF, Hurley JH. Structure of the GAT domain of human GGA1: A syntaxin amino- terminal domain fold in an endosomal trafficking adaptor. Proc Natl Acad Sci U S A. 2003;100:4451–4456. doi: 10.1073/pnas.0831133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. Embo J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. A Modular Polycistronic Expression System for Overexpressing Protein Complexes in Escherichia coli. Protein Expr Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr Sect D-Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks CM, Blessing RH, Miller R, Mungee R, Potter SA, Rappleye J, Smith GD, Xu H, Furey W. Towards automated protein structure determination: BnP, the SnB-PHASES interface. Zeitschrift Fur Kristallographie. 2002;217:686–693. [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Yamakami M, Yoshimori T, Yokosawa H. Tom1, a VHS domain-containing protein, interacts with tollip, ubiquitin, and clathrin. J Biol Chem. 2003;278:52865–52872. doi: 10.1074/jbc.M306740200. [DOI] [PubMed] [Google Scholar]
- Zhu GY, Zhai P, He XY, Terzyan S, Zhang RG, Joachimiak A, Tang J, Zhang XJC. Crystal structure of the human GGA1 GAT domain. Biochemistry. 2003;42:6392–6399. doi: 10.1021/bi034334n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



