Abstract
This study analyzed 28 Aspergillus strains belonging to the section Fumigati that were isolated from clinical samples in Spain. All isolates sporulated slowly and were unable to grow at 48°C. Phylogenetic analysis based on sequencing of partial sequences of the β-tubulin and rodlet A genes was used to classify the 28 strains into six different clades (Neosartorya hiratsukae, Neosartorya pseudofischeri, Aspergillus viridinutans, Aspergillus lentulus, Aspergillus fumigatiaffinis, and Aspergillus fumisynnematus). Antifungal susceptibility testing showed heterogeneous patterns and grouped the strains together by species. Most A. lentulus and A. fumigatiaffinis isolates showed high MICs of amphotericin B (geometric mean [GM] MICs, ≥4.5 μg/ml), itraconazole (GM MICs, ≥6 μg/ml), voriconazole (GM MICs, ≥3 μg/ml), and ravuconazole (GM MICs, ≥3 μg/ml); N pseudofischeri and A. viridinutans showed high MICs of itraconazole (GM MICs, ≥8 μg/ml), voriconazole (GM MICs, ≥3.33 μg/ml), and ravuconazole (GM MICs, ≥2 μg/ml); and N. hiratsukae and A. fumisynnematus were susceptible to all the antifungals tested. In conclusion, a number of different species whose morphological features resemble those of Aspergillus fumigatus could succeed in producing invasive infections in the susceptible host. In addition, some of them showed high MICs for most of the antifungals available for the treatment of patients infected with these organisms. The epidemiology and clinical relevance of these species should therefore be addressed.
The incidence of invasive aspergillosis continues to increase, due to the rising number of patients undergoing bone marrow or solid organ transplantation or corticosteroid treatment and those with hematological malignancies or pulmonary disease (18).
Invasive aspergillosis is mainly caused by Aspergillus fumigatus, although other species, such as Aspergillus terreus, Aspergillus niger, and Aspergillus flavus can also cause invasive infections (10, 15, 22). Furthermore, recent studies have reported on cases of aspergillosis caused by other Aspergillus species that belong to Aspergillus section Fumigati (4, 12, 14, 17).
Aspergillus section Fumigati has recently been reclassified by Samson et al. (36). It currently contains 25 different species, with 8 anamorphs (Aspergillus brevipes, Aspergillus duricaulis, A. fumigatus, Aspergillus fumigatiaffinis, Aspergillus lentulus, Aspergillus novofumigatus, Aspergillus unilateralis, Aspergillus viridinutans) and 17 telemorphs (Neorsartorya aurata, Neorsartorya aureola, Neorsartorya coreana, Neorsartorya fennelliae, Neorsartorya fischeri, Neorsartorya glabra, Neorsartorya lacinosa, Neorsartorya spinosa, Neorsartorya quadricincta, Neorsartorya stramenia, Neorsartorya spathulata, Neorsartorya hiratsukae, Neorsartorya pseudofischeri, Neorsartorya tetenoi, Neorsartorya mulplicata, Neorsartorya udagawae, and Neorsartorya sublevispora).
In the section Fumigati, besides A. fumigatus, other species, such as Neorsartorya fischeri, Neorsartorya pseudofischeri, Neorsartorya hiratsukae, and A. lentulus, have been reported to be human pathogens (3, 4, 12, 14, 17). This implies that in the appropriate human host, all of them could cause disease.
The conventional means of identification of A. fumigatus is based on its morphological characteristics and microscopic features. Several morphological characteristics for differentiation between species of the Aspergillus section Fumigati have been described. However, many species sporulate very slowly, and an extremely high level of expertise and long-term observation are also required to identify the species. Other species, such as those of the genus Neosartorya, are able to produce ascospores; however, a considerable length of time is usually required for the production of ascospores, and ascospore production is not a practical method of identification for clinical microbiology laboratories (14, 16, 17, 36). Although for the very experienced taxonomist the species included in the section Fumigati are not morphologically uniform, morphological observation is not sufficient to distinguish between them. This fact has led to species misidentification and also to the discarding of organisms as contaminants (6, 44).
In order to resolve this issue, a number of different techniques have been developed and used to identify the species belonging to this section. These include analysis of the profiles of secondary metabolites, isozyme electrophoretic pattern analysis (23, 27, 33, 38, 39, 40), and molecular data analysis (41, 44).
Although secondary resistance to azole drugs has been described in A. fumigatus strains (7, 8, 11, 25, 26, 28, 42), most A. fumigatus strains are susceptible to the antifungals available for the treatment of patients with invasive infections (13). Because few clinical laboratories routinely perform antifungal susceptibility testing of molds and resistance in the section Fumigati has already been reported (5, 24), the misidentification of these species is a matter of concern.
The aim of this study was to analyze clinical strains of Aspergillus section Fumigati. To date, we have analyzed a collection of 28 Aspergillus clinical strains that had previously been identified as atypical A. fumigatus isolates. We report on the results of the molecular identification by sequencing of both the β-tubulin and the rodlet A genes and the antifungal susceptibility testing profiles of the strains.
MATERIALS AND METHODS
Fungal strains and media.
A total of 37 strains were included in this study; 28 Aspergillus section Fumigati strains (Table 1) were independent clinical isolates from different patients and belong to the Mold Collection of the Centro Nacional de Microbiologia (CNM). Control strains included a set of five A. fumigatus strains belonging to the CNM (strains CNM-CM-3248, CNM-CM-3254, CNM-CM-3258, CNM-CM-3652, and CNM-CM-3722) and four strains obtained from the Centralbureau voor Schimmelcultures (CBS) (N. hiratsukae CNM-CM-4551 [CBS 109356] and CNM-CM-4554 [CBS 117067] and N. pseudofischeri CNM-CM-4487 [CBS 404.67] and CNM-CM-4488 [CBS 208.92]).
TABLE 1.
Origin, molecular identification, MICs, and MECs of different antifungal against clinical isolates of Aspergillus section Fumigati
| Isolate | Molecular identification
|
MIC (μg/ml)a
|
MEC (μg/ml)b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Originc | β-Tubulin | Rodlet A | AMB | ITC | VCZ | RVC | POS | TRB | CAS | MICA | |
| ATCC 2004305 | Reference strain | A. fumigatus | A. fumigatus | 0.25-1.0 | 0.125-1.0 | 0.25-1.0 | 0.25-1.0 | 0.03-0.25 | 2.0-8.0 | 0.12-0.5 | 0.03 |
| CNM-CM-1290 | Sputum | A. lentulus | A. lentulus | 4.5 | 9.25 | 4.5 | 3.5 | 0.40 | 1.33 | 0.28 | 0.05 |
| CNM-CM-3134 | Sputum | A. lentulus | A. lentulus | 10 | 10.25 | 6 | 6 | 0.56 | 1.75 | 0.38 | 0.07 |
| CNM-CM-3364 | BAL | A. lentulus | A. lentulus | 8.4 | 8 | 5.6 | 5.6 | 2 | 0.5 | 0.18 | 0.03 |
| CNM-CM-3537 | Sputum | A. lentulus | A. lentulus | 7 | 8.25 | 3.5 | 3 | 0.28 | 1 | 0.42 | 0.04 |
| CNM-CM-3538 | Sputum | A. lentulus | A. lentulus | 7.25 | 8.25 | 7.5 | 7 | 0.75 | 1 | 0.11 | 0.10 |
| CNM-CM-3583 | Sputum | A. lentulus | A. lentulus | 8 | 0.43 | 3.5 | 2 | 0.25 | 1.25 | 1 | 0.03 |
| CNM-CM-3599 | Sputum | A. lentulus | A. lentulus | 2.66 | 0.5 | 4 | 2 | 0.5 | 1 | 0.26 | 0.03 |
| CNM-CM-4330 | Sputum | A. lentulus | A. lentulus | 8 | 6 | 3 | 1.5 | 0.12 | 0.5 | 1.2 | 0.03 |
| CNM-CM-4370 | Sputum | A. lentulus | A. lentulus | 7.33 | 8 | 5.33 | 5.33 | 0.18 | 0.5 | 0.91 | 0.03 |
| CNM-CM-4387 | Sputum | A. lentulus | A. lentulus | 6 | 8 | 4 | 3 | 0.25 | 0.5 | 0.25 | 0.03 |
| CNM-CM-4415 | Sputum | A. lentulus | A. lentulus | 6 | 16 | 5.33 | 4.66 | 0.33 | 0.5 | 0.25 | 0.03 |
| CNM-CM-4420 | Nail | A. lentulus | A. lentulus | 10.66 | 8 | 5.33 | 4 | 0.33 | 1 | 0.33 | 0.03 |
| CNM-CM-4426 | Sputum | A. lentulus | A. lentulus | 12 | 6.66 | 5.33 | 5.33 | 0.20 | 0.83 | 0.22 | 0.03 |
| CNM-CM-4428 | Skin | A. lentulus | A. lentulus | 12 | 7 | 5 | 5 | 0.62 | 0.66 | 0.29 | 0.03 |
| CNM-CM-4063 | BAS | A. fumisynnematus | A. fumisynnematus | 1 | 0.25 | 0.83 | 0.83 | 0.12 | 1.66 | 0.5 | 0.03 |
| CNM-CM-2280 | Sputum | A. fumigatiaffinis | A. fumigatiaffinis | 16 | 6.66 | 6.66 | 6 | 1.16 | 1.66 | 1.3 | 0.04 |
| CNM-CM-3227 | BAL | A. fumigatiaffinis | A. fumigatiaffinis | 8 | 8 | 5.6 | 5 | 0.625 | 1.4 | 0.16 | 0.08 |
| CNM-CM-3303 | Skin | N. hiratsukae | N. hiratsukae | 0.5 | 0.12 | 0.5 | 0.375 | 0.06 | 0.09 | 0.34 | 0.03 |
| CNM-CM-3305 | Skin | N. hiratsukae | N. hiratsukae | 0.31 | 0.185 | 0.375 | 0.25 | 0.045 | 0.06 | 0.03 | 0.14 |
| CNM-CM-3764 | OPE | N. hiratsukae | N. hiratsukae | 1 | 0.33 | 1.66 | 1.33 | 0.16 | 0.25 | 0.26 | 0.04 |
| CNM-CM-3740 | OPE | N. hiratsukae | N. hiratsukae | 0.66 | 0.16 | 1 | 0.66 | 0.10 | 0.20 | 0.66 | 0.03 |
| CNM-CM-4328 | Corneal | N. hiratsukae | N. hiratsukae | 0.75 | 0.25 | 1.25 | 0.5 | 0.09 | 0.09 | 0.03 | 0.03 |
| CNM-CM-3769 | Sputum | N. pseudofischeri | N. pseudofischeri | 1 | 8 | 3.33 | 2 | 0.25 | 1.33 | 0.12 | 0.06 |
| CNM-CM-2270 | Sputum | N. pseudofischeri | N. pseudofischeri | 0.17 | 8 | 3 | 3.5 | 0.31 | 1 | 0.51 | 0.26 |
| CNM-CM-4060 | Sputum | N. pseudofischeri | N. pseudofischeri | 0.25 | 11 | 3.33 | 3.33 | 0.29 | 0.5 | 0.62 | 0.03 |
| CNM-CM-3914 | Nail | N. pseudofischeri | N. pseudofischeri | 0.25 | 16 | 6.66 | 4 | 0.5 | 0.5 | 1 | 0.03 |
| CNM-CM-3147 | OPE | A. viridinutans | A. viridinutans | 0.58 | 14.4 | 4 | 5.66 | 0.41 | 1.2 | 0.67 | 0.20 |
| CNM-CM-4518 | Nail | A. viridinutans | A. viridinutans | 0.37 | 16 | 4 | 4 | 0.25 | 0.75 | 1 | 0.03 |
The MICs of amphotericin B (AMB), itraconazole (ITC), voriconazole (VCZ), ravuconazole (RVC), posaconazole (POS), and terbinafine (TRB) are GMs. The MICs for the reference strain are ranges.
The MECs of caspofungin (CAS) and micafungin (MICA) are GMs. The CAS MEC for the reference strain is the range.
BAL, bronchoalveolar lavage fluid; BAS, bronchoalveolar aspirate; OPE, oropharyngeal exudate.
The fungi were grown at 37°C in potato dextrose agar (Oxoid, Madrid, Spain) or malt extract agar. The conidial stocks were preserved in sterile distilled water at 4°C (2).
Fungal morphology and growth conditions.
Fungal morphological features were examined by conventional methods (9). The differential temperature growth was determined by the presence or absence of growth at 37°C and 48°C for 3 days (4).
Antifungal susceptibility testing.
Broth microdilution susceptibility testing was performed as described in the CLSI (formerly NCCLS) reference method (29), with minor modifications. The modifications included the use of RPMI 1640 with l-glutamine buffered to pH 7 with 0.165 M morpholinepropanesulfonic acid and 10 M NaOH and supplemented with 2% glucose (Oxoid) and the use of an inoculum size of 1 × 105 to 5 × 105 CFU/ml (20, 31, 34).
The antifungal agents used in the study were amphotericin B (concentration range, 16 to 0.03 μg/ml; Sigma Aldrich Química), itraconazole (concentration range, 8 to 0.015 μg/ml; Janssen S.A., Madrid, Spain), voriconazole (concentration range, 8 to 0.015 μg/ml; Pfizer, S.A.), ravuconazole (concentration range, 8 to 0.015 μg/ml; Bristol-Myers, Squibb, Princeton, NJ), posaconazole (concentration range, 8 to 0.015 μg/ml; Schering-Plough Research Institute, Kenilworth, NJ), terbinafine (concentration range, 16 to 0.03 μg/ml; Novartis, Basel, Switzerland), caspofungin (concentration range, 16 to 0.03 μg/ml; Merck & Co, Inc., Rahway, NJ), and micafungin (concentration range, 16 to 0.03 μg/ml; Astellas Pharma Inc, Tokyo, Japan). Inoculum suspensions were prepared from fresh, mature (3- to 5-day-old) cultures by the use of a previously reported methodology (34). The plates were incubated at 35°C for 48 h in a humid atmosphere. Visual readings were performed with the help of a mirror. The endpoint for the MIC determination was the antifungal concentration that produced the complete inhibition of visual growth at 48 h. For the echinocandins, the minimal effective concentration (MEC) was used for endpoint determination. The MEC was defined as the minimal antifungal concentration that produced morphological alterations of hyphal growth at 48 h.
A. fumigatus ATCC 2004305 was used as the quality control strain to validate the MICs and MECs (reference values are shown in Table 1). Antifungal susceptibility testing was repeated at least three times on different days.
PCR amplification and sequencing.
Partial sequences of the β-tubulin and the rodlet A genes were amplified with primer set βtub3 (5′-TTCACCTTCAGACCGGT-3′) and βtub2 (4) and primer set RodA1 and RodA2, respectively (4).
All primers were synthesized by Sigma Genosys (Madrid, Spain). PCRs were carried out with a 50-μl volume containing 1× PCR buffer (Applied Biosystems, Madrid, Spain); 2 mM MgCl2 (Applied Biosystems); 250 μM each of dATP, dGTP, dCTP, and dTTP (Applied Biosystems); 1 μM of each primer; 2.5 U of Taq DNA polymerase (Applied Biosystems); and 25 to 50 ng of A. fumigatus genomic DNA. Amplification was performed in a thermal cycler (GeneAmp PCR system 9700; Applied Biosystems) for 1 cycle of 5 min at 94°C and then 30 cycles of 30 s at 94°C, 45 s at 55°C, and 1 to 2 min at 72°C, followed by 1 final cycle of 10 min at 72°C. The PCR products were analyzed by electrophoresis on 0.8% agarose gels and were visualized by transillumination (Gel Doc 2000; Bio-Rad Laboratories, Madrid, Spain) after they were stained with ethidium bromide (Sigma, Madrid, Spain).
The sequencing reactions were undertaken as described before (1) with primers βtub1 (4) and βtub4 (5′-AGCGTCCATGGTACCAGG-3′) for the β-tubulin gene and primers RodA3 (5′-AACGTCCGCTTCCCCGTTC-3′), RodA4 (5′-TACGGCATCGGAAGGAGAG-3′), and RodA5 (5′-TACGGCATCGGAGGGAGAG-3′) for the rodlet A gene.
Sequence analysis.
The sequences were assembled and edited with the SeqMan II and EditSeq software packages (Lasergene; DNAStar, Inc., Madison, WI). Sequence analysis was performed by comparing the DNA sequences with those of the control strains included in this study and with the sequences obtained from the GenBank database. Fourteen β-tubulin gene partial sequences and nine rodlet A gene partial sequences were used and are listed in Table 2.
TABLE 2.
GenBank sequences and accession numbers of the genes used in this study
| GenBank accession no. | Gene | Isolate |
|---|---|---|
| DQ094884 | β-Tubulin | A. fumigatiaffinis IBT 13131 |
| DQ094885 | β-Tubulin | A. fumigatiaffinis IBT 12703 |
| AB248076 | β-Tubulin | A. fumisynnematus IFM 42277 |
| AB249897 | Rodlet A | A. fumisynnematus IFM 42277 |
| AB248077 | β-Tubulin | A. fumisynnematus 90-BP-70 |
| AB249898 | Rodlet A | A. fumisynnematus 90-BP-70 |
| AB248078 | β-Tubulin | A. fumisynnematus 90-BP-177 |
| AB249899 | Rodlet A | A. fumisynnematus 90-BP-177 |
| AY738513 | β-Tubulin | A. lentulus FH 5 |
| AY738514 | Rodlet A | A. lentulus FH 5 |
| AY738517 | β-Tubulin | A. lentulus FH 4 |
| AY738519 | Rodlet A | A. lentulus FH 4 |
| AY738520 | β-Tubulin | A. lentulus FH 7 |
| AY738522 | Rodlet A | A. lentulus FH 7 |
| AY738523 | β-Tubulin | A. lentulus FH 220 |
| AY738525 | Rodlet A | A. lentulus FH 220 |
| DQ094886 | β-Tubulin | A. novofumigatus IBT 16806 |
| DQ094887 | β-Tubulin | A. novofumigatus IBT 16755 |
| AB248299 | β-Tubulin | A. viridinutans IFM 54303 |
| AY590130 | β-Tubulin | A. viridinutans MK284 |
| AB250103 | Rodlet A | A. viridinutans |
| AF057312 | β-Tubulin | A. clavatus H 522 |
| AF057322 | Rodlet A | A. clavatus H 522 |
Phylogenetic analysis.
All phylogenetic analyses were conducted with InfoQuest FP software (version 4.50; Bio-Rad Laboratories). The methodology used was maximum-parsimony clustering. Phylogram stability was assessed by parsimony bootstrapping with 2,000 simulations. The Aspergillus clavatus β-tubulin and rodlet A gene sequences were used as the outgroups (Table 2).
Nucleotide sequence accession numbers.
The following are the GenBank accession numbers for the β-tubulin and rodlet A gene fragment sequences from all the strains used in this work: for the β-tubulin gene fragments, CM-1290, EU310839; CM-2270, EU310840; CM-2280, EU310841; CM-3134, EU310842; CM-3147, EU310843; CM-3227, EU310844; CM-3248, EU310845; CM-3254, EU310846; CM-3258, EU310847; CM-3303, EU310848; CM-3305, EU310849; CM-3364, EU310850; CM-3537, EU310851; CM-3538, EU310852; CM-3583, EU310853; CM-3599, EU310854; CM-3652, EU310855; CM-3722, EU310856; CM-3740, EU310857; CM-3764, EU310858; CM-3769, EU310859; CM-3914, EU310860; CM-4060, EU310861; CM-4063, EU310862; CM-4328, EU310863; CM-4330, EU310864; CM-4370, EU310865; CM-4387, EU310866; CM-4415, EU310867; CM-4420, EU310868; CM-4426, EU310869; CM-4428, EU310870; and CM-4518, EU310871; for the rodlet A gene fragments, CM-1290, EU310806; CM-2270, EU310807; CM-2280, EU310808; CM-3134, EU310809; CM-3147, EU310810; CM-3227, EU310811; CM-3248, EU310812; CM-3254, EU310813; CM-3258, EU310814; CM-3303, EU310815; CM-3305, EU310816; CM-3364, EU310817; CM-3537, EU310818; CM-3538, EU310819; CM-3583, EU310820; CM-3599, EU310821; CM-3652, EU310822; CM-3722, EU310823; CM-3740, EU310824; CM-3764, EU310825; CM-3769, EU310826; CM-3914, EU310827; CM-4060, EU310828; CM-4063, EU310829; CM-4328, EU310830; CM-4330, EU310831; CM-4370, EU310832; CM-4387, EU310833; CM-4415, EU310834; CM-4420, EU310835; CM-4426, EU310836; CM-4428, EU310837; and CM-4518, EU310838.
RESULTS
Clinical isolate morphology and growth temperature.
The clinical origins of the 28 Aspergillus section Fumigati strains are shown in Table 1. These were isolated from 21 respiratory specimens, 1 ocular swab specimen, and 6 skin or nail samples.
All strains were first identified as atypical A. fumigatus strains by conventional macroscopic and microscopic morphological analysis methods. Identification to the Aspergillus genus level was straightforward; however, we were not able to discriminate between the different species of Aspergillus section Fumigati. While A. fumigatus grew at 37°C and 48°C, all the other strains analyzed in this study grew at 37°C but were not able to grow at 48°C.
Molecular identification of Aspergillus section Fumigati.
Partial DNA sequences of the β-tubulin and rodlet A genes were obtained and analyzed. Four CBS strains were amplified with the primers described above in order to compare the 28 atypical Aspergillus with members of the section Fumigati. We also included a set of β-tubulin and rodlet A sequences from members of Aspergillus section Fumigati available from GenBank (Table 2).
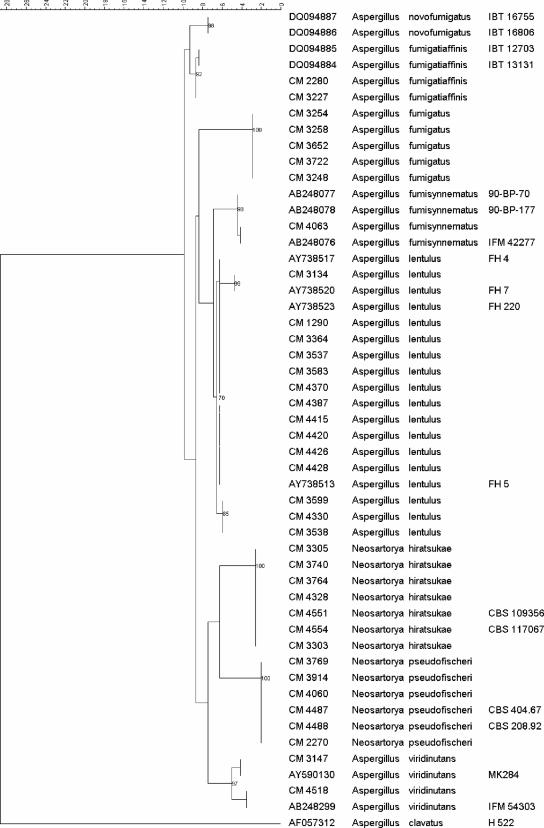
The phylogenetic tree produced by maximum parsimony of the β-tubulin sequences (Fig. 1) grouped the 28 clinical isolates into six different clades (N. hiratsukae, N. pseudofischeri, A. viridinutans, A. lentulus, A. fumigatiaffinis, and A. fumisynnematus).
FIG. 1.
Phylogenetic tree obtained by maximum-parsimony phylogenetic analysis with 2,000 bootstrap simulations on the basis of the β-tubulin sequences from all the strains included in the study.
On the basis of these results, 14 atypical A. fumigatus strains were identified as A. lentulus, supported by bootstrap values from 70% to 99%. Five isolates were identified as N. hiratsukae with a bootstrap value of 100%, four were identified as N. pseudofischeri (bootstrap value, 100%), two were identified as A. viridinutans (bootstrap value, 97%), two were identified as A. fumigatiaffinis (bootstrap value, 92%), and one was identified as A. fumisynnematus (bootstrap value, 98%). None of the Aspergillus section Fumigati sequences analyzed matched the A. fumigatus sequences.
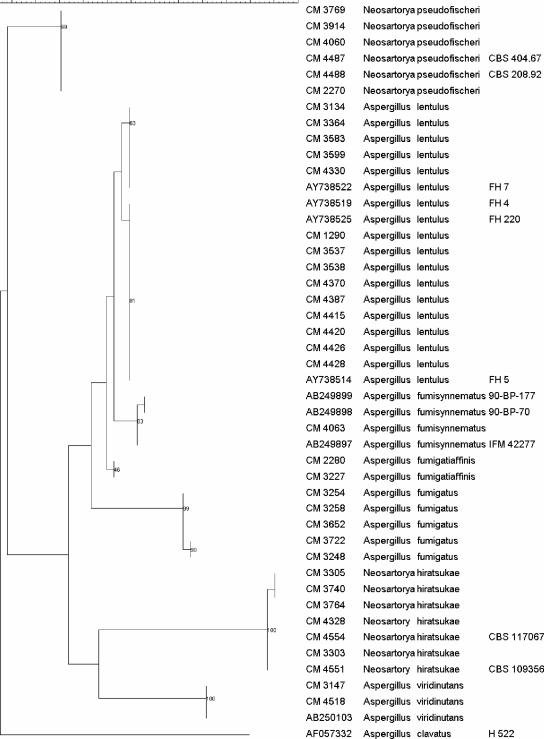
Figure 2 shows the results of a phylogenetic analysis obtained by maximum parsimony of the rodlet A gene sequences. According to those results, the 28 clinical strains fell into the same six clades. Fourteen samples were grouped with A. lentulus with bootstrap values of 63% and 81%. Five isolates were identified as N. hiratsukae with a bootstrap of 100%. Four isolates were identified as N. pseudofischeri, two were identified as A. viridinutans, two were identified as A. fumigatiaffinis, and one was identified as A. fumisynnematus, supported by bootstrap values of 99%, 100%, 46%, and 83%, respectively. All A. fumigatus sequences were divided into a single group according to their rodlet A sequences.
FIG. 2.
Phylogenetic tree obtained by maximum-parsimony phylogenetic analysis with 2,000 bootstrap simulations on the basis of the rodlet A sequences from all the strains included in the study.
The identities and classification results for the Aspergillus section Fumigati strains on the basis of their β-tubulin and rodlet A sequences are summarized in Table 1.
Antifungal susceptibility testing.
The MICs and MECs of the antifungal agents for the collection of clinical isolates are shown in Table 1 and are expressed as geometric means (GMs). The species were identified by β-tubulin and rodlet A gene sequencing. Analyses of the susceptibility phenotypes (Table 1) resulted in clear differences between Aspergillus species of the section Fumigati. We were able to differentiate three different antifungal phenotypes. Twelve of the 14 A. lentulus isolates and the two A. fumigatiaffinis isolates showed high MICs of amphotericin B (GM MICs, 4.5 μg/ml to 16 μg/ml), itraconazole (GM MICs, 6 μg/ml to 10.25 μg/ml), voriconazole (GM MICs, 3 μg/ml to 7.5 μg/ml), and ravuconazole (GM MICs, 1.5 μg/ml to 7 μg/ml). Although strains CNM-CM-3583 and CNM-CM-3599 did not follow this pattern for itraconazole (GM MICs, 0.43 μg/ml and 0.5 μg/ml, respectively), they retained the high MICs of amphotericin B (GM MICs, ≥2.66 μg/ml), voriconazole (GM MICs, ≥3.5 μg/ml), and ravuconazole (GM MICs, 2 μg/ml). A second profile was related to the species N. pseudofischeri and A. viridinutans. They showed high MICs of itraconazole (GM MICs, ≥8 μg/ml), voriconazole (GM MICs, ≥3.33 μg/ml), and ravuconazole (GM MICs, ≥2 μg/ml) but were susceptible to amphotericin B (GM MICs, ≤1 μg/ml). In contrast, strains identified as N. hiratsukae and A. fumisynnematus were more susceptible in vitro to all the antifungal compounds tested.
Among the azoles, posaconazole showed better activity in vitro (GM MICs, ≤0.75 μg/ml) against all clinical isolates analyzed in this study. Moreover, all strains were susceptible to terbinafine (GM MICs, ≤1.75 μg/ml) and the echinocandins, showing GM MECs of caspofungin and micafungin of ≤1.3 μg/ml and ≤0.26 μg/ml, respectively.
DISCUSSION
The present study highlights the limitations of phenotypic methods for the identification of some genera of molds. The use of molecular methods to partially sequence the β-tubulin and rodlet A genes enabled us to identify to the species level all clinical strains included in the study and previously classified as “atypical” A. fumigatus isolates. The majority of molecular methods use either specific probes or universal primers that are normally directed to conserved regions of the rRNA gene, particularly to the internal transcribed spacer regions (32, 35). However, internal transcribed spacer regions do not have enough phylogenetic strength to resolve the evolutionary relationship with strong bootstrap support for Aspergillus species from the section Fumigati (4, 5, 24, 41). Sequence analysis of both the β-tubulin and the rodlet A genes revealed that this method accurately differentiated the non-A. fumigatus isolates from the A. fumigatus isolates (4, 5). Therefore, all clinical isolates used in this study were identified to the species level by maximum-parsimony analysis of the β-tubulin and the rodlet A gene sequences.
On the basis of the identities of β-tubulin and rodlet A DNA sequences, the 28 strains were divided into six monophyletic clades supported by bootstrap values that differentiated the species (Fig. 1 and 2). The clades identified the isolates as belonging to the following species: 14 strains were A. lentulus, 5 strains were N. hiratsukae, 4 strains were N. pseudofischeri, 2 strains were A. viridinutans, 2 strains were A. fumigatiaffinis, and 1 strain was A. fumisynnematus.
A. lentulus has been isolated from soil and air, and it has also been isolated from patients with invasive infections (4). In this regard, most A. lentulus isolates recovered in this study were cultured from respiratory samples. This fact demonstrates the potential invasiveness of this pathogen in a susceptible host because A. fumigatus is able to colonize the human respiratory tract. A. viridinutans, N. hiratsukae, and N. pseudofischeri have also been isolated from humans (44), although only N. hiratsukae and N. pseudofischeri have been associated with human invasive fungal infections (3, 14, 17).
A. fumisynnematus was described as a separate taxon on the basis of the partial cytochrome b gene sequences of the species (43). However, this species was not included in the last classification of Aspergillus section Fumigati because the type strain of the species was not available for phenotypic characterization (36). To our knowledge, A. fumisynnematus and A. fumigatiaffinis had always been isolated from environmental sources. We describe for the first time the isolation of A. fumisynnematus and A. fumigatiaffinis strains from human hosts, which raises the intriguing issue of whether these species should be considered pathogenic fungi.
The common antifungal susceptibility phenotype of A. fumigatus is characterized by low MICs of the azole drugs, amphotericin B, and echinocandins. A. fumigatus strains showing resistance to azole drugs have been reported, although they have always maintained low amphotericin B MICs (11, 25, 26). In contrast, most A. lentulus and A. fumigatiaffinis isolates analyzed here showed extremely high MICs of amphotericin B, itraconazole, voriconazole, and ravuconazole.
Although 2 of the 14 A. lentulus isolates seemed to be susceptible to itraconazole, it is important to emphasize that we have not observed a uniform itraconazole susceptibility pattern, even after repeating the susceptibility testing more than eight times (data not shown). Due to the slow growth of most of these species, endpoint reading was easier at 72 h than at 48 h.
Since it has been demonstrated that elevated MICs of amphotericin B are associated with poor clinical outcomes (19, 30), the high MICs of amphotericin B for A. lentulus and A. fumigatiaffinis could have a remarkable clinical impact that merits research. In addition, it is well known that A. terreus strains have higher amphotericin B MICs than A. fumigatus strains, and this fact has been associated with a poorer response to amphotericin B in patients infected with this species (21, 37).
Even though A. fumisynnematus seems to be very closely related to A. lentulus, it had a different profile of susceptibility to all drugs tested. However, more isolates need to be analyzed in order to establish the antifungal susceptibility profile for this species.
Consistent with previous data (3, 44), the antifungal phenotypes of N. pseudofischeri and A. viridinutans showed that they had high itraconazole, voriconazole, and ravuconazole MICs.
All N. hiratsukae isolates were susceptible to all the antifungal drugs tested. There has been only one report of cerebral aspergillosis caused by N. hiratsukae (14), in which the strain cultured from that patient showed a pattern of MICs similar to the patterns described in this study.
Among the azoles, posaconazole showed the highest level of activity against all species analyzed. Terbinafine also had good activity in vitro against all clinical isolates compared with the MICs for A. fumigatus.
Finally, our antifungal susceptibility testing results for the echinocandins showed that all strains were susceptible to both caspofungin and micafungin (MECs, ≤1.3 μg/ml). This result contradicts the findings of other authors, but since a reference method for testing the susceptibilities of filamentous fungi to echinocandins has not yet been defined, conclusions about the echinocandin susceptibility or resistance of Aspergillus spp. should be made carefully.
This study emphasizes that molecular methods are needed for the correct identification of members of Aspergillus section Fumigati to the species level. Moreover, the members of this section have different antifungal susceptibility profiles. The identification of species or isolates showing high MICs of the antifungals used clinically is therefore mandatory. It should be noted that high in vitro MICs may not necessarily reflect decreased susceptibility in vivo, especially because these species have growth differences that could affect the results. Therefore, to ascertain the true clinical importance of these species, epidemiological studies must be performed together with in vitro-in vivo correlation studies. In the meantime, it is difficult to give practical advice for clinical laboratories, but we suggest that those isolates which appear to be A. fumigatus under the microscope but which have poor sporulation or slow growth be sent to reference laboratories. An alternative approach could be to send any A. fumigatus-like strain which does not grow at 48°C to a reference laboratory. Due to the shortage of data and the unpredictable susceptibility profiles of some isolates, we highly recommend that antifungal susceptibility testing be performed by a standardized methodology for all isolates associated with human infections.
Acknowledgments
This work was funded in part by grants MPY1175/06 from the Instituto de Salud Carlos III and SAF2005-06541 from the Ministerio de Educacion y Ciencia L. Alcazar-Fuoli held a postdoctoral contract from an EU-STREP project (LSHM-CT-2005-518199). A. Alastruey-Izquierdo held a predoctoral fellowship (grant FI05/00856) from the Fondo de Investigación Sanitaria (FIS, ISCIII).
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Alastruey-Izquierdo, A., M. Cuenca-Estrella, A. Monzon, and J. L. Rodriguez-Tudela. 2007. Prevalence and susceptibility testing of new species of Pseudallescheria and Scedosporium in a collection of clinical mold isolates. Antimicrob. Agents Chemother. 51:748-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcazar-Fuoli, L., E. Mellado, G. Garcia-Effron, M. J. Buitrago, J. F. Lopez, J. O. Grimalt, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2006. Aspergillus fumigatus C-5 sterol desaturases (Erg3A and Erg3B): role in sterol biosynthesis and antifungal drugs susceptibility. Antimicrob. Agents Chemother. 50:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee, S. A., J. Gribskov, M. Brandt, J. Ito, A. Fothergill, and K. A. Marr. 2005. Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. J. Clin. Microbiol. 43:5996-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balajee, S. A., J. L. Gribskov, E. Hanley, D. Nickle, and K. A. Marr. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balajee, S. A., D. Nickle, J. Varga, and K. A. Marr. 2006. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot. Cell 5:1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balajee, S. A., M. Weaver, A. Imhof, J. Gribskov, and K. A. Marr. 2004. Aspergillus fumigatus variant with decreased susceptibility to multiple antifungals. Antimicrob. Agents Chemother. 48:1197-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chryssanthou, E. 1997. In vitro susceptibility of respiratory isolates of Aspergillus species to itraconazole and amphotericin B. Acquired resistance to itraconazole. Scand. J. Infect. Dis. 29:509-512. [DOI] [PubMed] [Google Scholar]
- 8.Dannaoui, E., E. Borel, M. F. Monier, M. A. Piens, S. Picot, and F. Persat. 2001. Acquired itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 47:333-340. [DOI] [PubMed] [Google Scholar]
- 9.de Hoog, G. S., J. Guarro, C. S. Tan, R. G. F. Wintermans, and J. Gené. 1995. Hyphomycetes, p. 308-1007. In G. S. de Hoog and J. Guarro (ed.), Atlas of clinical fungi. Reus, Tarragona, Spain.
- 10.Denning, D. W. 2006. Aspergillus spp. and aspergillosis—progress on many fronts. Med. Mycol. 44(Suppl. 1):S1-S2. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber, J., J. Chomicki, J. W. Brandsberg, R. Jones, and K. J. Hammerman. 1973. Pulmonary aspergillosis caused by Aspergillus fischeri var. spinosus: report of a case and value of serologic studies. Am. J. Clin. Pathol. 60:861-866. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Lopez, A., G. Garcia-Effron, E. Mellado, A. Monzon, J. L. Rodriguez-Tudela, and M. Cuenca-Estrella. 2003. In vitro activities of three licensed antifungal agents against Spanish clinical isolates of Aspergillus spp. Antimicrob. Agents Chemother. 47:3085-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarro, J., E. G. Kallas, P. Godoy, A. Karenina, J. Gene, A. Stchigel, and A. L. Colombo. 2002. Cerebral aspergillosis caused by Neosartorya hiratsukae, Brazil. Emerg. Infect. Dis. 8:989-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedayati, M. T., A. C. Pasqualotto, P. A. Warn, P. Bowyer, and D. W. Denning. 2007. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153:1677-1692. [DOI] [PubMed] [Google Scholar]
- 16.Hong, S. B., H. S. Cho, H. D. Shin, J. C. Frisvad, and R. A. Samson. 2006. Novel Neosartorya species isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 56:477-486. [DOI] [PubMed] [Google Scholar]
- 17.Jarv, H., J. Lehtmaa, R. C. Summerbell, E. S. Hoekstra, R. A. Samson, and P. Naaber. 2004. Isolation of Neosartorya pseudofischeri from blood: first hint of pulmonary aspergillosis. J. Clin. Microbiol. 42:925-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21:161-172. [DOI] [PubMed] [Google Scholar]
- 19.Kontoyiannis, D. P., G. P. Bodey, H. Hanna, R. Hachem, M. Boktour, E. Girgaway, M. Mardani, and I. I. Raad. 2004. Outcome determinants of fusariosis in a tertiary care cancer center: the impact of neutrophil recovery. Leuk. Lymphoma 45:139-141. [DOI] [PubMed] [Google Scholar]
- 20.Lass-Florl, C., M. Cuenca-Estrella, D. W. Denning, and J. L. Rodriguez-Tudela. 2006. Antifungal susceptibility testing in Aspergillus spp. according to EUCAST methodology. Med. Mycol. 44(Suppl):319-325. [DOI] [PubMed] [Google Scholar]
- 21.Lass-Florl, C., K. Griff, A. Mayr, A. Petzer, G. Gastl, H. Bonatti, M. Freund, G. Kropshofer, M. P. Dierich, and D. Nachbaur. 2005. Epidemiology and outcome of infections due to Aspergillus terreus: 10-year single centre experience. Br. J. Haematol. 131:201-207. [DOI] [PubMed] [Google Scholar]
- 22.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda, H., S. Kohno, S. Maesaki, H. Yamada, H. Koga, M. Tamura, H. Kuraishi, and J. Sugiyama. 1992. Application of ubiquinone systems and electrophoretic comparison of enzymes to identification of clinical isolates of Aspergillus fumigatus and several other species of Aspergillus. J. Clin. Microbiol. 30:1999-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellado, E., L. Alcazar-Fuoli, G. Garcia-Effron, A. Alastruey-Izquierdo, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2006. New resistance mechanisms to azole drugs in Aspergillus fumigatus and emergence of antifungal drug-resistant A. fumigatus atypical strains. Med. Mycol. 44(Suppl.):367-371. [DOI] [PubMed] [Google Scholar]
- 25.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2004. Substitutions at methionine 220 in the 14α-sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48:2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, W. J. Melchers, P. E. Verweij, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell, C. G., J. Slight, and K. Donaldson. 1997. Diffusible component from the spore surface of the fungus Aspergillus fumigatus which inhibits the macrophage oxidative burst is distinct from gliotoxin and other hyphal toxins. Thorax 52:796-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosquera, J., and D. W. Denning. 2002. Azole cross-resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 46:556-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, approved standard. Document M38-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 30.Perfect, J. R., K. A. Marr, T. J. Walsh, R. N. Greenberg, B. Dupont, J. Torre-Cisneros, G. Just-Nubling, H. T. Schlamm, I. Lutsar, A. Espinel-Ingroff, and E. Johnson. 2003. Voriconazole treatment for less-common, emerging, or refractory fungal infections. Clin. Infect. Dis. 36:1122-1131. [DOI] [PubMed] [Google Scholar]
- 31.Petrikkou, E., J. L. Rodriguez-Tudela, M. Cuenca-Estrella, A. Gomez, A. Molleja, and E. Mellado. 2001. Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. J. Clin. Microbiol. 39:1345-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pryce, T. M., S. Palladino, I. D. Kay, and G. W. Coombs. 2003. Rapid identification of fungi by sequencing the ITS1 and ITS2 regions using an automated capillary electrophoresis system. Med. Mycol. 41:369-381. [DOI] [PubMed] [Google Scholar]
- 33.Rementeria, A., N. Lopez-Molina, A. Ludwig, A. B. Vivanco, J. Bikandi, J. Ponton, and J. Garaizar. 2005. Genes and molecules involved in Aspergillus fumigatus virulence. Rev. Iberoam. Micol. 22:1-23. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Tudela, J. L., E. Chryssanthou, E. Petrikkou, J. Mosquera, D. W. Denning, and M. Cuenca-Estrella. 2003. Interlaboratory evaluation of hematocytometer method of inoculum preparation for testing antifungal susceptibilities of filamentous fungi. J. Clin. Microbiol. 41:5236-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Tudela, J. L., T. M. Diaz-Guerra, E. Mellado, V. Cano, C. Tapia, A. Perkins, A. Gomez-Lopez, L. Rodero, and M. Cuenca-Estrella. 2005. Susceptibility patterns and molecular identification of Trichosporon species. Antimicrob. Agents Chemother. 49:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samson, R. A., S. B. Hong, and J. C. Frisvad. 2006. Old and new concepts of species differentiation in Aspergillus. Med. Mycol. 44(Suppl.):133-148. [DOI] [PubMed] [Google Scholar]
- 37.Steinbach, W. J., J. R. Perfect, W. A. Schell, T. J. Walsh, and D. K. Benjamin, Jr. 2004. In vitro analyses, animal models, and 60 clinical cases of invasive Aspergillus terreus infection. Antimicrob. Agents Chemother. 48:3217-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsitsigiannis, D. I., J. W. Bok, D. Andes, K. F. Nielsen, J. C. Frisvad, and N. P. Keller. 2005. Aspergillus cyclooxygenase-like enzymes are associated with prostaglandin production and virulence. Infect. Immun. 73:4548-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varga, J., E. Kevei, E. Rinyu, J. Teren, and Z. Kozakiewicz. 1996. Ochratoxin production by Aspergillus species. Appl. Environ. Microbiol. 62:4461-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varga, J., E. Rinyu, I. Kiss, B. Botos, and Z. Kozakiewicz. 1997. Carbon source utilization and isoenzyme analysis as taxonomic aids for toxigenic Neosartorya species and their relatives. Acta Microbiol. Immunol. Hung. 44:1-11. [PubMed] [Google Scholar]
- 41.Varga, J., Z. Vida, B. Toth, F. Debets, and Y. Horie. 2000. Phylogenetic analysis of newly described Neosartorya species. Antonie van Leeuwenhoek 77:235-239. [DOI] [PubMed] [Google Scholar]
- 42.Verweij, P. E., E. Mellado, and W. J. Melchers. 2007. Multiple-triazole-resistant aspergillosis. N. Engl. J. Med. 356:1481-1483. [DOI] [PubMed] [Google Scholar]
- 43.Wang, L., K. Yokoyama, M. Miyaji, and K. Nishimura. 2000. Mitochondrial cytochrome b gene analysis of Aspergillus fumigatus and related species. J. Clin. Microbiol. 38:1352-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaguchi, T., Y. Horie, R. Tanaka, T. Matsuzawa, J. Ito, and K. Nishimura. 2007. Molecular phylogenetics of multiple genes on Aspergillus section Fumigati isolated from clinical specimens in Japan. Nippon Ishinkin Gakkai Zasshi 48:37-46. [DOI] [PubMed] [Google Scholar]