Abstract
We conducted a quantitative structure-activity relationship (QSAR) study based on a database of 127 compounds previously tested against the liver stage of Plasmodium yoelii in order to develop a model capable of predicting the in vitro antimalarial activities of new compounds. Topological indices were used as structural descriptors, and their relation to antimalarial activity was determined by using linear discriminant analysis. A topological model consisting of two discriminant functions was created. The first function discriminated between active and inactive compounds, and the second identified the most active among the active compounds. The model was then applied sequentially to a large database of compounds with unknown activity against liver stages of Plasmodium. Seventeen drugs that were predicted to be active or inactive were selected for testing against the hepatic stage of P. yoelii in vitro. Antiretroviral, antifungal, and cardiotonic drugs were found to be highly active (nanomolar 50% inhibitory concentration values), and two ionophores completely inhibited parasite development. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed on hepatocyte cultures for all compounds, and none of these compounds were toxic in vitro. For both ionophores, the same in vitro assay as those for P. yoelii has confirmed their in vitro activities on Plasmodium falciparum. A similar topological model was used to estimate the octanol/water partition of each compound. These results demonstrate the utility of the QSAR and molecular topology approaches for identifying new drugs that are active against the hepatic stage of malaria parasites. We also show the remarkable efficacy of some drugs that were not previously reported to have antiparasitic activity.
Each year, the malaria parasite Plasmodium falciparum infects 300 to 660 million persons worldwide and causes several million deaths (25). New antimalarial drugs are urgently needed, especially considering the increasing prevalence of drug-resistant P. falciparum strains and the lack of effective vaccines and vector control measures. The Plasmodium liver stage is an interesting drug target, as it precedes the emergence of blood stages that cause the symptoms and complications of malaria. Drugs that inhibit parasite maturation within hepatocytes could be used for short-term prophylaxis in areas of endemicity (refugees and travelers, etc.). In addition, such drugs would be unlikely to select resistant strains, as the number of mitoses that occur during hepatic schizogony is many orders of magnitude smaller than that during the erythrocyte stage, thereby limiting the risk of mutant selection.
Atovaquone and primaquine, licensed drugs, are currently the only drugs known to be effective against liver stages. However, atovaquone does not act against dormant forms of Plasmodium vivax and Plasmodium ovale and tends to select resistant clones among blood stages of P. falciparum, while primaquine carries a high risk of acute hemolysis in glucose-6-phosphate dehydrogenase-deficient patients, limiting the prophylactic use of these drugs. The latter limitation also applies to other related synthetic 8-aminoquinolines such as bulaquine (2) and tafenoquine (27).
The identification and assessment of candidate antimalarial drugs that are effective against exoerythrocytic stages of Plasmodium are hampered by the lack of simple in vitro and in vivo tests. Several mathematical approaches have been proposed to facilitate the search for new active compounds. Equation systems linking quantitative structure-activity (QSAR) relationships are particularly relevant and can be applied to large libraries of compounds for virtual computational screening (6, 15). However, these models require good structural descriptors that reliably represent the molecular features responsible for the relevant biological activity. Molecular topology is one way of describing molecular structures. It follows a two-dimensional approach, taking into account the internal atomic arrangement. The structure of each molecule is represented by specific subsets of topological indices (TIs) (4). These indices, when well chosen, provide a unique way of characterizing a molecular structure (14). Moreover, they correlate with many physical, chemical, and biological properties of structurally heterogeneous groups of compounds and can be used to find new drugs (8). These models have already been used to predict the activities of candidate antimicrobial and antimalarial agents (5, 10-12, 18, 19, 21).
The aim of this study was to develop and assess QSAR models based on molecular topology in order to identify new compounds that are active on the liver stages of Plasmodium as well as predict physicochemical parameters influencing intestinal absorption.
MATERIALS AND METHODS
Use of molecular topology to obtain structural descriptors.
A database of 127 compounds with known in vitro activities against the liver stage of P. yoelii yoelii was constructed from a literature search of several medical databases and from our own experimental data. The database comprises 24 families of congeneric chemicals, naphthoquinones, amino-alcohols, amino-quinolines, acyclic and cyclic peroxides, quinolones and fluoroquinolones, cyclines, macrolides, lincosamides, penicillin, an herbal alkaloid, inhibitors of dihydrofolate reductase, iron chelators, acridine-type Mannich bases, vitamins, a sponge extract, anti-inflammatory drugs, colchicine, antiarrhythmic, antiseptic, and antifungal compounds, and insecticides. The plane chemical structure of each drug was described with the aid of the Chemdraw software package (version 2002). Each compound was characterized by a set of 62 TIs. We used Kier and Hall's connectivity indices (13), topological charge indices (9), quotients and differences between valence and nonvalence connectivity indices (mCt = mχt,/mχtv and mDt = mχt, − mχtv), PRn (number of pairs of ramifications at topological distance n, with n ranging from 0 to 4), Vn (number of vertices with topological valence n, with n being 3 or 4), and other graph-theoretical descriptors (not outlined here, as they were not selected for the final model). All descriptors were calculated with Desmol11 software (R. Garcia-Domenech, Unidad de Investigación de Diseno de Fármacos y Conectividad Molecular, Facultad de Farmacia, Universitat de Valencia, Valencia, Spain).
LDA.
Linear discriminant analysis (LDA) is a pattern recognition method providing a classification model based on the combination of variables that best predicts the category or group to which a given compound belongs. Database compounds were allocated to highly active, active, and inactive groups according to their 50% inhibitory concentrations (IC50) using a first cutoff of 25 μM to separate active compounds (IC50 < 25 μM) from inactive compounds (IC50 > 25 μM) and a second cut off of 1 μM to separate highly active compounds (IC50 < 1 μM) from weakly to moderately active compounds (1 μM < IC50 < 10 μM). The cutoff was applicable, as it clearly distinguished among inactive, active, and highly active drugs. LDA was then applied to these three groups in order to obtain two discriminant functions, DF1 and DF2, which classified a compound as being active/inactive or active/highly active, respectively. The independent variables in this study were the calculated TI, and the discriminatory property was the activity against the liver stage of Plasmodium. The discriminatory capacity was assessed as the percentage of correct classifications in each set of compounds. The classification criterion was the Mahalanobis minimal distance (distance of each case to the mean of all the cases in a category). The quality of the discriminant function was evaluated by using the Wilks parameter, λ, which was obtained by multivariate analysis of variance that tests the equality of group means for the variable in the discriminant model. The method used to select the descriptors was based on the Fisher-Snedecor parameter (F), which determines the relative importance of candidate variables. The software used for the LDA study was the BMDP New System 2 package, module 7M. The variables used to compute the linear classification function are chosen in s stepwise manner: at each step, the variable that makes the largest contribution to the separation of the groups is entered into the discriminant equation (or the variable that makes the smallest contribution is removed).
PDDs.
The pharmacological distribution diagram (PDD) is a graphical representation that provides a straightforward way of visualizing the regions of minimum overlap as well as the regions in which the probability of finding active compounds is at maximum. A PDD is a frequency distribution diagram of dependent variables in which the ordinate represents the expectancy (probability of active) and the abscissa represents the values of DF in the interval (7). For an arbitrary interval of values of a given function, an “expectancy of activity” can be defined as Ea = a/(i + 1), where a is the number of active compounds in the interval divided by the total number of active compounds and i is the number of inactive compounds in the interval divided by the total number of inactive compounds. The expectancy of inactivity is defined in a symmetrical way, as Ei = i/(a + 1). With these diagrams, we can visually determine the intervals in which there is a maximum probability of finding new active compounds and a minimum probability of finding inactive compounds.
Topological virtual screening.
The topological model selected and constructed with the DF1 and DF2 functions was used to find new antimalarial agents from among a database of 479 compounds listed in the Merck index. This database was composed of drugs belonging to several therapeutic categories (antineoplastics, antivirals, antifungals, and antibacterials, etc.). A first selection step was performed by using the discriminant function DF1. The DF2 function was then used to sort the compounds selected as being active by DF1. PDDs were used to assign thresholds to discriminate active from inactive compounds with the highest probability of success. The compounds predicted to be active by the two equations (DF1 and DF2) within the preestablished cutoffs were considered to be potentially active. Among these, several commercially available compounds were assayed in vitro against the liver stage of P. yoelii yoelii.
MLR.
Multilinear regression (MLR) analysis based on molecular topology was used to predict the octanol/water partition constant (P) of each molecule, as this parameter is closely related to the intestinal absorption of a molecule after oral administration. P is the ratio of the concentration of the compound in octanol to the concentration of the compound in water and provides information on hydrophobicity (1, 16). For this prediction, the correlation between the calculated TI and the observed log P for 57 compounds was determined by MLR. The experimentally determined log P values of these compounds were obtained from the ChemIDplus database (National Library of Medicine). MLR analysis was performed with the 9R module of the BMDP program, which estimates regression equations for the best subsets of predictor variables by means of the Furnival-Wilson algorithm and provides detailed residual analysis. The lower Mallows Cp was used to identify the best subsets (Mallows' Cp = RSS/s2 − n + 2p′, where RSS is the residual sum of squares for the best test subsets, p′ is the number of independent variables in the subsets [including the constant], n is the number of cases, and s2 is the residual mean square based on the regression using all independent variables). The stability of the selected mathematical model can be evaluated through leave-one-out cross-validation: to do this, one compound of the set is extracted, and the model is recalculated using the remaining N − 1 compounds as a training set. The property is then predicted for the removed element. This process is repeated for all the compounds of the set, obtaining a prediction for every one.
In vitro antimalarial activity against the liver stage of Plasmodium.
The in vitro activity of drugs was studied against hepatic stages of P. yoelii yoelii, and only drugs with complete parasite inhibition were examined for activity against P. falciparum. Pharmacological tests were performed as previously described (17). Briefly, mouse or human hepatocytes were isolated by collagenase perfusion, and 105 hepatocyte cells were seeded in each well of Lab-Teck chamber slides, allowing cell confluence. They were incubated at 37°C with 4% CO2-96% air for 24 h before being challenged with 9 × 104 P. yoelii yoelii (265 By strain) or P. falciparum (NF54 strain) sporozoites obtained by the dissection of the salivary glands of artificially infected Anopheles stephensi mosquitoes. At the same time, drugs were added to duplicate cultures at seven concentrations ranging from 1 × 10−6 to 30 μM. Each day, the drugs were removed during the incubation period in order to ensure stable drug concentrations despite hepatocyte metabolism. Cells were incubated with the drugs for 48 h with P. yoelii yoelii and for 96 h with P. falciparum and then fixed with methanol. Parasites were revealed with polyclonal HSPi72 serum followed by goat anti-mouse immunoglobulin conjugated to fluorescein isothiocyanate. Schizonts were counted to determine the IC50 by linear regression, as previously described (17).
Toxicity assay.
Toxicity was determined by using the colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (22). Briefly, mouse hepatocyte cultures were prepared in 96-well plates as described above. Drugs were tested in triplicate wells at eight concentrations ranging from 6.4 × 10−3 μM to 100 μM. One hundred microliters of 0.5 mg/ml MTT (5 mg/ml MTT in water diluted 10× in William's medium without fetal bovine serum) was added to each well, and the plates were incubated for 4 h (37°C, 5% CO2). The medium was discarded, and the cells were resuspended in ethanol and dimethyl sulfoxide (50/50, vol/vol). The plates were read in an enzyme-linked immunosorbent assay plate reader (550-nm absorbance wavelength and 620-nm reference wavelength). The results were expressed as the percent change in viability compared to drug-free control cultures. Viability data were used to plot concentration-effect curves, from which 50% toxic concentrations (TC50) were estimated by linear regression. Each drug and concentration was tested at least in triplicate. The selectivity index (SI) was defined as the ratio of the TC50 to the IC50.
RESULTS
Mathematical modeling.
The test compounds were classified according to their structures and their activities against Plasmodium based on a mathematical model consisting of two equations. The first equation was designed to discriminate between active and inactive compounds, and the second was designed to identify the most active of the active compounds. The first equation, designated DF1, distinguished compounds that were predicted to have no significant in vitro activity against liver stages of P. yoelii yoelii (arbitrarily, IC50 of >25 μM) from those likely to be active (IC50 < 25 μM). The high cutoff of 25 μM was chosen in order to avoid discarding drugs with potential activity in the first screening step.
This equation comprised 11 independent variables: DF1 = 3.17 + 1.000χv + 1.281χv − 14.04J1v − 22.94J3v + 96.23J4v − 65.98J5 + 1.880D − 23.534Dc + 0.294Cc − 0.51PR3 +0.38V3.
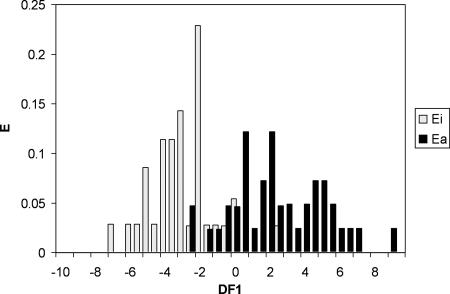
Statistical parameters accounting for the significance of this equation were as follows: N = 76, F = 7.95, and λ = 0.42. The quality of this discriminant function was evaluated by using Wilks parameter, λ. With this function, a compound is considered to be either active or inactive depending on its DF1 value. DF1 values of <0 and >0 predict that a compound will be inactive or active, respectively, within a 95% confidence interval. In the training group (76 compounds), 35 out of 41 experimentally active compounds were correctly classified (85.4% accuracy), and 32 out of 35 experimentally inactive compounds were correctly classified (91.4% accuracy). Cross-validation (jackknifed matrix) of the training group showed that 30 (85.7%) out of the 35 inactive compounds and 33 (80.5%) out of the 41 active compounds were correctly classified. The PDD (Fig. 1) showed that drugs with DF1 values between 0.5 and 10 were classified as being active and those with DF1 values between −8 and −0.5 were classified as being inactive. The classification was uncertain for drugs with values between −0.5 and 0.5. Drugs with values above 10 or below −8 were considered to be “unclassified”.
FIG. 1.
PDD of activity against the hepatic stage of Plasmodium from the discriminant function DF1 (Ea and Ei represent the expectancies or the probabilities of finding a compound to be active or inactive for each value of DF1, respectively). Shown is the PDD of antimalarial activity in the training group (76 compounds) obtained after LDA statistical treatment. Black bars, active drugs; gray bars, inactive drugs. The y axis represents the expectancy, and the x axis represents the value of DF1.
The second LDA equation, DF2, was used to discriminate weakly to moderately active compounds (1 μM < IC50 < 10 μM) from highly active compounds (IC50 < 1 μM). Once again, the 1 μM cutoff was arbitrarily chosen.
Six independent variables composed DF2, as follows: DF2 = 81.90 − 3.454χpv + 7.41G5v − 70.211C − 1.44PR1 − 1.21V3 + 2.44V4.
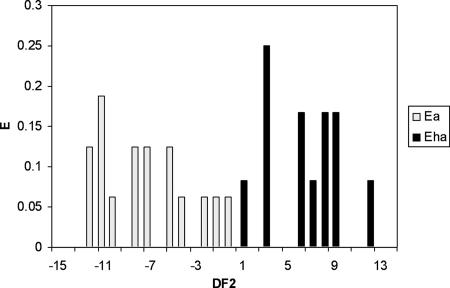
The statistical parameters were as follows: N = 28, F = 12.41, and λ = 0.21. By this function, a compound was considered to be active or highly active according the DF2 value. If DF2 was <0, the compound was predicted to be active, and if DF2 was >0, the compound was predicted to be highly active, with a 95% confidence interval. In the training set, consisting of 28 compounds with experimentally determined activities, 15 (93.8%) out of the 16 active compounds and 12 (100%) out of the 12 highly active compounds were correctly classified. Cross-validation of the training group showed that 14 (87.5%) out of the 16 active compounds and 12 (100%) out of the 12 highly active compounds were correctly classified. The corresponding PDD of this equation (Fig. 2) showed that compounds were classified as being highly active at DF2 values between 1 and 15 and as weakly or moderately active at values between −13 and 0. Drugs with values above 15 or below −13 were considered to be “unclassified”.
FIG. 2.
PDD of activity against the hepatic stage of Plasmodium from the discriminant function DF2 (Ea and Eha represent the expectancies or the probabilities of finding a compound to be active or highly active for each value of DF2, respectively). Shown is a PDD of antimalarial activity in the training group (28 compounds) obtained after LDA statistical treatment. Black bars, highly active drugs; gray bars, active drugs. The y axis represents the expectancy, and the x axis represents the value of DF2.
Topological virtual screening.
Based on the mathematical model, virtual topological screening was applied to a database of 479 heterogeneous drug molecules (antiviral, antineoplastic, antifungal, and cardiotonic, etc.). The model predicted that 62 (13%) of these compounds should be active against the liver stage of Plasmodium. Seventeen commercially available compounds (13 predicted to be active and 4 predicted to be inactive) were tested in vitro (see below) in order to assess the predictive capability of the model. The list of the 49 compounds predicted to be active and not tested in vitro is provided in Table S3 in the supplemental material. Table 1 illustrates the DF1 and DF2 values and the resulting classification of each compound on the basis of the PDD diagrams. Only one compound, delaverdine, could not be classified, because of an outlying DF1 value. Three compounds (monensin, nigericin, and vinblastine) were classified as being active by DF1 but had outlying DF2 values.
TABLE 1.
Predicted drug activity on liver stage of P. yoelii yoelii
| Drug (therapeutic category) | DF1a (class) | DF2b (class) | IC50 expc (nM) | TC50 expd (nM) | SI | Log Pe |
|---|---|---|---|---|---|---|
| Active drugs | ||||||
| Monensin (antibacterial/ionophore) | 4.22 (A) | −21.62 (NC) | <10−3 | 3.20 × 104 | >3.2 × 107 | 2.77 |
| Nigericin (ionophore) | 3.36 (A) | −24.22 (NC) | <10−3 | 1.32 × 104 | >1.32 × 107 | 3.78 |
| Delaverdine (antiviral) | −0.21 (NC) | 6.75 (HA) | 0.846 | >1 × 105 | >1.18 × 105 | 3.65 |
| Mibefradil (antihypertensive) | 7.59 (A) | 6.66 (HA) | 0.873 | 6.79 × 104 | 7.7 × 104 | 3.47 |
| Licochalcone A (estrogenic flavonoid) | 8.26 (A) | 7.99 (HA) | 0.927 | 6.34 × 104 | 6.83 × 104 | 2.57 |
| Miconazole (antifungal) | 1.46 (A) | 1.77 (HA) | 2.03 | 7.04 × 104 | 3.47 × 104 | 5.94 |
| Dobutamine (cardiotonic) | 5.74 (A) | 1.52 (HA) | 3.7 | >1 × 105 | >2.70 × 104 | 1.57 |
| Ritonavir (antiviral) | 8.80 (A) | −5.38 (A) | 34.2 | 6.73 × 104 | 1.96 × 103 | 6.71 |
| Saquinavir (antiviral) | 9.14 (A) | −6.76 (A) | 35.2 | 6.37 × 104 | 1.80 × 103 | 4.96 |
| Epoximicin (antineoplastic) | 8.17 (A) | 7.92 (HA) | 3.95 × 103 | 4.73 × 104 | 11.97 | 2.38 |
| Indinavir (antiviral) | 8.89 (A) | −10.55 (A) | 5 × 103 | 8.77 × 103 | 1.754 | 5.94 |
| Vinblastine (antineoplastic) | 1.21 (A) | 38.71 (NC) | 7.95 × 103 | 5.45 × 104 | 6.85 | 3.55 |
| Nordihydroguaiaretic acid (antineoplastic) | 9.03 (A) | 2.88 (HA) | 3 × 104 | >1 × 105 | >3.33 | 1.93 |
| Inactive drugs | ||||||
| Fenbendazole (antihelminthic) | −3.33 (I) | 3 × 104 | >1 × 105 | >3.33 | 3.63 | |
| Quinacrine (anthelminthic/antimalarial) | −0.93 (I) | 3 × 104 | >1.16 × 104 | >0.38 | 3.74 | |
| Rimandine (antiviral) | −2.69 (I) | 35.6 | >1 × 105 | >3.33 | 3.85 | |
| Thiophanote (anthelminthic) | −4.04 (I) | 3 × 104 | >1 × 105 | >3.33 | 1.93 | |
| Reference drugs | ||||||
| Atovaquone | 9.37 (A) | 3.63 (HA) | 57 | 16.5 × 103 | 289 | 3.67 |
| Primaquine | 0.91 (A) | 1.79 (HA) | 75.7 | 12.5 × 103 | 165 | 1.73 |
From discriminant function DF1.
From discriminant function DF2. IC50 exp, experimental IC50; TC50 exp, experimental TC50; log P calc, calculated log P.
Evaluation of antimalarial activity in vitro against the liver stage of P. yoelii yoelii as described previously by Mahmoudi et al. (17, 18).
Assessment in vitro by MTT assay.
Value of log P calculated with the MLR equation.
Prediction of the octanol/water partition constant (log P) by MLR analysis.
Given that log P is representative of the hydrophobicity and oral absorption of a molecule, we developed a topological equation capable of predicting log P. The equation was built up from a set of 57 structurally heterogeneous compounds for which experimental log P values were available. Results were validated using the leave-one-out cross-validation.
The best predictive equation was as follows: log P = 13.67 + 9.79J1 − 0.520D − 14.340C + 0.144Cc + 0.29V3.
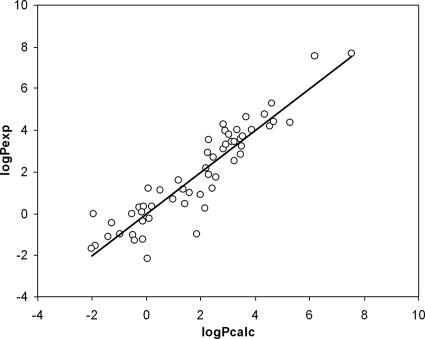
The statistical parameters were as follows: r2 = 0.85, r2cv, = 0.79, standard error = 0.92, F-stat = 59.0, and P (significance) < 0.00001. Figure 3 shows the good correlation obtained between experimental and calculated log P values. Detailed results for experimental and predicted log P values of these 57 compounds are provided in Table S2 in the supplemental material. Using this equation, we estimated log P values for selected antimalarial compounds. Values ranged between 1.57 and 6.71, reflecting a wide range of hydrophobicity among effective compounds.
FIG. 3.
Experimental versus calculated log P values obtained from MLR analysis. Shown is a comparison between experimental (y axis) and calculated (x axis) log P values for 57 compounds.
In vitro antimalarial activity against the liver stage of Plasmodium.
Table 1 shows the in vitro activities of the 17 compounds selected for in vitro testing. Twelve out of the 13 compounds predicted to be active showed an inhibitory effect on parasite growth, with IC50 values ranging between 1 picomolar and 7.95 μM. The IC50 values of primaquine and atovaquone, used as references, were 75.7 and 57 nM, respectively. Monensin and nigericin showed complete parasite inhibition, with picomolar IC50 values. Seven compounds had IC50 values below 50 nM (delaverdine, midefradil, dobutamine, licochalcone A, miconazole, saquinavir, and ritonavir), and three compounds had IC50 values below 8 μM (indinavir, vinblastine, and epoximicin). Only one compound (nordihydroguaiaretic acid) that was predicted to be active was found to be inactive (IC50 > 30 μM). Among the four compounds predicted to be inactive, three were completely inactive in vitro (IC50 > 30 μM) (fenbendazole, quinacrine, and thiophanote), and one, rimantadine, was active (IC50 of 35.6 nM). The most active compounds, monensin and nigericin, were examined for activity against P. falciparum. The same profile of activity was found with a complete inhibitory effect at picomolar concentrations (IC50 < 10−3 nM).
Toxicity assay.
The drugs tested for in vitro activity on P. yoelii yoelii were also examined for their toxicity on hepatocyte cells (MTT assay). TC50 values are shown in Table 1. None of the 13 compounds with antimalarial activity was toxic for hepatocytes (TC50 > 5 μM). An SI was then calculated as TC50/IC50 (Table 1). Ten of the 13 active compounds had an SI above 1,000. Three compounds had SI values between 1.77 and 12 (indinavir, vinblastine, and epoximicin). The primaquine and atovaquone SIs were 165 and 289, respectively.
DISCUSSION
The aim of this study was to build up a QSAR model suitable for screening a large molecular database for compounds that are active on Plasmodium liver stages.
Based on molecular topology, a mathematical model was built up from a training set of 127 compounds whose activities against hepatic stages of Plasmodium had previously been determined in vitro. This training set was comprised of heterogeneous molecular structures and a larger number of inactive than active molecules. The precise experimental IC50 values of several compounds were not available from published sources. Because of this numerical imbalance between active and inactive compounds and the lack of firm IC50 values, we had to build a stepwise model, as no single equation could reliably select active compounds.
The first equation (DF1) discriminated between inactive and active compounds by using a cutoff of 25 μM, while the second equation (DF2) selected highly active compounds among all active compounds (IC50 < 1 μM). The discriminant function DF1 classified 83% of compounds in the training group correctly. There was little overlap between the active and inactive groups, but several compounds had DF1 values between −0.5 and 0.5, a range where no firm conclusions could be drawn. The second equation, DF2, classified about 97% of compounds in the training set correctly. Again, there was little overlap between the groups, confirming the quality of the discriminant function.
The significant role played by connectivity and topological charge indices in the different discriminant functions achieved must be emphasized.
The mathematical model was then applied to a database of 479 drugs with unknown activity against hepatic stages of Plasmodium. Despite a wide diversity of molecular structures and activities, the model predicted that 62 drugs (13%) would be active. Nine of the 13 compounds chosen for in vitro testing showed remarkable antimalarial activity. By comparison to the reference drugs primaquine and atovaquone, which had IC50s of 75.7 nM and 57 nM, respectively, these nine compounds were 10- to 1,000-fold more potent in vitro. The most active compounds against the hepatic stages of P. yoelii yoelii and P. falciparum were monensin and nigericin, with IC50s of <10−3 nM. These results demonstrate the effectiveness of the topological model described herein for identifying new potential antimalarial drugs. However, the mathematical prediction was confirmed by the in vitro results only, and a confirmation of the in vivo activity will permit the validation of the mathematical model.
We have also examined the cellular toxicities of these drugs and calculated an SI based on the ratio between activity (IC50) and toxicity (TC50). Ten compounds had TC50 values above those of primaquine and an SI above 103, compared to 165 for primaquine and 289 for atovaquone.
We have also designed a mathematical model capable of predicting the octanol/water partition constant of these molecules, as this parameter is indicative of hydrophobicity. MLR analysis yielded an equation capable of predicting the log P value. The predicted values were indicative of good intestinal absorption with most of the compounds tested (log P between 1.5 and 3.5).
A search of the literature conducted after the experimental phase showed that none of these compounds had previously been shown to have antimalarial properties against the hepatic stage.
Molecular topology has some drawbacks; for example, it takes into account only the two-dimensional molecular structure, and it does not distinguish stereoisomers. However, based on our results and the known modes of action of several drugs that we found to inhibit Plasmodium, several targets can be highlighted, such as aspartic proteases (inhibited by human immunodeficiency virus [HIV] protease inhibitors); K+, Na+, and Ca2+ channels (inhibited by nigericin, monensin, and mibefradil, respectively); microtubules (inhibited by vinblastine); the proteasome (inhibited by epoximicin); and fumarate reductase (inhibited by licochalcone A).
Some of these drugs were previously shown to be active on the erythrocytic stage of Plasmodium. Licochalcone A was tested in vitro against blood stages of P. falciparum and in vivo against P. yoelii (28). The latter experiment used synchronous cultures and strongly suggested that the main effect of licochalcone A is to inhibit erythrocytic invasion by merozoites and/or to inhibit the initial growth of internalized merozoites. Intravenous licochalcone A administration reduced parasitemia in mice infected by P. yoelii.
Adovelande and Schrevel (3) demonstrated that monensin and nigericin exhibited intrinsic antimalarial activities at picomolar levels in vitro and in vivo. Our results with monensin and nigericin suggest that these drugs could be used to block transmission, as we obtained 100% inhibition of the hepatic stage of Plasmodium in vitro. Recently, Skinner-Adams et al. (24) reported that HIV protease inhibitors such as saquinavir, ritonavir, and indinavir directly inhibited the growth of erythrocytic stages of chloroquine-sensitive and chloroquine-resistant P. falciparum strains in vitro at clinically relevant concentrations. Further studies are required to determine the activities of HIV protease inhibitors against malaria in vivo. It would be interesting to examine the possible impacts of these drugs on HIV-infected patients living in areas of endemicity.
In conclusion, a combination of structural description by using TIs and statistical treatment by LDA can reliably select new compounds that are effective against the liver stages of Plasmodium. The predictive model thus obtained can readily be applied to large databases of drugs in order to identify active structures. These results confirm the utility of molecular topology as a powerful tool in the search for new antimalarial drugs.
The in vitro validation of the model was performed using cultures of P. yoelii yoelii, but we can reasonably assume that such results can be extrapolated to P. falciparum (most of the molecules known so far to prevent hepatic development are efficient in both rodent and human Plasmodium infections) (17, 26). Furthermore, we have tested two ionophores in vitro (monensin and nigericin) against the P. falciparum liver stage, and complete inhibition of parasite development was observed. However, there are exceptional examples of discrepancies between human and rodent species while inhibiting the development of P. yoelii in vitro and in vivo (20); for instance, doxycycline has never been shown to have any effect on the hepatic multiplication of P. falciparum in humans in areas of endemicity (23).
Supplementary Material
Acknowledgments
Nassira Mahmoudi was financially supported by the Ministère de l'Education Nationale, de la Recherche, et de la Technologie and by the Raoul Follereau Association. We acknowledge financial support from the following sources: Cooperation Franco-Espagnole en Recherche Médicale, accord INSERM-CSIC projet conjoint-2002-2005; Projet PAL and Ministère de la Recherche, Spanish Red de Investigacion de Centros de Enfermedades Tropicales, RICET, (C03/04); and project SAF2005-PI052128 of the Fondo de Investigación Sanitaria, Ministerio de Sanidad, Spain, and Impact Malaria, Sanofi-Aventis.
We thank Enrico Lazaro for providing the crambesicine analogue.
Footnotes
Published ahead of print on 22 January 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Abraham, M. H., and Y. H. Zhao. 2004. Determination of solvation descriptors for ionic species: hydrogen bond acidity and basicity. J. Org. Chem. 69:4677-4685. [DOI] [PubMed] [Google Scholar]
- 2.Adak, T., N. Valecha, and V. P. Sharma. 2001. Plasmodium vivax polymorphism in a clinical drug trial. Clin. Diagn. Lab. Immunol. 8:891-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adovelande, J., and J. Schrevel. 1996. Carboxylic ionophores in malaria chemotherapy: the effects of monensin and nigericin on Plasmodium falciparum in vitro and Plasmodium vinckei petteri in vivo. Life Sci. 59:PL309-PL315. [DOI] [PubMed] [Google Scholar]
- 4.Balaban, A. T. 1983. Topological indices based topological distances in molecular graphs. Pure Appl. Chem. 55:199-206. [Google Scholar]
- 5.de Gregorio-Alapont, C., R. García-Domenech, J. Gálvez, M. J. Ros, S. Wolski, and M. D. García. 2000. Molecular topology: a useful tool for the search of new antibacterials. Bioorg. Med. Chem. Lett. 10:2033-2036. [DOI] [PubMed] [Google Scholar]
- 6.de Julian-Ortiz, J. V., J. Galvez, C. Munoz-Collado, R. Garcia-Domenech, and C. Gimeno-Cardona. 1999. Virtual combinatorial syntheses and computational screening of new potential anti-herpes compounds. J. Med. Chem. 42:3308-3314. [DOI] [PubMed] [Google Scholar]
- 7.Galvez, J., R. Garcia-Domenech, C. de Gregorio Alapont, J. V. de Julian-Ortiz, and L. Popa. 1996. Pharmacological distribution diagrams: a tool for de novo drug design. J. Mol. Graph. 14:272-276. [DOI] [PubMed] [Google Scholar]
- 8.Galvez, J., R. Garcia-Domenech, J. V. de Julian-Ortiz, and R. Soler. 1995. Topological approach to drug design. J. Chem. Inf. Comput. Sci. 35:272-284. [DOI] [PubMed] [Google Scholar]
- 9.Galvez, J., R. Garcia-Domenech, M. T. Salabert, and R. Soler. 1994. Charge indices. New topological descriptors. J. Chem. Inf. Comput. Sci. 34:520-525. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Garcia, A., J. Galvez, J. V. de Julian-Ortiz, R. Garcia-Domenech, C. Munoz, R. Guna, and R. Borras. 2004. New agents active against Mycobacterium avium complex selected by molecular topology: a virtual screening method. J. Antimicrob. Chemother. 53:65-73. [DOI] [PubMed] [Google Scholar]
- 11.Gozalbes, R., M. Brun-Pascaud, R. Garcia-Domenech, J. Galvez, P. M. Girard, J. P. Doucet, and F. Derouin. 2000. Anti-Toxoplasma activities of 24 quinolones and fluoroquinolones in vitro: prediction of activity by molecular topology and virtual computational techniques. Antimicrob. Agents Chemother. 44:2771-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gozalbes, R., M. Brun-Pascaud, R. Garcia-Domenech, J. Galvez, P. M. Girard, J. P. Doucet, and F. Derouin. 2000. Prediction of quinolone activity against Mycobacterium avium by molecular topology and virtual computational screening. Antimicrob. Agents Chemother. 44:2764-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kier, L. B., and L. H. Hall. 1983. General definition of valence delta-values for molecular connectivity. J. Pharm. Sci. 72:1170-1173. [DOI] [PubMed] [Google Scholar]
- 14.Kier, L. B., and L. H. Hall. 1986. Molecular connectivity in structure-activity analysis. John Wiley & Sons, Letchworth, England.
- 15.Lahana, R. 1997. Chimie combinatoire virtuelle. Sci. Am. Ed. Française 241:56-58. [Google Scholar]
- 16.Leo, G., and G. Wersig. 1971. Specialty-specific marked reports in anamnesis—development and initial experiences. Methods Inf. Med. Suppl. 5:219-229. (In German.) [PubMed] [Google Scholar]
- 17.Mahmoudi, N., L. Ciceron, J.-F. Franetich, K. Farhati, O. Silvie, W. Eling, R. Sauerwein, M. Danis, D. Mazier, and F. Derouin. 2003. In vitro activities of 25 quinolones and fluoroquinolones against liver and blood stage Plasmodium spp. Antimicrob. Agents Chemother. 47:2636-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmoudi, N., J. V. de Julian-Ortiz, L. Ciceron, J. Galvez, D. Mazier, M. Danis, F. Derouin, and R. Garcia-Domenech. 2006. Identification of new antimalarial drugs by linear discriminant analysis and topological virtual screening. J. Antimicrob. Chemother. 57:489-497. [DOI] [PubMed] [Google Scholar]
- 19.Marrero-Ponce, Y., R. Medina-Marrero, F. Torrens, Y. Martinez, V. Romero-Zaldivar, and E. A. Castro. 2005. Atom, atom-type, and total nonstochastic and stochastic quadratic fingerprints: a promising approach for modeling of antibacterial activity. Bioorg. Med. Chem. 13:2881-2899. [DOI] [PubMed] [Google Scholar]
- 20.Marussig, M., A. Motard, L. Renia, D. Baccam, J. Lebras, G. Charmot, and D. Mazier. 1993. Activity of doxycycline against preerythrocytic malaria. J. Infect. Dis. 168:1603-1604. [DOI] [PubMed] [Google Scholar]
- 21.Mishra, R. K., R. Garcia-Domenech, and J. Galvez. 2001. Getting discriminant functions of antibacterial activity from physicochemical and topological parameters. J. Chem. Inf. Comput. Sci. 41:387-393. [DOI] [PubMed] [Google Scholar]
- 22.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 23.Shmuklarsky, M. J., E. F. Boudreau, L. W. Pang, J. I. Smith, I. Schneider, L. Fleckenstein, M. M. Abdelrahim, C. J. Canfield, and B. Schuster. 1994. Failure of doxycycline as a causal prophylactic agent against Plasmodium falciparum malaria in healthy nonimmune volunteers. Ann. Intern. Med. 120:294-299. [DOI] [PubMed] [Google Scholar]
- 24.Skinner-Adams, T. S., J. S. McCarthy, D. L. Gardiner, P. M. Hilton, and K. T. Andrews. 2004. Antiretrovirals as antimalarial agents. J. Infect. Dis. 190:1998-2000. [DOI] [PubMed] [Google Scholar]
- 25.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434:214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahel, E., D. Mazier, A. Guillouzo, F. Miltgen, I. Landau, S. Mellouk, R. L. Beaudoin, P. Langlois, and M. Gentilini. 1988. Iron chelators: in vitro inhibitory effect on the liver stage of rodent and human malaria. Am. J. Trop. Med. Hyg. 39:236-240. [DOI] [PubMed] [Google Scholar]
- 27.Walsh, D. S., C. Eamsila, T. Sasiprapha, S. Sangkharomya, P. Khaewsathien, P. Supakalin, D. B. Tang, P. Jarasrumgsichol, C. Cherdchu, M. D. Edstein, K. H. Rieckmann, and T. G. Brewer. 2004. Efficacy of monthly tafenoquine for prophylaxis of Plasmodium vivax and multidrug-resistant P. falciparum malaria. J. Infect. Dis. 190:1456-1463. [DOI] [PubMed] [Google Scholar]
- 28.Ziegler, H. L., H. S. Hansen, D. Staerk, S. B. Christensen, H. Hagerstrand, and J. W. Jaroszewski. 2004. The antiparasitic compound licochalcone a is a potent echinocytogenic agent that modifies the erythrocyte membrane in the concentration range where antiplasmodial activity is observed. Antimicrob. Agents Chemother. 48:4067-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.