Abstract
Norwalk virus (NV) is the prototype strain of a group of noncultivable human caliciviruses responsible for epidemic outbreaks of acute gastroenteritis. The capsid protein VP1 is synthesized from a subgenomic RNA that contains two open reading frames (ORFs), ORF2 and ORF3, and the 3′ untranslated region (UTR). ORF2 and ORF3 code for the capsid protein (VP1) and a small structural basic protein (VP2), respectively. We discovered that the yields of virus-like particles (VLPs) composed of VP1 are significantly reduced when this protein is expressed from ORF2 alone. To determine how the 3′ terminus of the NV subgenomic RNA regulates VP1 expression, we compared VP1 expression levels by using recombinant baculovirus constructs containing different 3′ elements. High VP1 levels were detected by using a recombinant baculovirus that contained ORF2, ORF3, and the 3′UTR (ORF2+3+3′UTR). In contrast, expression of VP1 from constructs that lacked the 3′UTR (ORF2+3), ORF3 (ORF2+3′UTR), or both (ORF2 alone) was highly reduced. Elimination of VP2 synthesis from the subgenomic RNA by mutation resulted in VP1 levels similar to those obtained with the ORF2 construct alone, suggesting a cis role for VP2 in upregulation of VP1 expression levels. Comparisons of the kinetics of RNA and capsid protein expression levels by using constructs with or without ORF3 or the 3′UTR revealed that the 3′UTR increased the levels of VP1 RNA, whereas the presence of VP2 resulted in increased levels of VP1. Furthermore, VP2 increased VP1 stability and protected VP1 from disassembly and protease degradation. The increase in VP1 expression levels caused by the presence of VP2 in cis was also observed in mammalian cells.
The Norwalk virus (NV) is a member of the genus Norovirus in the family Caliciviridae. The recent accessibility of molecular diagnostic assays has demonstrated that noroviruses are the leading cause of outbreaks of nonbacterial gastroenteritis worldwide (17). In addition, caliciviruses have been recently classified as category B biodefense pathogens (http://www.niaid.nih.gov/biodefense/). The NV 7.7-kb positive-strand RNA genome is organized into a 5′ untranslated region (UTR), three open reading frames (ORFs), a 3′UTR, and a poly(A) tail (19). The NV predicted subgenomic RNA contains ORF2, ORF3, and the 3′UTR. ORF2 and ORF3 code for the major capsid protein (VP1) and a small basic structural protein (VP2), respectively. The ORF3 protein (VP2) has been detected in NV virions and virus-like particles (VLPs) (15). VP2 or a VP2-equivalent protein has been detected in the virions of other members of the Caliciviridae family such as the rabbit hemorrhagic disease virus (RHDV) (34) and feline calicivirus (FCV) (54). VP2 is not essential for the formation of NV VLPs because baculovirus recombinants expressing VP1 alone are able to produce NV VLPs (5, 58). The highly basic VP2 may serve a function similar to the basic N terminus of plant virus capsid proteins, which are involved in RNA binding, since the N terminus of NV VP1 is acidic and is not thought to interact directly with the viral genome (15, 27, 49).
Few studies on the molecular biology of human caliciviruses have been performed, in part because of the lack of a replication system. Studies with cultivable animal caliciviruses (FCV, RHDV, vesicular exanthema swine virus, and San Miguel sea lion virus) have found two different RNA molecules, 7.5 and 2.3 kb each, in infected cells (6, 13, 39). In the case of FCV, infected cells produce a polyadenylated genomic RNA and a bicistronic subgenomic RNA encoding ORF2 and ORF3 (39). The expression level of VP2 from the bicistronic NV RNA containing ORF2 and ORF3 is very low (15). Proteins, like VP2, made in small amounts may have regulatory functions if very little protein is needed to perform their function efficiently. Mechanisms of termination-reinitiation or internal initiation have been suggested for the synthesis of FCV VP2 based on in vitro studies in rabbit reticulocyte lysates (28).
Positive-strand viral RNA genomes frequently contain cis-acting elements necessary for gene regulation via interaction with viral or cellular proteins (29, 35, 43, 53, 56). Thus far, the role of cis-acting elements such as the poly(A) tail or the 3′UTR in NV gene expression or replication remains unclear due to the lack of an infectious clone or a tissue culture system for this virus. However, the baculovirus expression system has been a useful model for studying NV biology, including virion assembly and structure (5, 25, 31, 49, 57). During the course of studies in NV assembly, we noticed that the expression of VP1 in insect cells was enhanced by the presence of the NV 3′UTR and ORF3 in baculovirus recombinants. We hypothesized that these sequences at the 3′ end of the NV genome contain cis-acting regulatory elements required for gene expression or translational regulation. To address this hypothesis, we compared the expression levels of VP1 from different baculovirus recombinants that contained or lacked the ORF3 gene or the 3′UTR. We present data here showing that regulatory elements downstream of the ORF2 gene are involved in the control of ORF2 gene expression and that the 3′UTR increases the level of ORF2 RNA, whereas VP2 increases VP1 expression and stabilizes the capsid. We propose and provide evidence supporting a model for a mechanism by which VP2 regulates the stability of VP1.
MATERIALS AND METHODS
Cells.
Spodoptera frugiperda 9 (Sf9) cells, obtained from the American Type Culture Collection (Rockville, Md.) were grown in a 1.5-liter bioreactor unit (Celligen Cell Culture System; New Brunswick Scientific, Edison, N.J.) and maintained on TNM-FH medium (Gibco-BRL/Life Technologies, Grand Island, N.Y.) containing 10% fetal bovine serum (FBS; Summit Biotechnology, Fort Collins, Colo.). Sf9 suspension cell cultures were grown in 2-liter spinner culture flasks (Corning Glass Works, Corning, N.Y.) at 27°C with constant stirring.
Human embryonic kidney (HEK) 293T cells immortalized with adenovirus type 5 (Ad5; strain F2853-5b) and transformed with simian virus 40 (SV40) T antigen were cultured in Dulbecco modified Eagle minimum essential medium (DMEM; Gibco-BRL) with high glucose (4,500 mg/ml), supplemented with 10% FBS, 0.2 mM nonessential amino acids (Sigma Chemical Co., St. Louis, Mo.), 10 mM HEPES (Gibco-BRL), 1 mM sodium pyruvate (Gibco-BRL), 100 U of penicillin (Gibco-BRL)/ml, and 100 μg of streptomycin (Gibco-BRL)/ml.
Cloning and construction of baculovirus recombinants and GFP-NV fusion constructs.
Ten baculovirus recombinants and two green fluorescent protein (GFP)-VP1 fusion contructs were produced to monitor the expression levels of the NV VP1 in Sf9 insect cells and HEK 293T mammalian cells, respectively. The NV baculovirus recombinants, luciferase baculovirus recombinants, and GFP-VP1 fusion protein constructs produced in the present study, along with the primers used in their construction, are listed in Table 1.
TABLE 1.
Names, sequences, polarities, and restriction enzyme sites of primers used in this study to generate all baculovirus recombinants for expression in insect cells and the GFP constructs for expression in mammalian cells
| Name | Sequence (5′ to 3′)a | Sequence locationa | Sense | Restriction enzyme site | Constructs generated |
|---|---|---|---|---|---|
| NV132 | GGCCTGATCAATGATGATGGCGTCTAAGGAC | 5357-5385 | + | BclI | ORF2, ORF2+3, ORF2+3+3′UTR, ORF2+3′UTR |
| NV-3′UTR-Xho | CCCGCTCGAGAACATCAAATTAAACCTAAT | 7624-7637 | − | XhoI | ORF2+3+3′UTR, ORF3+3′UTR |
| NV133 | CCCGCTCGAGCATCGCCTATTATTTGCGAAT | 7687-7666 | − | XhoI | ORF2+3 |
| Luc-5′ | CCCGCTGGATCCATGACTTCGAAAGTTTATGATCCA | 1-24 | + | BamHI | Luc-Fast, Luc-3′UTR-Fast |
| Luc-3′ | CCCGCTCGAGGAATTATTGTTCATTTTTGAGAACTC | 745-775 | − | XhoI | Luc-Fast |
| NV131 | CCCGCTCGAGTTATCGGCGCAGACCAAGCCT | 6928-6950 | − | XhoI | ORF2 |
| ORF2+3′UTR | CCCGCTCGAGAACATCAAATTAAACCTAATTAAACCTAA TTAAATGAATATGATGCCCACATTTCATATTACAACATT ATCGGCGCAGACCAAGCCT | 6929-6952/7611-7654 | − | XhoI | ORF2+3′UTR |
| Luc-3′UTR NV | CCCGCTCGAGAACATCAAATTAAACCTAATTAAACCTAA TTAAATGAATATGATGCCCACATTTCATATTACAACAGA ATTATTGTTCATTTTTGAGAACTCGCT | 7611-7654/745-775 | − | XhoI | Luc-3′UTR-Fast |
| AUG-ORF3-5 | GGCTTGGTCTGCGCCGATAAGGATCCATGGCCCAAGCC ATAATTG | 6929-6970 | + | None | Site-directed mutagenesis-AUG-ORF3 |
| AUG-ORF3-3 | GCACCAATTATGGCTTGGGCCGTTATCGGCGCAGACCA AGCC | 6929-6973 | − | None | Site-directed mutagenesis-AUG-ORF3 |
| GFP-NV-5 | GGGCCTCGAGGCGCGATGATGATGGCGTCTAAGGAC | 5357-5385 | + | XhoI | GFP-ORF2+3+3′UTR NV |
| GFP-NV-3 | CCCGGGAACATCAAATTAAACCTAATT | 7637-7654 | − | XmaI | GFP-ORF2+3+3′UTR NV |
| ORF3-5-BamHI | GCGGGGGGATCCATGATGATGGCCCAAGCCATAA TTGGT | 6950-6973 | + | BamHI | ORF3+3′UTR |
Sequence locations for each primer on Norwalk sequence (gi9630803) and Renilla luciferase (shown in italics [gi28864243]).
(i) NV baculovirus recombinants.
Constructs containing different regions of the NV subgenomic RNA (Fig. 1A) were generated by cloning into the pFastBac1 expression vector (Gibco-BRL) by PCR amplification with a clone containing the entire NV subgenomic RNA(pVLNV-2-3 [30]) as a template. For the present study, we have retained the names for the genes in the constructs as ORF2 and ORF3, and we refer to their protein products as VP1 and VP2, respectively. Recombinant baculovirus transfer plasmids containing the entire or truncated NV subgenomic RNAs were generated by inserting PCR-amplified fragments flanked by BclI and XhoI restriction enzyme sites into BamHI and XhoI restriction enzyme sites in the multiple cloning site of the pFastBac1 transfer vector as described elsewhere (5). The dye termination method was used to sequence the clones. The recombinant baculoviruses were generated by using the Bac-to-Bac baculovirus expression system (Gibco-BRL) according to the manufacturer's protocol. Recombinant baculoviruses were plaque purified twice prior to infection of Sf9 cells, and two clones for each recombinant baculovirus were confirmed to contain the specific NV DNA sequences by PCR with the same primers used for the cloning procedure. For each construct, four recombinants were plaque purified. VP1 expression levels were equivalent for each of the recombinants, and only one of the recombinant baculoviruses was used for further studies.
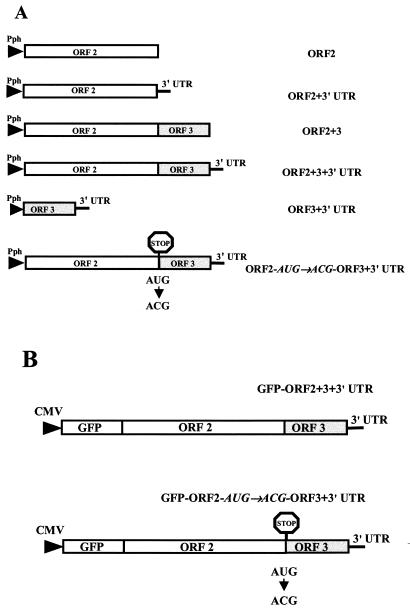
FIG. 1.
Schematic representation of recombinant baculoviruses and GFP contructs generated in the present study to monitor the expression levels of NV VP1 in insect cells and mammalian cells. (A) Baculovirus recombinants to analyze the expression of VP1 in Sf9 insect cells were generated by cloning into the pFastBac1 expression vector (Gibco-BRL). Since the same restriction enzyme site (BamHI) was used to generate all baculovirus recombinants, all seven constructs possess an identical 5′-terminal sequence. In addition, all of these constructs encode the poly(A) signal from SV40. These recombinant baculoviruses were used to study the effect of the different 3′-terminal elements of the NV subgenomic RNA on NV VP1 capsid protein expression. The stop signal indicates the site targeted by directed mutagenesis to block VP2 expression. (B) Constructs generated to analyze the effect of ORF3 or VP2 expression on VP1 expression in mammalian cells. The entire NV subgenomic RNA was placed, in frame, downstream of the GFP gene in the pEGFP-C1 vector (Clontech), and VP1 was expressed as a fusion protein with the GFP protein. The stop signal indicates a point mutation generated to block the expression of VP2. Abbreviations: Pph, polyhedron promoter; UTR, untranslated region; ORF, open reading frame.
(ii) Luciferase baculovirus recombinants.
Recombinant baculovirus transfer vectors containing the Renilla luciferase gene alone (Luc-Fast) and the luciferase gene with the NV 3′UTR (Luc-3′UTR-Fast) were generated by inserting PCR-amplified fragments flanked by BamHI and XhoI into the multiple cloning site of the pFastBac1 transfer vector. The template used for PCR was plasmid pRL-CMV (Promega, Madison, Wis.) that contains the Renilla luciferase gene. Three recombinant baculoviruses containing the different luciferase constructs were produced by using the Bac-to-Bac baculovirus expression system according to the manufacturer's protocol. After plaque purification, stocks of the luciferase containing recombinant baculoviruses were prepared and titered. Two clones for each recombinant baculovirus were confirmed to contain the specific luciferase-NV DNA sequences by PCR with the same primers used for the cloning procedure.
(iii) GFP-VP1 fusion protein constructs.
To analyze whether the regulation of the level of VP1 by the VP2 could be reproduced in mammalian cells, the NV subgenomic RNA (ORF2+3+3′UTR) was placed, in frame, downstream of GFP in the pEGFP-C1 vector (Clontech, Palo Alto, Calif.) (Fig. 1B). The GFP-VP1 fusion construct, GFP-ORF2+3+3′UTR, contains a linker of three amino acids (Arg-Gly-Ala) between the GFP and the ORF2 gene to allow efficient expression and folding of both proteins.
Site-directed mutagenesis.
The generation of a point mutation to change the start codon of the ORF3 gene from AUG to ACG (Met→Thr) was performed by using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's protocol. The template used for the mutagenesis was the pFastBac1 clone that contains the entire NV subgenomic RNA (ORF2+3+3′UTR), and the primers used to generate the mutations in the NV baculovirus recombinants (ORF2-AUG→ACG-ORF3+3′UTR) (Fig. 1A) and in the GFP-VP1 fusion protein construct (GFP-ORF2-AUG→ACG-ORF3+3′UTR) (Fig. 1B) are listed in Table 1. All mutations were confirmed by sequencing by using the dye termination method.
Expression and analysis of recombinant NV (rNV) in insect cells.
Expression of VP1 was evaluated in cell lysates and the medium of Sf9 cultures. The Sf9 cells, grown in monolayers as described previously (10), were infected at a multiplicity of infection (MOI) of 10 with the appropriate recombinant baculovirus, and the cells were collected 36 h postinfection (hpi). The expression of the recombinant VP1 capsid protein in the medium was analyzed by infection of a 200-ml insect cell culture at a density of 3 × 106 cells/ml (6 × 108 total cells per spinner flask) with an MOI of 5. The supernatant was collected at 5 days postinfection, and particles were purified as described previously (5). The amount of VLP protein obtained was quantitated by BCA protein assay reagent (Pierce, Rockford, Ill.) according to the manufacturer's instructions. To quantitate the VP1 produced by the different constructs, an antigen enzyme-linked immunosorbent assay (ELISA) specific for NV capsid protein was performed (5, 31). In addition, VP1 expression was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli (36) with modifications as described previously (5). The proteins separated by SDS-PAGE were transferred onto nitrocellulose membranes (Hybond-C; Amersham Life Sciences, Little Chalfont, England) as described previously (5). VP1 was detected by using a rabbit-hyperimmune anti-rNV serum.
RNA purification and Northern blot.
Total RNA was extracted by using guanidine isothiocyanate (37). Briefly, insect cells (3 × 106), infected at an MOI of 10 with the different recombinant baculoviruses expressing NV genes, were harvested at 36 hpi and the cells were washed with phosphate-buffered saline (PBS) and collected in 3 ml of 4 M guanidine isothiocyanate solution (GSCN solution; 4 M guanidine isothiocyanate, 1 M Tris, 0.5 M EDTA, 0.5% sarcosyl, 1% β-mercaptoethanol, 30% antifoam A [pH 7.5]). The cells were homogenized immediately for 1 min in a Cole-Parmer PCR homogenizer (Cole-Parmer Instrument Company, Vernon Hills, Ill.). Then, 3-ml portions of homogenized cells were layered over 2 ml of a 5.7 M CsCl cushion, followed by centrifugation at 36,000 rpm for 20 h at 25°C in an SW55 Beckman rotor. The supernatant was removed by aspiration, the tubes were drained, and the bottoms of the tubes were cut off to avoid contamination of the RNA pellet with residual DNA and proteins from the tube wall. The RNA pellets were suspended in water and then precipitated with ethanol. After the pellets were washed with 70% ethanol to eliminate residual salts, they were suspended in water, and the RNA concentration in the samples was determined spectrophotometrically (DU Series 60 spectrophotometer; Beckman, Fullerton, Calif.) at 260 nm.
The same amounts (5 μg) of total RNA for each recombinant were loaded onto a denaturing agarose gel, transferred to a Nytran membrane (Schleicher & Schuell, Kenne, N.H.) by downward capillary transfer, and analyzed by Northern blotting as described previously (38). Briefly, a radiolabeled [32P]ORF2 antisense RNA probe was generated by in vitro transcription of the previously constructed plasmid pGEM-ORF2 by using the Maxiscript transcription in vitro system (Ambion, Inc., Austin, Tex.) (57). Hybridization with the [32P]ORF2 antisense probe proceeded overnight at 65°C in PerfectHyb hybridization buffer (Sigma, St. Louis, Mo.). After hybridization, the membrane was washed once in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% (wt/vol) SDS for 20 min at room temperature and then three times in 0.2× SSC-0.1% (wt/vol) SDS for 20 min at 65°C. The membrane was then air dried, wrapped in cellophane, and exposed to BIOMax film (Kodak, Rochester, N.Y.).
Phosphorimaging analysis.
To measure the levels of ORF2 RNA detected by Northern blot analysis and of VP1 detected by immunoprecipitation, the bands from each sample were quantitated by phosphorimaging. This phosphorimaging was accomplished by using the Molecular Dynamics Storm system (Sunnyvale, Calif.), and ImageQuant NT software (also from Molecular Dynamics) was used for phosphorimaging data analysis.
Densitometric analysis of GFP-VP1 expression in mammalian cells.
The GFP expression intensity for the wild-type NV subgenomic RNA (GFP-ORF2+3+3′UTR) and the mutant that did not express the ORF3 protein (GFP-ORF2-AUG→ACG-ORF3+3′UTR) was determined in HEK 293T cells by using both densitometry and flow cytometry analysis. Mammalian HEK 293T cells were transfected as 80% confluent monolayers grown on 24-well plates (Corning Costar Corp.) by using Lipofectamine 2000 (Gibco-BRL) according to the manufacturer's instructions. In addition to the NV constructs, a reporter vector, pGL2 (Promega), containing the firefly luciferase gene under the SV40 promoter was transfected as an internal control for determining transfection efficiency. The transfected cells were incubated for 20 to 24 h. Before the cells were analyzed for protein expression, the presence of GFP was verified by using an Olympus IX70 fluorescent microscope (Olympus America, Inc., Melville, N.Y.). The luciferase values were quantitated by using the luciferase assay system (Promega). Each well of the transfected cells was suspended in the same volume of reporter lysis buffer (Promega), and equal volumes for each construct were analyzed by Western blotting by using a rabbit hyperimmune serum against the NV VP1 capsid protein. The bands obtained in Western blots that corresponded to VP1 were analyzed by densitometry using a Molecular Dynamics personal densitometer SI, and the data were analyzed with the ImageQuant NT software. The obtained densitometric quantitative data were normalized by using the luciferase activity value for each sample, and then the relative amounts of VP1 were calculated.
Flow cytometric analysis of GFP-VP1 expression.
In addition to the densitometric analysis, HEK 293T cells transfected with the plasmid expressing the GFP-ORF2+3+3′UTR or the GFP-ORF2-AUG→ACG-ORF3+3′UTR were analyzed for the levels of GFP by flow cytometry. Control HEK 293T cells were transfected with the pGL2 control reporter vector containing a firefly luciferase gene under the SV40 promoter (Promega). HEK 293T cells were first washed with 0.01 M PBS (pH 7.0) and then incubated in 200 μl of 2 mM EDTA in 0.01 M PBS (pH 7.0) for 15 min at room temperature to gently detach the cells from the plate. The cells were sedimented at 2,000 rpm for 5 min (IEC Micromax [Needham Heights, Mass.] microcentrifuge) and then suspended in 3% FBS in 0.01 M PBS. Cell GFP intensity was measured by using a Profile I flow cytometer (Coulter Electronics, Hialeah, Fla.). At least 10,000 forward-scatter gated events were collected per specimen.
Pulse-chase labeling.
Pulse-chase experiments to test the protein stability of the VP1 capsid protein were performed according to the protocol described for recombinant proteins expressed in baculovirus (5, 44, 45).
(i) Insect cells.
Briefly, 1.2 × 107 Sf9 cells growing in suspension were infected in a 15-ml tube at an MOI of 20. At 36 hpi, the medium was removed, and then the cells were suspended and cultivated in methionine-free Grace's medium (Gibco-BRL) for 1 h before [35S]methionine (40 μCi/ml) was added to the mix. After 15 min, the medium was replaced with fresh methionine-free Grace's medium with a 20-fold excess of unlabeled methionine (Gibco-BRL). Aliquots (1 ml), equivalent to 3 × 106 cells, were collected every 12 h postlabeling for 48 h, lysed by radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Triton X-100, 10 mM Tris-HCl [pH 7.2], 1% Trasylol), and frozen at −20°C until analyzed by immunoprecipitation as previously described (5) with a rabbit hyperimmune serum against the NV VP1 capsid protein.
(ii) Mammalian cells.
At 24 h posttransfection, HEK 293T cells were incubated in DMEM without l-methionine or l-cysteine (Gibco-BRL) for 1 h. Then, 40 μCi of [35S]methionine/ml was added to the medium. After 30 min, the medium was replaced with fresh DMEM supplemented with unlabeled methionine (Gibco-BRL). The cultures were harvested at 0, 6, 12, and 24 h postlabeling by lysing the cells with RIPA buffer. The lysed cells were stored at −20°C until analyzed by immunoprecipitation as described before (5).
Protease cleavage analysis.
To compare the sensitivity of VP1 to protease degradation when the protein was in the soluble form or in purified NV VLPs (with VP1 only or with VP1+VP2), samples were diluted in 50 mM Tris-HCl (pH 6.7) to maintain VLP integrity or in 50 mM Tris-HCl (pH 8.9) to promote particle disassembly before treatment with 200 μg of trypsin (Sigma)/ml or 5 μg of pancreatin 4× USP (Gibco-BRL)/ml for 2 h at 37°C in PBS. The proteins were separated by SDS-12% PAGE and analyzed by Western blotting as described above. The particles used in this analysis were produced by using the previously described constructs pVL-NV (VP1+VP2) (31) and pVL-ORF2 (VP1 alone) (57).
DLS.
Purified NV VLPs containing only VP1 (VP1 VLPs) or VLPs containing both VP1 and VP2 (VP1/VP2 VLPs) were diluted to 0.5 μg/μl in Tris-HCl (pH 6.7) to maintain VLP integrity. The particles were analyzed by dynamic light scattering (DLS) by using a DynaPro-801 (Protein Solutions, Charlottesville, Va.) with a miniature solid state laser (wavelength, λ = 829.5 nm) at 26°C. The time-average scattered intensity, [I(q)]T, and its time correlation were obtained at a scattering angle of 90°. Prior to DLS analysis, VLP samples were filtered through a 0.22-μm-pore-size polyvinylidene difluoride membrane (Millipore Corp., Bedford, Mass.). For each sample, 25 readings were recorded. In the present study, we determined the hydrodynamic radius, RH, of the rNV capsid protein from the equation: RH = κT/6πηTmax, where κT denotes the Boltzmann energy, η is the solvent viscosity, and Tmax is the value at the point where the intensity correlation function was maximal.
Statistics.
Statistical analyses were performed by using the SPSS version 7.5 for Windows (SPSS, Inc., Chicago, Ill.). Differences in protein expression were compared by using the Kruskal-Wallis test, followed by the Mann-Whitney U test or by use of Pearson correlation coefficients. Comparisons with a P value of <0.05 were considered statistically significant.
RESULTS
The level of NV VP1 capsid protein expressed in Sf9 insect cells depends on the 3′ terminus of the NV genome.
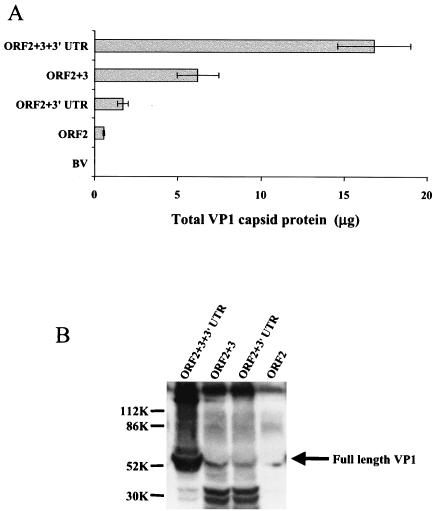
While making baculovirus constructs in the pFastBac1 transfer vector to rapidly produce and analyze VP1 mutants (5), we noted that inclusion of the ORF3 gene and the 3′UTR in the baculovirus recombinant construct resulted in higher levels of expression of the VP1 capsid protein. To confirm and elucidate the role of the ORF3 gene and the 3′UTR on VP1 expression, we compared VP1 expression from recombinant baculoviruses that contained only NV ORF2, NV ORF2+3, NV ORF2+3′UTR, and the entire NV subgenomic RNA (ORF2+3+3′UTR). The total amount of VP1 in supernatants or cell lysates was measured by ELISA after infection of the same number of Sf9 cells (3 × 106 cells/ml) (Fig. 2). The VP1 levels in cell lysates correlated with the levels of protein in supernatants. The level of VP1 expressed in cell lysates with the ORF2 alone construct was relatively low (0.5 μg; Fig. 2A). The inclusion of the NV 3′UTR or ORF3 gene (ORF2+3′UTR construct or ORF2+3 construct, respectively) downstream of ORF2 significantly increased (P ≤ 0.023) the expression of VP1 by 3- and 11-fold, respectively; however, a dramatic increase of >30-fold (P < 0.001) was observed in the expression level of VP1 from the construct that contained the entire subgenomic RNA (ORF2+3+3′UTR; Fig. 2A). No recombinant VP1 was detected in cells infected with a control wild-type baculovirus.
FIG. 2.
VP1 expression levels obtained with the baculovirus recombinants containing NV ORF2 only, NV ORF2+3, NV ORF2+3′UTR, or the entire NV subgenomic RNA (ORF2+3+3′UTR). (A) VP1 yield from 3 × 106 Sf9 insect cells infected with each recombinant baculovirus measured at 36 hpi by ELISA from the cell lysate. Purified rNV VLPs were used as the positive antigen control at concentrations ranging from 0.5 to 62 ng. The values shown are the arithmetic mean of at least four independent experiments. Error bars represent one standard error of the mean. (B) Expression of VP1 analyzed by Western blotting with a rabbit hyperimmune serum specific to VP1. Infected cell lysates (3 × 106 cells) were harvested at 36 hpi in SDS-PAGE sample buffer and analyzed by SDS-12% PAGE and Western blotting. To compare the relative levels of VP1 expression of all recombinants, the same volume of lysate was loaded for each recombinant baculovirus. The arrow indicates the bands that correspond to the uncleaved VP1. Molecular weight markers are indicated on the left. Two approximately 30K cleavage products were present in all of the samples but in larger amounts in the lysates from cells infected with the ORF2+3 and ORF2+3′UTR recombinant baculoviruses. The Western blotting and the ELISA analysis were performed with the same antibody; therefore, these 30K VP1-related degradation products were detected in the total protein quantification done by ELISA. BV, wild-type baculovirus.
The same samples described above were analyzed by Western blot by loading the same amounts of cell lysate for each construct onto 12% polyacrylamide gels. When samples were analyzed by Western blot with a rabbit hyperimmune serum to NV VLPs, the relative levels of NV VP1 capsid protein obtained for each recombinant baculovirus concurred with that obtained by ELISA (Fig. 2B). In addition, the yield of NV VLPs purified from the supernatant of cells (6 × 108) was higher for the ORF2+3+3′UTR recombinant (20 mg), whereas the yield of VLPs was lower from the recombinants that lacked the 3′UTR (ORF2+3; 10 mg) or ORF3 (ORF2+3′UTR; 2 mg) (data not shown). The lowest yield (0.6 mg) of VLPs was obtained from the infection with the recombinant that lacked both ORF3 and the 3′UTR. Therefore, both the 3′UTR and the ORF3 and/or VP2 are required for efficient expression of VP1.
The synthesis of VP2 is required for the increased yields of VP1.
To analyze whether the VP2 protein or the ORF3 RNA was involved in the regulation of VP1 expression, a mutant, ORF2-AUG→ACG-ORF3+3′UTR, was generated by site-directed mutagenesis in the backbone of the construct that contained the entire subgenomic RNA (Fig. 1A). The ORF2-AUG→ACG-ORF3+3′UTR recombinant mutant baculovirus was designed to replace the ORF3 gene start (AUG) codon with a threonine codon (ACG). The ORF2-AUG→ACG-ORF3+3′UTR recombinant baculovirus did not express VP2 or any VP2-related protein, as determined by Western blotting with two different peptide sera (15, 16) (data not shown), but it expressed VP1 and contained the entire subgenomic RNA. The levels of VP1 expression were analyzed by ELISA and compared to the wild-type levels by using the same lysate volume from cultures with an equal number (3 × 106) of infected Sf9 cells. The VP1 level from the ORF2-AUG→ACG-ORF3+3′UTR recombinant mutant baculovirus was significantly reduced (12-fold less, P ≤ 0.002) compared to the VP1 produced by using the ORF2+3+3′UTR recombinant baculovirus (see Fig. 3B). The data from the ORF2-AUG→ACG-ORF3+3′UTR mutant suggested VP2 synthesis is required for the efficient expression of VP1.
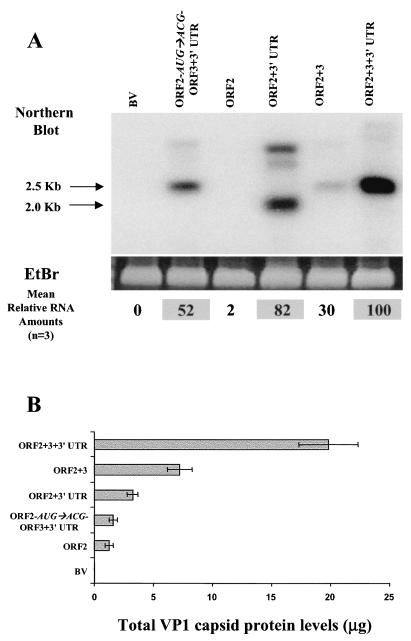
FIG. 3.
Comparison of steady-state levels of ORF2 mRNA and VP1 expressed from different constructs. (A) The steady-state levels of NV ORF2 mRNA were assessed at 36 hpi in 3 × 106 cells infected with the different recombinant baculoviruses by Northern blotting with a 32P-labeled probe specific for the NV ORF2 gene. Ethidium bromide (EtBr) staining of the total amount of RNA run was used as a loading control. The ethidium bromide-stained bands shown correspond to the rRNA. The bands that corresponded to the expected molecular sizes, 2.0 kb for ORF2 alone and 2.5 kb for the entire subgenomic RNA, were quantitated by phosphorimaging. The values shown at the bottom of each lane of the gel are the average of three independent experiments and represent the relative amounts of RNA with respect to the amount obtained with the entire subgenomic RNA construct (ORF2+3+3′UTR). The highest values are shaded. The band corresponding to the RNA for the ORF2-alone construct was not high enough to be seen on this exposure of the gel, but it was detected by the phosphorimager. (B) The level (μg) of VP1 from the same samples was determined by analysis of duplicate samples by ELISA. Purified rNV VLPs were used as the positive antigen control at concentrations ranging from 0.5 to 62 ng. Each bar shows the arithmetic means of three independent experiments. Error bars represent one standard error of the mean.
Is the effect of VP2 and 3′UTR at the protein or RNA level?
Both the 3′UTR and VP2 expression affected the levels of VP1 expression. To determine the mechanism by which expression of VP1 was regulated by VP2 or the 3′UTR, ORF2 RNA and protein levels in Sf9 insect cells (3 × 106) infected with the different recombinant baculoviruses were analyzed in parallel. The ORF2 RNA was analyzed by Northern blot with a probe specific for the ORF2 gene, and the VP1 levels were assessed by ELISA (Fig. 3).
The Northern blot analysis showed a 2.5-kb band corresponding to the expected size for the entire subgenomic RNA in cells expressing the ORF2+3, ORF2+3+3′UTR, and ORF2-AUG→ACG-ORF3+3′UTR constructs. In addition, a smaller band corresponding to the size of the ORF2 RNA (2.0 kb) was observed in cells expressing the ORF2+3′UTR. The ORF2 alone construct did not synthesize enough ORF2 mRNA to be detected visually at 36 hpi, although it could be detected at 48 hpi (data not shown). Some higher-molecular-weight bands, possibly extended RNAs from the constructs, were also present (Fig. 3A). The absence of an ORF2 RNA band in the wild-type baculovirus-infected cell RNA showed the specificity of the probe and the stringency of the hybridization conditions. Bands were quantitated by densitometric analysis and the highest level of RNA, seen with the ORF2+3+3′UTR recombinant baculovirus, was set at 100%. The RNA values for the other recombinant baculoviruses are presented as a relative amount with respect to the ORF2+3+3′UTR recombinant. The average for three experiments is shown at the bottom of the Northern blot (Fig. 3A). Steady-state levels of NV ORF2 mRNA increased 40-fold when the construct contained the 3′UTR downstream of ORF2 (ORF2+3′UTR) but only 15-fold when the ORF3 gene was present (ORF2+3) over that of the ORF2 gene alone (Fig. 3A). The presence of both the ORF3 and 3′UTR (ORF2+3+3′UTR) increased the steady-state levels of ORF2 RNA 50-fold. Therefore, these data indicate that the 3′UTR of NV increases the steady-state levels of ORF2 RNA. However, when VP2 protein expression was blocked by mutating the ORF3 start codon (ORF2-AUG→ACG-ORF3+3′UTR), the level of ORF2 RNA was reduced twofold, even though the 3′UTR was still present in the construct; this reduction in the amount of ORF2 RNA may be due to changes in the stability of the RNA caused by the mutation. This construct (ORF2-AUG→ACG-ORF3+3′UTR) produced significantly higher RNA levels than any of the constructs that lacked the 3′UTR, so the twofold reduction in RNA levels produced by the ORF2-AUG→ACG-ORF3+3′UTR recombinant compared to the parental ORF2+3+3′UTR recombinant cannot account for the 50-fold decrease in the level of VP1 expression.
Concurrently, at the protein level, the recombinant baculovirus containing the entire NV subgenomic RNA produced significantly higher levels (6- to 15-fold; P ≤ 0.002) of VP1 than those recombinants that lacked VP2 expression (ORF2, ORF2-AUG→ACG-ORF3+3′UTR, and ORF2+3′UTR) (Fig. 3B). The VP1 expression level was equivalent (P = 0.740) between the ORF2 and the ORF2-AUG→ACG-ORF3+3′UTR recombinant baculoviruses, but inclusion of the 3′UTR (ORF2+3′UTR) or ORF3 (ORF2+3) increased (P ≤ 0.043) the level of VP1 expression by 2.5- or 6-fold, respectively, over that produced by the ORF2 alone recombinant baculovirus (Fig. 3B). Taken together, the comparison of the steady-state levels of ORF2 mRNA with the corresponding amount of VP1 synthesized under identical conditions suggests that the NV 3′UTR upregulates VP1 expression at the RNA level, whereas VP2 increases VP1 protein levels.
VP2 expressed in trans does not restore VP1 expression levels.
Since synthesis of VP2 was required for the optimal expression of VP1, we next examined whether it was possible to restore the levels of VP1 by coinfecting each of the recombinant baculoviruses expressing VP1 alone with a recombinant baculovirus that only expresses VP2 (ORF3 + 3′UTR). Coinfection of the recombinant baculovirus ORF3+3′UTR with each of the baculoviruses ORF2, ORF2+3′UTR, or ORF2-AUG→ACG-ORF3+3′UTR resulted in a similar (P ≥ 0.756) level of VP1 expressed compared to cells infected with each of the corresponding recombinant baculoviruses alone (data not shown). Since in every case the expression level of VP1 was not restored when VP2 was expressed in trans, these results suggest that VP2 acts in cis to affect the levels of VP1 protein expression.
VP2 expression affects VP1 stability.
One mechanism by which VP2 may increase expression of VP1 is by stabilizing the expressed protein. To determine whether VP2 increases VP1 stability, a pulse-chase experiment was performed. VP1 was pulse-labeled in recombinant baculovirus-infected cells, and the radiolabeled VP1 was immunoprecipitated with an antibody specific for VP1 at different times postlabeling. After SDS-PAGE analysis, the bands corresponding to VP1 were quantitated by phosphorimaging and are represented as a relative amount of VP1 present at each different time relative to the amount of VP1 present immediately after labeling (0 h postlabeling) (Fig. 4A). The half-life of VP1 was determined by comparing the relative amount of VP1 over time for the different constructs (Fig. 4B). The half-life of VP1 (24 h) was >2-fold higher for the constructs containing VP2 (P ≤ 0.003) compared to the half-life of VP1 (10 h) expressed from a recombinant baculovirus that lacked VP2 (ORF2+3′UTR). Therefore, the presence of VP2 increases the stability of the VP1 protein.
FIG. 4.
Analysis of VP1 stability. (A) For pulse-chase experiments performed in insect cells, cells were infected with the recombinant baculoviruses containing the entire NV subgenomic RNA (ORF2+3+3′UTR) or lacking the 3′UTR (ORF2+3) or the ORF3 gene (ORF2+3′UTR). After a 30-min pulse at 36 hpi, chase periods of 6, 12, and 24 h were analyzed. The samples from each of the indicated time points were immunoprecipitated with a rabbit hyperimmune serum against the NV VP1 capsid protein in RIPA buffer, and the samples were then analyzed by autoradiography on an SDS-12% polyacrylamide gel. The arrows indicate the bands that correspond to the uncleaved VP1. The asterisk indicates a VP1-related band that is not present in the NV VLPs and has not been detected previously. This band was not included in the VP1 quantitation and was not present in the wild-type baculovirus (BV WT). The band was not detected when a monoclonal antibody was used to immunoprecipitate the protein; however, the epitope that the monoclonal antibody used may not be present in this VP1-related protein. (B) Phosphorimaging was performed to quantitate each band, and the amount of VP1 present at the zero time point for each recombinant baculovirus was set to 100%. The amount of VP1 at additional time points was represented relative to the amount of protein at 0 h after pulse-labeling. The values shown are the arithmetic means of three independent experiments. The error bars represent one standard error of the mean. The half-life of VP1 expressed from each of the recombinant baculoviruses tested was calculated from the curves obtained.
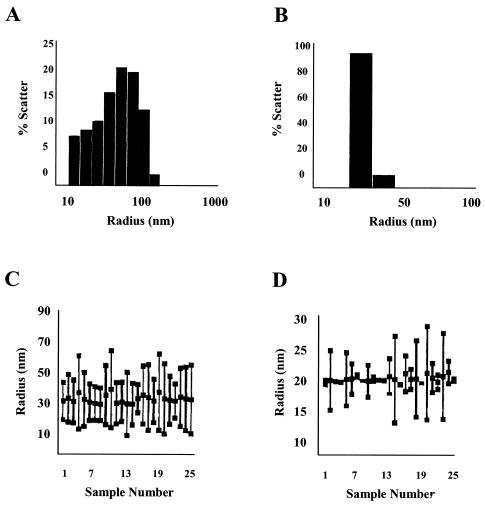
VP2 increases NV VLP particle stability and size homogeneity.
Since VP2 interacts with VP1 in a specific manner and VP2 is present in virus particles (15), we wondered whether VP2 allows the formation of more stable particles, thus protecting the VP1 capsid protein in particles from disruption or protease degradation. To analyze whether the presence of VP2 stabilizes the particles, we performed DLS analysis of NV VLPs produced with VP1 alone or with both VP1 and VP2. VLP preparations were analyzed in Tris-HCl (pH 6.7), which does not affect particle stability (58). Three different preparations of independently purified VLPs were analyzed. The particle preparations formed by VP1 and VP2 contained a significantly (P = 0.021) larger number of intact particles (85%) than particles composed entirely of VP1 (∼15%) (Fig. 5A and B). The particles that contained VP2 in addition to VP1 were more monodisperse and homogeneous in size than the ones that lacked VP2 (Fig. 5C and D). These results suggest that the presence of VP2 in NV VLPs increases the stability and size homogeneity of the particles.
FIG. 5.
DLS analysis of the homogeneity of intact particles in preparations of VLPs composed of only VP1 or of both VP1and VP2 proteins (VP1/VP2). (A and B) Graphs show the percentage of intact particles from either VP1 VLP or VP1/VP2 VLP preparations, respectively. The particles were analyzed in conditions known to keep the particles assembled (Tris-HCl [pH 6.7]). Histograms of the percentage of scatter for VP1 (A) or VP1/VP2 particles (B) particles measured at a 90° scattering angle are shown. Because the total protein concentration was similar for all of the preparations, the percentage of scatter represents the molecular mass and effective protein concentration for each particle size. (C and D) In addition, the particle size distributions for VP1 (C) and VP1/VP2 (D) particles are shown. The vertical lines in panels C and D represent the distributions of different particle sizes in the sample for each reading. The top square represents the largest particle found in the sample (nanometers of radius), the middle square represents the average particle size, and the bottom square the smallest particle size found in the sample. A monodispersed, homogeneous sample is expected to have only one size of particle, depicted as one square and no line. Twenty-five readings were recorded for each sample. Three different samples for each type of particles (VP1 or VP1/VP2) were analyzed. The data from one representative reading are shown.
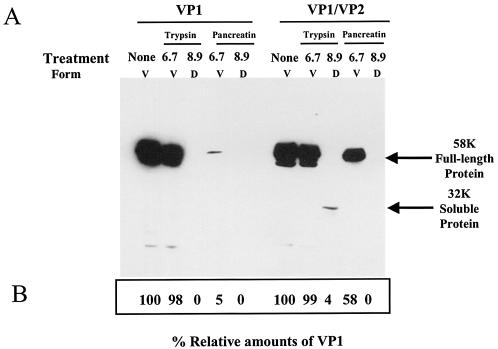
VP2 increases the stability of VP1 and protects VP1 from protease degradation.
To further analyze the effect of VP2 on VP1 stability, we subjected VP1 or VP1/VP2 NV VLPs to protease treatment (Fig. 6). Intact VP1 or VP1/VP2 NV VLPs in Tris-HCl (pH 6.7), or particles suspended in Tris-HCl (pH 8.9), to promote disassembly (58), were treated with trypsin or pancreatin, and cleavage products were analyzed by Western blotting. In order to observe a difference in sensitivity to protease treatment between the two types of particles, the particles were treated for an extended time with a high concentration of protease. In the case of the protein originating from the VP1/VP2 particles (right panel), most (99%) of the VP1 from the intact VLPs, in Tris-HCl (pH 6.7), remained protected from trypsin digestion. In contrast, for the VP1/VP2 (pH 8.9) disassembled VLPs, a significant amount of VP1 was cleaved by trypsin, as indicated by the loss of the 58K band and the appearance of a smaller band (32K) that corresponds in apparent molecular weight to the protruding domain of the protein (Fig. 6) (26). NV VLPs were more sensitive to pancreatin digestion than to trypsin digestion. Although some VP1 remained protected from pancreatin treatment in the intact VP1/VP2 VLPs (58%), treatment of the disassembled VP1/VP2 VLPs with pancreatin completely degraded VP1. Treatment of the particles containing only VP1 (left panel) showed that VP1 was sensitive to trypsin and pancreatin degradation when VP1 was disassembled at pH 8.9. When intact particles at pH 6.7 were treated with pancreatin, the VP1 from VP1 particles was also more sensitive to pancreatin treatment than VP1 from VP1/VP2 particles. These results indicate that VP1 is less sensitive to protease treatment when assembled into VLPs. In addition, these data suggest that when VP2 is associated with particles, VP1 is more resistant to protease degradation. Thus, VP2 appears to influence the conformation or stability of VP1 in VLPs.
FIG. 6.
Analysis of the sensitivity of intact or partially denatured rNV VLPs to protease degradation. VLPs composed of only the VP1 capsid protein (VP1 VLPs) or composed of both VP1 and VP2 proteins (VP1/VP2 VLPs) were kept assembled (Tris-HCl [pH 6.7]) or disassembled by high-pH treatment (Tris-HCl [pH 8.9]) prior to protease digestion. (A) After protease digestion, the proteins were separated in SDS-12% polyacrylamide gels, and the bands were analyzed by Western blotting with a rabbit serum specific to VP1 (30). The arrows indicate the bands that correspond to the uncleaved 58K full-length VP1 and the 32K soluble protein formed by the protruding domain (26). V, VLPs; D, disassembled form of the NV VP1 capsid protein. (B) The arithmetic means of the densitometric values corresponding to the VP1 bands are shown for three experiments.
The increase of VP1 expression in the presence of VP2 is also observed in mammalian cells.
To determine whether VP2 also enhances the expression of VP1 in mammalian cells, we analyzed the effect of VP2 on VP1 levels in HEK 293T cells. A clone containing the entire NV subgenomic RNA downstream of the GFP under the cytomegalovirus promoter was constructed (GFP-ORF2+3+3′UTR) (Fig. 1B), so VP1 was expressed as a fusion protein with GFP. In addition, another construct was generated by site-directed mutagenesis by using the GFP-ORF2+3+3′UTR as a template to mutate the ORF3 initiation codon (AUG→ACG) to block VP2 expression (Fig. 1B).
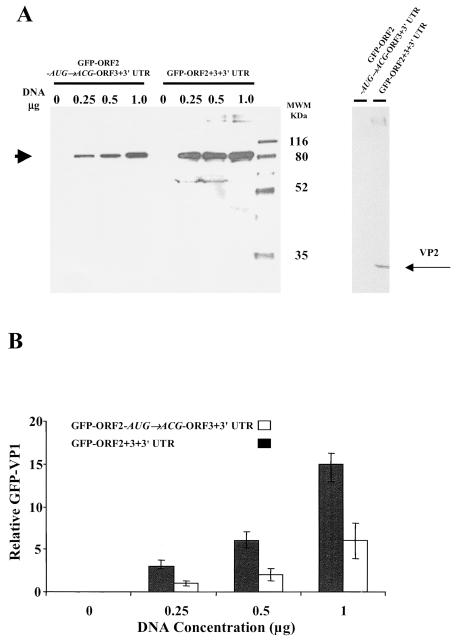
The levels of GFP-VP1 expression were compared after transfection of HEK 293T cells with either the construct expressing the entire subgenomic NV RNA (GFP-ORF2+3+3′UTR) or the mutated construct (GFP-ORF2-AUG→ACG-ORF3+3′UTR) by both densitometry and flow cytometry analyses. Cotransfection with luciferase was used as an internal control for transfection efficiency. In both cases, the same numbers of cells were transfected with the same amount of DNA from each construct, and GFP-VP1 expression was analyzed 24 h posttransfection. The same volume of each cell lysate was analyzed by Western blot with an antibody specific for the NV capsid protein (Fig. 7A), and the bands corresponding to the GFP-VP1 fusion protein were quantitated by densitometry and normalized to luciferase values (Fig. 7B). Over a range of amounts of DNA (0.25 to 1.0 μg) used for the transfections, the construct containing the entire subgenomic NV RNA reproducibly expressed significantly higher amounts (threefold; P ≤ 0.002) of the GFP-VP1 fusion protein than the construct that did not express VP2 (GFP-ORF2-AUG→ACG-ORF3+3′UTR). The amount of GFP-VP1 fusion protein detected in HEK 293T cells correlated (P = 0.001; r2 = 0.978 [Pearson correlation coefficient]) with the amount of DNA used to transfect the HEK 293T cells.
FIG. 7.
Regulatory effect of VP2 on expression of VP1 in mammalian cells. (A) Western blot analysis of a GFP-VP1 fusion protein expressed in HEK 293T cells after transfection with constructs containing the entire NV subgenomic RNA (GFP-ORF2+3+3′UTR) or lacking the ability to express VP2 (GFP-ORF2-AUG→ACG-ORF3+3′UTR). After transfection of cells with the indicated amount (in micrograms above each lane) of DNA into 293T cells, the same amount of cell lysate was used to analyze the expressed proteins. The NV VP1 capsid protein was detected by Western blotting with a rabbit serum specific to VP1. The sizes are shown in kilodaltons, and the arrow indicates the band that corresponds to the uncleaved GFP-VP1 fusion protein. The Western blot shown in the right panel was performed with rabbit sera specific to VP2 (15, 16). The band corresponding to the VP2 protein is indicated with an arrow and was only observed when the GFP-ORF2+3+3′UTR recombinant was expressed. (B) The bands corresponding to the expected molecular weight for the GFP-VP1 fusion protein in panel A were analyzed by densitometry. The densitometric values were normalized by using luciferase activity values and the relative amounts of GFP-VP1 are shown. Each bar shows the arithmetic means of three independent experiments performed with different DNA preparations. The error bars represent one standard error of the mean.
The same results were obtained when the cells were analyzed by flow cytometry (data not shown). The level of GFP-VP1 intensity was twofold higher (200.6 versus 123.6) when the construct expressed VP2. Similar transfection efficiencies were obtained with the GFP-ORF2+3+3′UTR (41%) and the GFP-ORF2-AUG→ACG-ORF3+3′UTR (31%) constructs. Therefore, the increase in VP1 expression due to VP2 observed in insect cells is reproduced in mammalian cells, suggesting that the effects exerted by NV VP2 are relevant in a system that more closely mimics natural NV host cells.
DISCUSSION
We report here the novel observation that expression and stability of the NV VP1 capsid protein are regulated by elements at the 3′ end of the NV genome. We determined that the level of VP1 expression in insect and mammalian cells is regulated by the expression of NV VP2 and the presence of the 3′UTR. Understanding the mechanism by which these elements regulate VP1 expression provides insight into NV RNA expression, as well as practical implications for expression of calicivirus capsid proteins for vaccine development and further structural studies. Importantly, understanding the requirements for optimal NV gene expression may enable the cultivation of the virus, a task that has been the focus of great effort for more than 30 years.
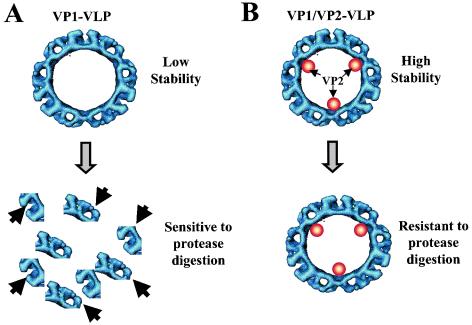
We have found that one of the functions of VP2, a small structural protein present in all caliciviruses examined to date, is to regulate the stability and therefore the levels of VP1 capsid protein during its expression in insect cells. Constructs in which the expression of VP2 was blocked showed a marked decrease in VP1 capsid protein level and stability. A working model is proposed to explain the effect of VP2 on the stability of the VP1 capsid protein (Fig. 8). VP2 may stabilize the VP1 capsid building blocks to produce a particle that is resistant to protein degradation and disassembly. In agreement with this model, NV VLPs that possess VP2 (VP1/VP2 VLPs) were more resistant to protease degradation and showed more integrity and homogeneity by DLS than NV VLPs that lacked VP2 (VP1 VLPs). The effect of the VP2 protein stabilizing the assembled capsid is reminiscent of the adenovirus small polypeptide pIX (7, 9, 51). Protein IX acts to reinforce the adenovirus virion keeping the capsid assembled and protecting it from protease degradation (14). Although there have not been other published studies on the involvement of the calicivirus VP2 in VP1 gene regulation or protein expression, blocking FCV VP2 expression by inclusion of a premature stop codon in the FCV ORF3 gene of a FCV infectious clone has been reported to block FCV replication (20).
FIG. 8.
Proposed model for the mechanism of the VP2 stabilizing effect on NV VP1 capsid protein. Particles composed of both VP1 and VP2 proteins are more stable than those composed only of the VP1 capsid protein. Therefore, particles composed only of VP1 (A) could be more sensitive to various treatments (such as pressure, heat, pH, or changes in ionic concentration; represented by the shaded arrow) that cause disassembly of the particles compared to particles that also contain VP2 (B). Once the particles disassemble, the soluble VP1 capsid protein may be more sensitive to protease degradation (solid arrows) because of the exposure of more cleavage sites.
In addition to the stability of VP1, the rate of synthesis of the VP1 capsid protein may be affected by the presence of VP2. Indeed, VP2 may also affect the expression rate of VP1 because the level of VP1 synthesized in a 30-min pulse was at least three times higher for the ORF2+3 recombinant baculovirus than for VP1 made by a recombinant that contained ORF2+3′UTR. This observation, which was reproduced at least five times, is not related to the degradation of the protein because, even in the absence of VP2, the half-life of VP1 was 10 h. Analysis of the involvement of VP2 in the regulation of VP1 translational initiation is not possible for NV since it would require an infectious clone. However, studies with the FCV infectious clone with mutations to block VP2 expression may facilitate determination of the mechanism involved in the regulation of VP1 expression by VP2.
The effect of VP2 is only seen when the protein is present in cis. This may be because the level of VP2 needs to be regulated, either in a temporal or a spatial manner. The expression level of VP2 from the bicistronic ORF2+3 subgenomic RNA is very low, whereas VP2 is expressed in high yields when a recombinant baculovirus generated with a construct containing only ORF3+3′UTR is used to infect insect cells (15). Proteins made in stochastic amounts, such as VP2, may have regulatory roles because very little protein is needed to perform their function(s) efficiently. If VP2, as it is believed, is inside the capsid and in close proximity to the VP1 capsid protein, then the introduction of the protein in trans, using a recombinant baculovirus that expresses VP2 in high yields, may overfill the particles, rendering them susceptible to disassembly and therefore to protease degradation. In fact, VLPs obtained from dual infections with individual ORF2 and ORF3 baculovirus constructs were previously noted to be unstable (15). Although NV VLPs are able to form in insect cells dually infected with independent baculovirus recombinants that express VP1 and VP2 separately, it is likely that only a limited number of VP2 molecules need to be and can be incorporated in stable particles (15). When overexpressed in trans, VP2 also might not be present at the correct place at the right time to function properly. If a low amount of VP2 is needed to perform its function efficiently, the mechanism by which it is synthesized from the subgenomic RNA should be regulated. A mechanism of termination-reinitiation or internal initiation has been suggested for synthesis of the VP2-like protein of FCV based on in vitro studies in rabbit reticulocyte lysates (28). Consistent with the idea of a termination-reinitiation mechanism for the synthesis of VP2, we found that a mutant with a premature stop codon located in ORF2 120 nucleotides upstream of the ORF3 AUG still synthesized VP2 (data not shown).
An alternative explanation for seeing the VP2 effect over VP1 expression only when the protein is present in cis, other than the requirement for a balanced equilibrium between the VP1/VP2 protein ratios, is that ORF3 must be translated in cis for successful protein function. A requisite for translation in cis could result from the preferential cis action of VP2. VP2 may be necessary in a newly synthesized form to bind the ORF2 RNA to establish it as a template for translation. A similar mechanism has been described for the synthesis of the β-tubulin protein that acts in cis to affect its own translational levels (3, 55, 59). Tubulin synthesis is controlled by a cotranslational mechanism that involves the binding of the tubulin mRNA to a nascent beta-tubulin peptide as it emerges from the ribosome (3, 55). Another possibility is that the function of ORF3 may require the act of the ribosomal passage itself in cis through the ORF3 RNA. Likewise, there is a cis-translation region in the poliovirus genome that is present in a coding region that is indispensable for the genome to replicate, even when viral proteins are provided in trans from a helper genome (18, 41, 50). Unfortunately, the lack of a replication system for NV continues to hamper studies of the involvement of VP2 in replication. Further studies are required to establish the details of the mechanism for the requirement of NV VP2 synthesis in cis. However, recognition that ORF3 may be part of a cis-translation regulatory system may help guide studies trying to establish infectious clones of NV or studies with infectious clones of cultivable caliciviruses, since mutation or deletion of this region may lead to lethal mutations.
Multiple forms of VP2 have been observed in NV virions and VLPs by Western blot analysis, including 23K and 35K forms (15). Recent sequence analysis by mass spectrometry of the 35K form of VP2 confirmed that it is composed only of phosphorylated VP2 (16). Do the two forms of VP2 have different roles during the viral life cycle? The 23K form of VP2 is found in NV VLPs (ORF2+3+3′UTR), whereas the 35K form is found primarily in NV virions that contain RNA. In some NV virions, the 23K VP2 form was found and electron microscopic analysis of these particles showed that they were mostly empty (15). The 23K form of VP2 seems to be sufficient to affect VP1 capsid protein stability since the VLPs analyzed by DLS and protease digestion contain only the 23K form of VP2. These results suggest that the phosphorylated 35K form of VP2 may have different functions from the 23K form. Other small, phosphorylated viral proteins have been reported to enhance translation. For example, NSP3, a nonstructural phosphorylated protein from rotaviruses, has been shown to enhance translation of viral RNAs by binding as a dimer to the 3′ end of the viral RNAs and likely circularizing the RNA (46-48). It has also been proposed that the 35K form of VP2 is involved in encapsidation of viral RNA. Further progress in dissecting the properties of these different forms of ORF3 will require the ability to purify each form of the protein in high yields, a challenge that has recently been achieved (16).
Importantly, the effect of VP2 on the expression levels of VP1 is also observed in mammalian HEK 293T cells. However, the different effects on the levels of expression in mammalian cells between the construct that expresses VP2 and the one that does not are not as remarkable (2- to 3-fold increase) as in insect cells (12-fold increase). One possible explanation for the lower effects seen in mammalian cells is that fusion of GFP to VP1 in the two constructs may have influenced the results. For example, the half-life of VP1 alone when fused to GFP was increased in mammalian cells (data not shown), and this could have reduced the detected stabilizing effect of VP2 on VP1. However, since both of the constructs have GFP and the only difference between them is the expression or lack of expression of VP2, the difference in levels of GFP-VP1 expression is not dependent on the presence of GFP but on the presence or absence of VP2. In addition, the effect of VP2 on VP1 expression in a replication-competent vector, such as baculoviruses, may be different than when the proteins are expressed from a cytomegalovirus promoter-driven expression plasmid.
The enhancement of VP1 expression levels by VP2 was not observed in a reticulocyte lysate system (data not shown). The reason for this may be the requirement for certain cellular factors absent in the reticulocyte lysates or the necessity for assembly of particles that in turn may require higher levels of protein expression than can be achieved in such a cell-free system.
In addition to the VP2 protein, the presence of the 3′UTR downstream of the NV ORF2 gene increases the steady-state levels of NV ORF2 mRNA. However, it does not directly affect the level of VP1 synthesis. The reason for the increased RNA levels in constructs that have the 3′UTR is not simply due to an increased length of the RNA because, even though the ORF2+3 recombinant produced a longer RNA than the ORF2+3′UTR, the steady-state levels of ORF2+3 RNA were lower. This raises the question of what regulatory elements are present in the NV 3′UTR that affect the steady-state levels of NV ORF2 mRNA. The 3′UTRs of positive-stranded RNA viruses such as hepatitis C, hepatitis E, and turnip crinkle viruses are the sites for binding of the RNA-dependent RNA polymerase complexes (1, 53). Several elements have been described at the 3′ end of RNA viral genomes that regulate viral gene expression (1, 12). Poly(A) tails, for example, are known to be intimately involved in the control of mRNA turnover by degradation (2, 11). In addition, AU-rich elements present at the 3′ end of the RNAs are bound by a number of cellular and viral proteins that alter the stability and translation rate of the RNAs (8). Most of the cis-acting signals in viral RNAs that are involved in gene expression are formed by secondary structures. Although there is little known about the presence of RNA structures at the 3′ end of calicivirus genomes, a predicted stem-loop at the 3′UTR of the RHDV and FCV genomes has been reported (52). In addition, the NV 3′UTR increases luciferase expression in insect cells when located downstream of the luciferase gene (data not shown), suggesting that, like other 3′UTR elements (12), the NV 3′UTR effect is not limited to NV genes. Although further analysis of the effect of the NV 3′UTR on foreign genes is needed, this observation may be of practical value; the NV 3′UTR could potentially be used to enhance the protein expression of other foreign genes in insect cells.
The results of the present study also have other practical applications. For example, although high yields of NV VLPs are obtained when VP1 is expressed in baculovirus constructs that contain the entire subgenomic RNA (32), expression of other calicivirus capsid proteins has resulted in low yields or unstable VLPs (4, 21-24, 42, 32, 33). Most of these reports used recombinant constructs that lacked the VP2 protein, the 3′UTR, or both or have used different baculovirus transfer vectors. Reanalysis of expression levels in insect or mammalian cells with new constructs that contain the entire subgenomic RNA may result in higher levels of more stable VLPs representing the capsids of a wide range of genera of human caliciviruses needed for antigenic, biological, structural, and vaccine studies.
Acknowledgments
We thank Carl Q.-Y. Zeng for assistance with insect cell cultures and Richard E. Sutton for providing the HEK 293T cells. We also thank Jeffrey M. Scott (Center for AIDS Research [CFAR], Immunology Core, Department of Immunology, Baylor College of Medicine) and Sarah E. Blutt for help in flow cytometric analyses and Sugoto Chakravarty and B. V. Venkataram Prasad for assistance with the DLS analysis. In addition, we thank Tracy D. Parker and Vivian Ruvolo for critically reviewing the manuscript.
This work was supported by Public Health Service grants AI46581, CA-09197, and DK-56338, which supports the Texas Gulf Coast Digestive Diseases Center.
REFERENCES
- 1.Agrawal, S., D. Gupta, and S. K. Panda. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282:87-101. [DOI] [PubMed] [Google Scholar]
- 2.Arora, R., C. Priano, A. B. Jacobson, and D. R. Mills. 1996. cis-Acting elements within an RNA coliphage genome: fold as you please, but fold you must!! J. Mol. Biol. 258:433-446. [DOI] [PubMed] [Google Scholar]
- 3.Bachurski, C. J., N. G. Theodorakis, R. M. Coulson, and D. W. Cleveland. 1994. An amino terminal tetrapeptide specifies cotranslational degradation of beta-tubulin but not alpha-tubulin mRNAs. Mol. Cell. Biol. 14:4076-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belliot, G., J. S. Noel, J.-F. Li, S. Yoshiyuki, Humphrey, T. M., Ando, T., Glass, R. I., and Monroe, S. S. 2001. Characterization of capsid genes, expressed in the baculovirus system, of three new genetically distinct strains of “Norwalk-like viruses.” J. Clin. Microbiol. 39:4288-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolotti-Ciarlet, A., L. J. White, R. Chen, B. V. Prasad, and M. K. Estes. 2002. Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 76:4044-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boga, J. A., M. S. Marin, R. Casais, M. Prieto, and F. Parra. 1992. In vitro translation of a subgenomic mRNA from purified virions of the Spanish field isolate AST/89 of rabbit hemorrhagic disease virus (RHDV). Virus Res. 26:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulanger, P., P. Lemay, G. E. Blair, and W. C. Russell. 1979. Characterization of adenovirus protein IX. J. Gen. Virol. 44:783-800. [DOI] [PubMed] [Google Scholar]
- 8.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colby, W. W., and T. Shenk. 1981. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J. Virol. 39:977-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford, S. E., M. Labbe, J. Cohen, M. H. Burroughs, Y. J. Zhou, and M. K. Estes. 1994. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J. Virol. 68:5945-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decker, C. J., and R. Parker. 1995. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr. Opin. Cell Biol. 7:386-392. [DOI] [PubMed] [Google Scholar]
- 12.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 13.Ehresmann, D. W., and F. L. Schaffer. 1979. Calicivirus intracellular RNA: fractionation of 18-22S RNA and lack of typical 5′-methylated caps on 36S and 22S San Miguel sea lion virus RNAs. Virology 95:251-255. [DOI] [PubMed] [Google Scholar]
- 14.Furcinitti, P. S., J. van Oostrum, and R. M. Burnett. 1989. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 8:3563-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glass, P. J., C. Q.-Y. Zeng, and M. K. Estes. 2003. Two non-overlapping domains on the Norwalk virus ORF3 protein are involved in the formation of the phosphorylated 35K protein and ORF3-capsid protein interactions. J. Virol. 77:3569-3577. in press. [DOI] [PMC free article] [PubMed]
- 17.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181(Suppl. 2):S254-S261. [DOI] [PubMed] [Google Scholar]
- 18.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, Barclay, W., and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 20.Green, K. Y., S. Sosnovtsev, and S. Sosnovtseva. 2002. The feline calicivirus (FCV) VP2 protein is essential for replication, p. 22. International Congress of Virology, Paris, France.
- 21.Green, K. Y., A. Z. Kapikian, J. Valdesuso, S. Sosnovtsev, J. J. Treanor, and J. F. Lew. 1997. Expression and self-assembly of recombinant capsid protein from the antigenically distinct Hawaii human calicivirus. J. Clin. Microbiol. 35:1909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo, M., Y. Qian, K.-O. Chang, and L. Saif. 2001. Expression and self-assembly in baculovirus of porcine enteric calicivirus capsids into virus-like particles and their use in an enzyme-linked immunosorbent assay for antibody detection in swine. J. Clin. Microbiol. 39:1487-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hale, A. D., S. E. Crawford, M. Ciarlet, J. Green, C. Gallimore, D. W. Brown, X. Jiang, and M. K. Estes. 1999. Expression and self-assembly of Grimsby virus: antigenic distinction from Norwalk and Mexico viruses. Clin. Diagn. Lab. Immunol. 6:142-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy, M. E., S. F. Kramer, J. J. Treanor, and M. K. Estes. 1997. Human calicivirus genogroup II capsid sequence diversity revealed by analyses of the prototype Snow Mountain agent. Arch. Virol. 142:1469-1479. [DOI] [PubMed] [Google Scholar]
- 25.Hardy, M. E., T. N. Tanaka, N. Kitamoto, L. J. White, J. M. Ball, X. Jiang, and M. K. Estes. 1996. Antigenic mapping of the recombinant Norwalk virus capsid protein using monoclonal antibodies. Virology 217:252-261. [DOI] [PubMed] [Google Scholar]
- 26.Hardy, M. E., L. J. White, J. M. Ball, and M. K. Estes. 1995. Specific proteolytic cleavage of recombinant Norwalk virus capsid protein. J. Virol. 69:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, S. C. 1989. Common features in the design of small RNA viruses, p. 3-19. In M. B. A. Oldstone and A. Notkins (ed.), Concepts in viral pathogenesis. Springer-Verlag, New York, N.Y.
- 28.Herbert, T. P., I. Brierley, and T. D. Brown. 1996. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 77:123-127. [DOI] [PubMed] [Google Scholar]
- 29.Hwang, Y. K., and M. A. Brinton. 1998. A 68-nucleotide sequence within the 3′ noncoding region of simian hemorrhagic fever virus negative-strand RNA binds to four MA104 cell proteins. J. Virol. 72:4341-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, X., J. Wang, D. Y. Graham, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang, X., D. Matson, G. M. Ruiz-Palacios, J. Hu, J. Treanor, and L. K. Pickering. 1995. Expression, self-assembly, and antigenicity of a snow mountain agent-like calicivirus capsid protein. J. Clin. Microbiol. 33:1452-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi, S., K. Sakae, Y. Suzuki, H. Ishiko, K. Kamata, K. Suzuki, K. Natori, T. Miyamura, and N. Takeda. 2000. Expression of recombinant capsid proteins of Chitta virus, a genogroup II Norwalk virus, and development of an ELISA to detect the viral antigen. Microbiol. Immunol. 44:687-693. [DOI] [PubMed] [Google Scholar]
- 34.Konig, M., H. J. Thiel, and G. Meyers. 1998. Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J. Virol. 72:4492-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhn, R. J., Z. Hong, and J. H. Strauss. 1990. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J. Virol. 64:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli, U. K., F. Beguin, and G. Gujer-Kellenberger. 1970. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J. Mol. Biol. 47:69-85. [DOI] [PubMed] [Google Scholar]
- 37.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Ming, Y. Z., X. Di, E. P. Gomez-Sanchez, and C. E. Gomez-Sanchez. 1994. Improved downward capillary transfer for blotting of DNA and RNA. BioTechniques 16:58-59. [PubMed] [Google Scholar]
- 39.Neill, J. D., and W. L. Mengeling. 1988. Further characterization of the virus-specific RNAs in feline calicivirus infected cells. Virus Res. 11:59-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neill, J. D. 1992. Nucleotide sequence of the capsid protein gene of the two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among caliciviruses capsid proteins. Virus Res. 24:211-222. [DOI] [PubMed] [Google Scholar]
- 41.Novak, J. E., and K. Kirkegaard. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 8:1726-1737. [DOI] [PubMed] [Google Scholar]
- 42.Numata, K., M. E. Hardy, S. Nakata, S. Chiba, and M. K. Estes. 1997. Molecular characterization of morphologically typical human calicivirus Sapporo. Arch. Virol. 142:1537-1552. [DOI] [PubMed] [Google Scholar]
- 43.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 44.Oker-Blom, C., R. F. Pettersson, and M. D. Summers. 1989. Baculovirus polyhedrin promoter-directed expression of rubella virus envelope glycoproteins, E1 and E2, in Spodoptera frugiperda cells. Virology 172:82-91. [DOI] [PubMed] [Google Scholar]
- 45.Oker-Blom, C., and M. D. Summers. 1989. Expression of Sindbis virus 26S cDNA in Spodoptera frugiperda (Sf9) cells, using a baculovirus expression vector. J. Virol. 63:1256-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piron, M., T. Delaunay, J. Grosclaude, and D. Poncet. 1999. Identification of the RNA-binding, dimerization, and eIF4GI-binding domains of rotavirus nonstructural protein NSP3. J. Virol. 73:5411-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poncet, D., C. Aponte, and J. Cohen. 1996. Structure and function of rotavirus nonstructural protein NSP3. Arch. Virol. Suppl. 12:29-35. [DOI] [PubMed] [Google Scholar]
- 49.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 50.Rieder, E., A. V. Paul, D. W. Kim, J. H. Van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosa-Calatrava, M., L. Grave, F. Puvion-Dutilleul, B. Chatton, and C. Kedinger. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 75:7131-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seal, B. S., J. D. Neill, and J. F. Ridpath. 1994. Predicted stem-loop structures and variation in nucleotide sequence of 3′ noncoding regions among animal calicivirus genomes. Virus Genes 8:243-247. [DOI] [PubMed] [Google Scholar]
- 53.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:6-14. [DOI] [PubMed] [Google Scholar]
- 54.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193-203. [DOI] [PubMed] [Google Scholar]
- 55.Theodorakis, N. G., and D. W. Cleveland. 1992. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol. Cell. Biol. 12:791-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiley, L. S., M. Hagen, J. T. Matthews, and M. Krystal. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68:5108-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White, L. J., J. M. Ball, M. E. Hardy, T. N. Tanaka, N. Kitamoto, and M. K. Estes. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70:6589-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White, L. J., M. E. Hardy, and M. K. Estes. 1997. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J. Virol. 71:8066-8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yen, T. J., P. S. Machlin, and D. W. Cleveland. 1988. Autoregulated instablility of β-tubulin mRNAs by recognition of the nascent amino terminus of β-tubulin. Nature 334:580-585. [DOI] [PubMed] [Google Scholar]