Abstract
The switch from latent to lytic infection of Kaposi's sarcoma-associated herpesvirus is initiated by the immediate early transcriptional activator protein Rta/open reading frame 50 (ORF50). We examined the transcriptional regulation of the ORF50 core promoter in response to lytic cycle stimulation. We show that the ORF50 promoter is highly responsive to sodium butyrate (NaB) and trichostatin A (TSA), two chemicals known to inhibit histone deacetylases. The NaB and TSA responsive element was mapped to a 70-bp minimal promoter containing an essential GC box that binds Sp1/Sp3 in vitro and in vivo. Micrococcal nuclease mapping studies revealed that a nucleosome is positioned over the transcriptional initiation and the Sp1/3 binding sites. Stimulation with NaB or TSA increased histone acetylation and restriction enzyme accessibility of the ORF50 promoter transcription initiation site. Chromatin immunoprecipitation assay was used to demonstrate that the ORF50 promoter is associated with several different histone deacetylase proteins (including HDAC1, 5, and 7) in latently infected cells. NaB treatment led to the rapid association of Ini1/Snf5, a component of the Swi/Snf family of chromatin remodeling proteins, with the ORF50 promoter. Ectopic expression of the CREB-binding protein (CBP) histone acetyltransferase (HAT) stimulated plasmid-based ORF50 transcription in a HAT-dependent manner, suggesting that CBP recruitment to the ORF50 promoter can be an initiating event for transcription and viral reactivation. Together, these results suggest that remodeling of a stably positioned nucleosome at the transcriptional initiation site of ORF50 is a regulatory step in the transition from latent to lytic infection.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is a gamma-2 herpesvirus (also referred to as human herpesvirus 8) that persists as a multicopy episome in latently infected B lymphocytes (reviewed in references 1, 12, 20, 25, 32, and 60). KSHV was originally isolated from KS lesions, and numerous studies support the role of the virus as the causative agent of this endothelial neoplasm, including the isolation of KSHV DNA from endothelial and tumor-derived spindle cells and the ability to produce KS-like endothelial malignancy in transgenic animals (9, 29). KSHV has also been associated with two other lymphoproliferative disorders, primary effusion lymphomas (PELs) and multicentric Castleman's disease (13, 16, 62). Like Epstein-Barr virus (EBV), KSHV has distinct lytic and latent DNA replication and gene expression programs in lymphocytes (7, 55). Both latent and lytic cycle replication are essential for the long-term persistence of the virus, and gene products from both expression programs have been implicated in the pathogenesis of KSHV-associated disease (12, 32).
Several cell lines carrying latent episomal genomes of KSHV have been isolated from PEL or KS tumors, and these have been useful tools for the study of viral latent and lytic replication and gene expression (11, 26, 27, 55). During latency in PEL cells, KSHV transcription is restricted to a few viral genes, including the latency-associated nuclear antigen, v-FLIP, v-CycD, kaposin, and vIRF-2 (10, 24, 34, 46, 54, 57). The latency-associated viral products have been shown to have growth-transforming and cell cycle-deregulating properties likely to contribute to KSHV-associated malignancies (12, 20, 35). Lytic cycle replication and gene expression have also been implicated in KSHV-associated disease. Lytic infection can be detected in subpopulations of KS endothelial cells (64), and lytic cycle genes encode several proteins with homology to cellular cytokines that are likely to contribute to KSHV-associated pathology (6, 8, 44, 49, 50). Furthermore, drugs that inhibit KSHV lytic replication, like foscarnet and ganciclovir, limit clinical KS development (40, 45). An increase in KSHV viral load correlates with the progression to KS (5, 70), and it is likely that the lytic amplification increases the risk of KSHV endothelial cell infections.
Lytic cycle replication of KSHV in B lymphocytes can be initiated by ectopic expression of the open reading frame 50 (ORF50)-encoded immediate early protein Rta (28, 39, 66). KSHV Rta, like its homolog in EBV, is a potent transcriptional activator with sequence-specific DNA binding potential (38, 58, 61). KSHV Rta has been reported to autoactivate its own expression (22) through association with an octamer binding site (58) and to interact with several cellular proteins, including the CREB-binding protein (CBP) and RBP-Jk (30, 36). In cultured cells, KSHV reactivation can be initiated with pleiotropic agents, like phorbol esters and sodium butyrate (NaB) (14, 22, 42, 43, 55), or a DNA demethylating agent, 5′-azacytidine (18). The physiological signals leading to the activation of the ORF50 promoter regulating Rta transcription have not been clearly defined, although reactivation of KSHV latently infected PEL cells has been stimulated by coinfection with human immunodeficiency virus (68), incubation with human immunodeficiency virus-infected cell supernatants (69), and treatment with the inflammatory cytokines gamma interferon and oncostatin M (15).
In KSHV latently infected cells, ORF50 gene expression is highly restricted, suggesting that transcriptional repression of ORF50 is required to maintain the latent state (33). Chromatin modification and nucleosome positioning have been shown to play significant roles in the maintenance of stable gene expression patterns, especially in the heritable repression of transcription (reviewed in references 4 and 67). To explore the potential role of chromatin in the maintenance of KSHV latency, we investigated the transcriptional regulation and chromatin organization of ORF50 in latently infected and lytically stimulated cells. We found that ORF50 is highly responsive to histone deacetylase inhibitors in KSHV-positive and -negative cells and that a nucleosome is positioned directly over the transcriptional initiation site. We found that cellular factors Sp1/Sp3 can bind to the sequences just upstream of the transcription initiation site and within the boundary of the nucleosomal DNA. Our results raise the possibility that Sp1/Sp3 contributes to stable nucleosome positioning and transcriptional repression in latently infected PEL cells and that modification and remodeling of this nucleosome is required for ORF50 transcription and KSHV lytic cycle initiation.
MATERIALS AND METHODS
Cells and culture conditions.
293 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and penicillin-streptomycin (50 U/ml). The lymphoblastoid cell lines BCBL-1, JSC-1, and DG75 were maintained in RPMI containing 10% fetal bovine serum and penicillin-streptomycin (50 U/ml). Cells were stimulated with 3 mM NaB (Sigma), or 1 μg of trichostatin A (TSA; Upstate Biotechnology)/ml, or 50 ng of phorbol-12-tetradecanoate-13-acetate (TPA; Sigma)/ml.
Plasmids.
P50 was constructed by cloning a PCR-generated DNA fragment of the ORF50 promoter sequence into the pGL3-Basic vector. The PCR was performed with primers P50-1 (5′-ATTACCATGGTTGTGGCTGCCTGGACAGT-3′) and P50-2 (5′-ATAACTCGAGCTTCCACGTTGATCCGGCTT-3′). The PCR product was digested with NcoI and XhoI and cloned into NcoI/XhoI-digested pGL3-Basic vector (Promega). The restriction sites in these oligonucleotides are underlined. The insert is 1,038 bp long, corresponding to KSHV genomic positions 70561 to 71598 (nuclear numbers are according to Russo et al.) (56).
The ORF50 promoter deletion mutant −298 was constructed by digesting the original P50 with KpnI followed by religation. The −298 ORF50 contained a 298-bp insert corresponding to genomic positions 71301 to 71598. All other deletion mutants were generated by PCR with Vent DNA polymerase and oligonucleotide primers with KpnI sites for 5′ amplification and NcoI sites for 3′ amplification. Primers were initiated at sequences relative to the ORF50 ATG (genomic position 71598) as indicated (see Fig. 3). Site-directed mutations were generated by using the QuikChange method (Stratagene, Inc.), inserting either a BamHI or EcoRI site into the sequences as indicated (see Fig. 3B). The parental plasmids for the substitution mutation were the 5′ −146 deletion and the 5′ −298 ORF50 deletion mutants. The results shown in Fig. 3 were obtained with 5′ −146, but similar results were observed with 5′ −298. The K8 immediate early promoter (PK8-IE500) was constructed by cloning a PCR-generated 505-bp fragment from KSHV genomic positions 74125 to 74629. The DNA fragment was cloned into SacI/SmaI-digested pGL3-Basic vector. The K8 delayed early promoter (pK8-DE250) was constructed by cloning a PCR-generated 254-bp fragment from KSHV genomic positions 74596 to 74849 into KpnI/SmaI-digested pGL3-Basic vector.
FIG. 3.
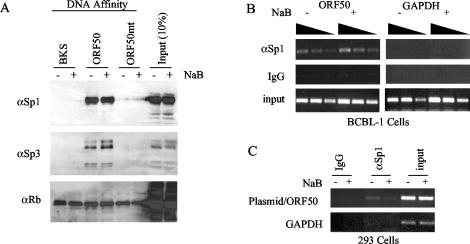
Sp1/3 binds to the ORF50 promoter in vitro and in vivo. (A) BCBL-1-derived nuclear extracts from NaB-treated (+) or untreated (−) cells were subject to DNA affinity purification with DNA derived from pBKSII (BKS), ORF50 (−298/+1), or the GC-box substitution mutant of ORF50 (−114) (ORF50mt). Bound proteins were assayed by Western blotting of SDS-polyacrylamide gel electrophoresis gels with antibodies specific for Sp1 (αSp1), Sp3 (αSp3), or Rb (αRb) as indicated to the left of each panel. (B) NaB-treated (+) or untreated (−) cells were analyzed by ChIP with antibodies specific for Sp1 or control IgG. Promoter fragments from the ORF50 promoter (left panel) or for the coding sequences of GAPDH (right panel) were generated by PCR. Input DNA is indicated in the lower panel. Threefold dilutions of input DNA were used to determine the linearity of PCRs. (C) 293 cells transfected with ORF50-Luc plasmid were assayed by ChIP for association with Sp1 before and after NaB treatment.
The BZLF1-luciferase reporter plasmid has been described previously (17) and consists of the sequences −220 to +12 cloned into the NheI-HindIII sites of pGL3-Basic. Cytomegalovirus (CMV)-Flag-CBP, CMV-Flag-CBP-ΔHAT (histone acetyltransferase), and CMV-p300 were gifts of G. Blobel (Children's Hospital of Pennsylvania, Philadelphia, Pa.) and have been described previously (17). CMV-TIP60 has been described previously (21). CMV-hGCN5 was described previously (41). CMV-P/CAF was a gift from Y. Nakatani (Harvard) (73).
RT-PCR.
RNA was obtained from BCBL-1 cells either untreated or treated with TSA (1 μg/ml), NaB (3 mM), or TPA (50 ng/ml) for 24 h by using the RNeasy kit (Qiagen). After treatment with DNase I, the OneStep reverse transcription (RT)-PCR kit (Qiagen) was used to analyze RNA. Reverse transcribed DNA was denatured at 94°C for 1 min, annealed with primers at 52°C for 1 min, and extended at 72°C for 1 min for 20 cycles. The products were analyzed on a 1.2% agarose gel.
Transfections and luciferase assay.
All cells were transfected with Lipofectamine 2000 (Invitrogen, Inc.) according to the manufacturer's specifications. Typical transfections of BCBL-1 or 293 cells involved the introduction of 0.5 μg of plasmid effector and reporter DNA for 105 cells. Cells were treated with 1 mM NaB until 24 h posttransfection and then harvested at 48 h posttransfection. Luciferase activity was assayed by using the Promega system. All data points were the averages of at least three independent transfections.
DNA affinity purification assay.
Biotinylated DNA was generated with a biotin-conjugated oligonucleotide and PCR amplification of sequences derived from either the ORF50 promoter (−298 to +1), ORF50 substitution mutant −114, or from pBSKII+ polylinker region. Biotinylated DNA was conjugated to streptavidin-linked magnetic beads (Dynal) essentially as described previously (23). DNA affinity resin was incubated with nuclear extracts from BCBL-1 cells dialyzed in D150 buffer (20 mM HEPES [pH 7.9], 20% glycerol, 1 mM EDTA, 150 mM KCl, 5 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and with 400 μg of competitor sonicated salmon sperm DNA/ml. After 45 min of incubation at 25°C, the resin was washed three times with D150 containing 0.05% IPEGAL (Sigma) and then eluted with D1000 buffer (identical to D150, except containing 1 M KCl). Eluted proteins were analyzed by Western blotting.
Western blotting and antibodies.
All Western blots were preblocked with 5% dried milk in Tris-saline (20 mM Tris [pH 8.0], 200 mM NaCl), incubated with antibody in the blocking agent (diluted from 1:500 to 1:1,000), and washed four times with Tris-saline containing 0.05% IPEGAL. Secondary antibodies were diluted 1:2,000 in blocking agent, washed as above, and then detected by enhanced chemiluminescence (Amersham). Antibodies to Sp1, Sp3, Rb, and Ini1/Snf5 were purchased from Santa Cruz Biotech. Antibodies for histone deacetylases 1 to 7 (HDAC1 to 7) were purchased from Upstate Biotechnology.
ChIP assay.
Chromatin immunoprecipitation (ChIP) protocol was essentially as described previously (52). PEL cell lines (BCBL-1 and JSC1) or transfected 293 cells were treated with or without NaB (3 mM) for 4 h and then treated with formaldehyde to a final concentration of 1% for 15 min at 25°C. Glycine was added to a final concentration of 0.125 M, and the mixture was incubated for an additional 5 min. The cells were washed in phosphate-buffered saline and resuspended in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.0], 167 mM NaCl, 1 mM PMSF, and protease inhibitor cocktail for mammalian cells [Sigma]). Lysates were sonicated to produce an average DNA fragment length of ∼300 bp. Extracts were then diluted 10-fold with immunoprecipitation (IP) dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.0], 167 mM NaCl, 1 mM PMSF, and protease inhibitor cocktail [Sigma]). Twenty microliters of the diluted sample was used to isolated total input controls. For IP, 1 ml of diluted sonicated extract was incubated with 2.5 μg of the appropriate antibody and incubated overnight at 4°C. For some antibodies, bovine serum albumin was added to a concentration of 100 μg/ml for each reaction mixture. Protein G-Sepharose beads were preblocked overnight at 4°C in IP buffer (0.1% sodium deoxycholate, 0.5% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM PMSF, 1× protease inhibitor cocktail [Sigma]) containing 500 μg of heat-denatured salmon sperm DNA/ml and 500 μg of bovine serum albumin/ml. To collect the immune complex, 50 ml of blocked protein G bead mixture was added to each reaction mixture and the mixture was rotated for 2 to 3 h at 4°C. Beads were centrifuged and washed for 10 min at 4°C with each of the following: 2× IP buffer, 2× IP buffer plus 600 mM NaCl, 1× LiCl wash (250 mM LiCl, 0.5% NP-40, 0.1% Na deoxycholate, 5 mM EDTA, 10 mM Tris-Cl [pH 8.0], 1 mM PMSF, 1× protease inhibitor cocktail [Sigma]), 2× Tris-EDTA (TE) buffer (Tris-Cl [pH 8.0], 1 mM EDTA). The immune complexes were eluted by incubation in 150 ml of TE-1% SDS at 65°C for 15 min, and supernatants were isolated and further incubated overnight at 65°C to reverse cross-linking. Total input controls were treated in the same manner at this point. After reverse cross-linking, 150 μl of TE and proteinase K (final concentration, 250 μg/ml) was added and the mixture was incubated for 2 to 3 h at 50°C. DNA was deproteinized by phenol-chloroform extraction and ethanol precipitation in the presence of 20 μg of glycogen. DNA was washed in 70% ethanol, dried, and resuspended in 25 ml of TE. For a typical PCR, 0.5 to 4 μl of IP DNA was assayed for 22 to 25 cycles and visualized by ethidium bromide staining of agarose gels. Primers for ORF50 were from genomic position 71301 (GGTACCGAATGCCACAATCTGTGCCCT) to the ATG at genomic position 71598 (ATGGTTTGTGGCTGCCTGGACAGTATTC). Primers for ORF72 were oPL821 (AATACAACCTAGAACCTAACGTGGTCG) and oPL823 (GAAGTGACGTCCGTCGCTAAGA), and primers for ORF73 were oPL824 (CCAGACTCTTCAACACCTATGCG) and oPL825 (GGATGATCCCACGTAGATCGG). Primers for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) coding sequence were 5′-TCACCACCATGGAGAAGGCT and 5′-GCCATCCACAGTCTTCTGGG.
Nucleosome mapping assays. (i) Primer extension.
JSC-1 cells or transfected 293 cells were untreated or treated with NaB (3 mM) for 6 or 24 h. Nucleus extraction and primer extension were performed as described previously (37) with some modifications. Briefly, the cells were harvested and disrupted in a Dounce homogenizer in lysis buffer (0.3 M sucrose, 2 mM magnesium acetate, 3 mM CaCl2, 1% Triton X-100, and 10 mM HEPES [pH 7.9]). The lysate was then spun through a pad (25% glycerol, 5 mM magnesium acetate, 0.1 mM EDTA, and 10 mM HEPES [pH 7.4]) at 1,000 × g for 15 min at 4°C. Then the nuclei were overdigested with micrococcal nuclease (Mnase) (500 U/ml) at 37°C for 20 min in the digestion buffer (25 mM KCl, 4 mM MgCl2, 1 mM CaCl2, 50 mM Tris [pH 7.4], and 12.5% glycerol). The reaction was stopped by the addition of an equal volume of stop buffer (2% SDS, 0.2 M NaCl, 10 mM EDTA, 10 mM EGTA, 50 mM Tris [pH 8.0], and proteinase K [100 μg/ml]) for 2 h at 50°C. Then the cDNA was extracted by phenol-chloroform and ethanol precipitation. The 150-bp DNA was then purified from a 1.5% agarose gel and purified by electroelution. Primer extension was performed on the purified DNA with Vent (exo-) DNA polymerase (New England Biolabs) and 32P-labeled sequence-specific primers. The product was analyzed on a 7% sequencing gel. Primer 1 (Pr1) initiated leftward at the KSHV sequence genomic position 71591 (5′-GTGGCTGCCTGGACAGTATTCTCACAAC), and primer 2 (Pr2) initiated rightward at the KSHV sequence genomic position 71491 (5′-CCAGAAACCAGTAGCTGGGTGGCAA).
(ii) Southern blotting.
Nuclei prepared as described above were partially digested with Mnase (50 U/ml, 37°C for 2 min). DNA purified as described above was then subjected to digestion with Asp718 (50 U/ml, 37°C overnight), electrophoresed on a 2% agarose gel followed by transfer to Zeta-Probe blotting membranes (Bio-Rad), and hybridized with a digoxigenin-labeled probe specific for the ORF50 promoter (Roche). The membrane was developed by using a digoxigenin detection kit (Roche).
(iii) Restriction enzyme accessibility assay.
Cell nuclei were prepared as described above. Then nuclei were resuspended in the New England Biolabs restriction digestion buffer for enzyme digestion. The total volume was 50 μl. Restriction enzyme digestion was performed at 37°C for 30 min with 20 U of restriction enzyme (HincII or Asp718) and stopped by the addition of stop buffer. After incubation at 50°C for 2 h with proteinase K, DNA was extracted and cut with another two restriction enzymes (NcoI and EcoRV) and then analyzed by Southern blotting.
RESULTS
KSHV ORF promoter is highly responsive to NaB treatment.
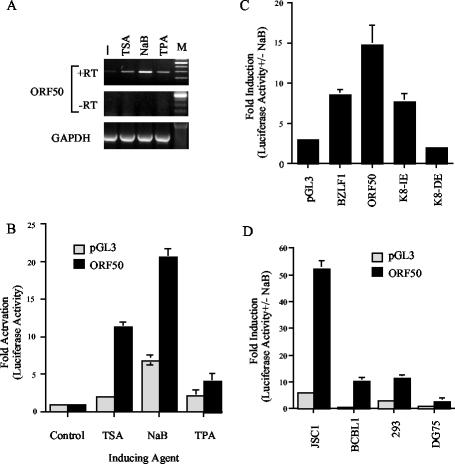
Latently infected PEL cells have been reported to undergo lytic cycle gene expression and DNA replication when treated with NaB or phorbol esters. We compared the effects on ORF50 RNA levels in BCBL-1 cells treated with either 3 mM NaB, 1 μg of TSA/ml, or 50 ng of TPA/ml for 24 h (Fig. 1A). We found that ORF50 RNA levels were increased most significantly when BCBL-1 cells were treated with NaB and TSA, suggesting that HDAC inhibition plays a significant role in lytic reactivation. To determine whether a plasmid-borne ORF50 promoter retained responsiveness to these HDAC inhibitors, we assayed an ∼1-kb region upstream of the translational start site of ORF50 (KSHV coordinate 71598) in a luciferase reporter plasmid-based transient transfection assay. The plasmid-borne ORF50 promoter behaved similarly to the viral ORF50 in response to these activating reagents (Fig. 1B). In particular, we found that NaB could stimulate the ORF50 promoter over 20-fold in transfected BCBL-1 cells while TSA produced a 12-fold activation and TPA produced an ∼3.5-fold activation.
FIG. 1.
Chemical induction of ORF50 promoter. (A) BCBL-1 cells were treated with TSA, NaB, or TPA and analyzed by RT-PCR for expression levels of ORF50 (top panel) or GAPDH (lower panel). Controls lacking reverse transcriptase (−RT) are indicated for ORF50. (B) The ORF50 promoter fused to luciferase (p50) or the pGL3-Basic control plasmid were transfected into BCBL-1 cells and treated with TSA, NaB, or TPA. (C) BCBL-1 cells were transfected with luciferase reporter plasmid pGL3-Basic or promoters derived from BZLF1, ORF50 (p50), the K8 immediate early promoter (K8-IE), or the K8 delayed early promoter (K8-DE). (D) p50 and pGL3-Basic control plasmids were transfected into JSC-1, BCBL-1, 293, or DG75 cells. Luciferase activity was measured as induction (n-fold) upon treatment with 3 mM NaB for 24 h.
The promoter specificity of the NaB response was determined by comparing several KSHV promoter regions as well as the promoter region for the EBV immediate early BZLF1 gene to ORF50 (Fig. 1C). We found that the ORF50 promoter was ∼7-fold more responsive to NaB than pGL3-Basic and ∼2-fold more responsive than the BZLF1 promoter of EBV. We also compared the NaB responsiveness of the KSHV K8 immediate early and delayed early promoters (59). Interestingly, NaB stimulated the K8 immediate early promoter but had no effect on the delayed early promoter, further suggesting that plasmid-borne promoters respond similarly to virus-borne promoters in response to HDAC inhibitors and that specific promoter DNA sequences confer responsiveness to NaB (59, 74).
ORF50 has been reported to be autostimulated by Rta (22). To eliminate the concern that NaB treatment of latently infected PEL cells was leading to Rta-mediated activation of ORF50, we compared the effect of NaB in various cell types, including KSHV-positive PEL cell lines JSC-1 and BCBL-1 and KSHV-negative cell lines 293 and DG75 (an EBV-negative Burkitt lymphoma-derived cell line) (Fig. 1D). We found that the ORF50 promoter was stimulated to significantly higher levels than pGL3-Basic in all cell types tested. Interestingly, KSHV-negative 293 cells supported NaB stimulation to levels similar to and higher than KSHV-positive BCBL-1 cells. We found that JSC1 cells, which are coinfected with KSHV and EBV, were the most responsive to NaB, with 50- to 60-fold activation. In contrast, NaB treatment of DG75 cells supported only ∼3-fold activation of ORF50, which was still significantly greater than that of pGL3-Basic (1.0-fold). We conclude that ORF50 promoter sequences have NaB-responsive elements and that these are independent of additional KSHV gene products, as might be expected by Rta autoactivation.
Identification of an ORF50 promoter element responsive to HDAC inhibitors.
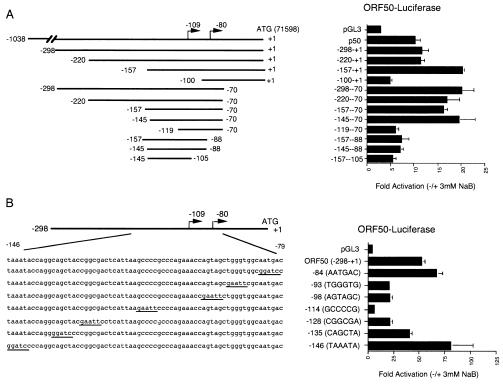
To determine whether a minimal promoter region could be identified which retained responsiveness to NaB treatment, a series of ORF50 promoter deletions were assayed by transient transfection for their response to NaB in PEL cells (Fig. 2). We found that 5′ deletions from −1038 to −220 (relative to the ATG +1) had little effect on NaB responsiveness while deletion of sequences −220 to −157 had a slight enhancement of the NaB response. Further 5′ deletions to −119 yielded a significant loss of NaB-induced activity. We also found that 3′ deletions to −70 (relative to the ATG) retained NaB responsiveness, but responsiveness was lost with deletions to −88. The transcriptional initiation site of ORF50 was mapped to nucleotide −109 (59) and to −80 (22) relative to the ATG. Combining the 5′ −145 and 3′ −70 deletion also retained significant NaB responsiveness, suggesting that this constituted a minimal 75-bp NaB response element.
FIG. 2.
Mutational analysis of ORF50 promoter response to NaB. (A) 5′ and 3′ deletion mutants of ORF50 were transfected into BCBL-1 cells and assayed for luciferase activity in the presence or absence of NaB. Luciferase activity is displayed as activation (n-fold) in response to NaB. Schematic diagrams of deletion mutants are shown to the left. The ATG start site is indicated as +1 and corresponds to position 71598 in the KSHV genome. (B) Substitution mutants of ORF50 (−298/+1) were assayed for NaB induction in JSC1 cells. Substitution mutations were indicated by underlined sequences in the schematic to the left.
To further characterize NaB response elements, we generated a series of seven substitution mutants distributed throughout the minimal promoter sequences mapped by deletion mutagenesis above. We found that substitution mutants −93, −98, and −128 reduced NaB responsiveness significantly (Fig. 2B). However, substitution of the GC-rich element at −114 completely abolished NaB responsiveness. This GC box encompasses the −109 transcriptional initiation site. Although basal levels were reduced, transcription levels were still measurably above those of pGL3-Basic (data not shown). Interestingly, we did not find any evidence for a TATA-like element within this promoter. A TATA-like element (TAAATA at −146) was proposed (22); however, substitution of this TAAATA element led to a significant increase in basal and NaB-stimulated levels, suggesting that ORF50 has a TATA-less promoter regulated largely through the GC-box element.
The NaB-responsive GC-box element binds Sp1/Sp3 in vitro and in vivo.
To determine which, if any, transcription factors bound to the GC box of the ORF50 promoter, we used DNA affinity methods to isolate specific proteins that could then be identified by Western blotting (Fig. 3). Wild-type and GC-box mutant ORF50 promoter DNA fragments were biotinylated and bound to streptavidin magnetic beads, incubated with BCBL-1 nuclear extract, washed extensively, and then assayed by Western blotting for candidate proteins. We found that the GC-box binding proteins Sp1 and Sp3 were specifically associated with wild-type ORF50 promoter DNA but did not bind to control pBSKII+ polylinker region DNA or to GC-box mutant (−114) ORF50 DNA. Several additional antibodies were tested in this assay (e.g., anti-Rb), but only Sp1 and Sp3 demonstrated specificity for the ORF50 wild-type promoter. These results indicate that Sp1 and Sp3 can bind to the ORF50 GC box in vitro.
To determine whether Sp1 could also bind to the ORF50 promoter in vivo, we used the ChIP assay (52). Formaldehyde cross-linked cellular extracts were subjected to IP with either Sp1-specific or immunoglobulin G (IgG) control antibody. DNA was isolated from these immunoprecipitates and then assayed by PCR for the sequences specific to ORF50 or to control sequences specific to the GAPDH coding sequence. We found that Sp1 chromatin immunoprecipitates were specifically enriched for viral ORF50 promoter DNA (Fig. 3B) as well as to the ORF50 sequences of transfected plasmid reporter DNA (Fig. 3C), suggesting that Sp1 binds to the ORF50 promoter in vivo. Similar experiments with Sp3 antibody indicated that Sp3 also bound to ORF50 but at reduced levels compared to Sp1 (data not shown). Taken together, these results suggest that Sp1 and Sp3 bind to the ORF50 GC-box element required for NaB-responsive transcription.
Nuclease mapping of nucleosomes on the ORF50 promoter.
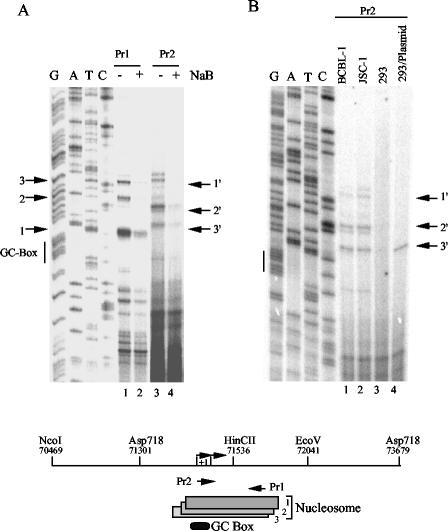
Based on our data, we hypothesized that the endogenous ORF50 promoter was associated with positionally phased nucleosomes and that these nucleosomes were altered in response to NaB stimulation. To examine this hypothesis, we first used a low-resolution Mnase assay to determine whether nucleosomes were positioned over the ORF50 regions that correlated with NaB responsiveness (Fig. 4A). Nuclei from PEL cells (BCBL-1) were treated with limiting amounts of Mnase for ∼2 min, and then isolated deproteinized DNA was cleaved with Asp718 restriction endonucleases. Cleaved DNA was then analyzed by Southern hybridization and indirect end-labeling with a probe covering the sequences between nucleotides 71301 (Asp718) and 71598. Southern blots revealed an Mnase-generated ladder indicative of chromatin-associated DNA. We also observed an Mnase hypersensitive site at ∼300 nucleotides in untreated BCBL-1 cells. In NaB-treated cells, this hypersensitive site was altered by ∼100 bp to produce a 200-bp fragment in this experimental analysis. Our interpretation of these results is that an Mnase-hypersensitive site exists at a position 300 bp from Asp718 (centered at ∼71601) in latently infected cells and that the hypersensitive site moves 100 bp closer to Asp718 (centered at ∼71501) in NaB-treated cells (Fig. 4C). These data suggest that a nucleosome is located near the transcriptional initiation site of ORF50 and that treatment with NaB alters the nucleosome organization in this region.
FIG. 4.
Nuclease mapping of ORF50 promoter chromatin. (A) Partial Mnase digestion of JSC1 cells treated with (+) or without (−) NaB was assayed by Southern blotting and probed with ORF50 sequences (positions 71301 to 71598). Hypersensitive sites are indicated by the arrows, and molecular size standards are indicated to the right. (B) Endonuclease sensitivity assay of ORF50 promoter region in JSC1 cells treated with (+) or without (−) NaB. Nuclei were incubated with Asp718 (left panel) or HincII (right panel), and then isolated DNA was cleaved with NcoI/EcoRV and analyzed by Southern blotting with an ORF50-specific probe. Asp718 (left panel) and HincII (right panel) cleavage products are indicated. A schematic of ORF50 restriction sites, GC box, and presumptive nucleosomes is indicated below. (C) Schematic indicating restriction sites and probe used for indirect end-labeling.
To further examine NaB-induced changes in the ORF50 promoter-associated nucleosome, we used a restriction endonuclease accessibility assay (Fig. 4B). HincII cleaves at position 71536 within the mapped nucleosome boundaries and could be used to probe the relative access of DNA near the ORF50 transcription initiation site. Asp718 cleaves at position 71301 upstream of the mapped nucleosome. JSC-1 nuclei were incubated with either HincII or Asp718 for 30 min. DNA was isolated and then cleaved with NcoI/EcoRV and then assessed for endonuclease cleavage by Southern blotting with an ORF50 probe. We found that cells treated with NaB were significantly more sensitive to HincII cleavage, suggesting that the nucleosome positioned at ORF50 was either eliminated in the presence of NaB or subject to chromatin remodeling activity that increases DNA accessibility of nucleosome-associated DNA. We did not observe significant changes in the accessibility of Asp718, indicating that DNA-associated factors and chromatin at this position were not altered in response to NaB.
To more precisely map the position of a nucleosome in the region surrounding the transcription start site, we used high-resolution Mnase mapping (Fig. 5A). Nuclei were treated with high levels of Mnase, and mononucleosome-sized fragments of ∼150 bp were isolated from agarose gels. Isolated mononucleosomal DNA was then assayed by primer extension with various oligonucleotide primers throughout the ORF50 region. We found that only primers located within the promoter region from −105 to −70 relative to the ATG produced detectable extension products, indicating that other regions were overdigested by Mnase treatment (data not shown). Using primers directed downstream from position 71490 (Pr2) and upstream from position 71590 (Pr1), we were able to map a nucleosome boundary to three overlapping positions, covering nucleotides 71482 to 71625 (nuc1), 71469 to 71610 (nuc 2), and 71462 to 71605 (nuc 3). These results could be accounted for by different nucleosome positions in different cells or on different viral DNA molecules within single cells. We next compared the effect of NaB on the positioning of this nucleosome and found that the qualitative mapping of the nucleosome was not altered significantly, but rather the relative abundance of nucleosome protection was reduced. We also found that this nucleosome positioning over ORF50 was indistinguishable in two different PEL cell lines, BCBL1 and JSC1, and the 3′ position could be clearly detected in 293 cells transfected with ORF50 containing reporter plasmid (Fig. 5B). These results suggest that ORF50 promoter sequences contain a strong nucleosome-positioning element that may contribute to stable transcription repression during latency.
FIG. 5.
Primer extension mapping of Mnase I nuclease cleavage sites in ORF50. (A) Mononucleosomal fragments from Mnase-treated nuclei were gel purified and analyzed by primer extension with primers Pr1 and Pr2, as indicated above the gel. Sequence reactions were generated with Pr1, and the GC box is indicated to the left. Numbered arrows indicate the major primer extension products that correlate with Mnase I cleavage sites and nucleosome boundaries. (B) Primer extension with Pr2 was used to compare the mononucleosomal fragments derived from BCBL-1, JSC-1, and 293 cells and 293 cells transfected with ORF50-Luc plasmid DNA. Sequencing lanes were generated with Pr1. The schematic at the bottom indicates the nucleosome positions mapped by the above primer extension and analysis.
Histone acetylation of ORF50 during reactivation.
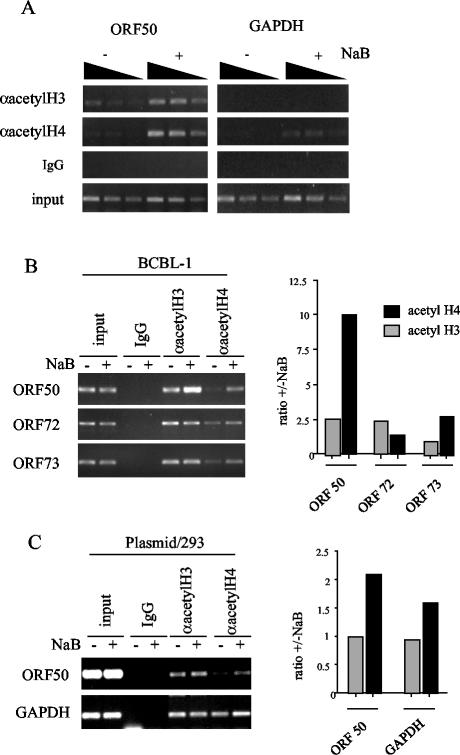
NaB is thought to function by inhibition of HDACs. To directly test whether NaB treatment led to a specific increase in histone acetylation of ORF50 promoter-associated nucleosomes, we used the ChIP assay with antibodies specific for hyperacetylated histones H3 and H4. We found that NaB led to a three- to sixfold increase in histone H3 and H4 acetylation at the ORF50 promoter (Fig. 6A). In contrast, acetylated histones were not significantly enriched at the coding sequence of GAPDH (Fig. 6A), and comparable increases in acetylation were not found on KSHV promoters for latently expressed genes ORF72 and ORF73 (Fig. 6B). Importantly, we were also able to show that histone acetylation also increases on transfected plasmid-based ORF50 promoter sequences after treatment with NaB (Fig. 6C). These results indicate that transcription activation of ORF50 correlates with an increase in histone H3 and H4 acetylation.
FIG. 6.
Chromatin modifications and modifying enzymes at ORF50. (A) A ChIP assay with antibodies specific for hyperacetylated histone H3 (αacetylH3) or H4 (αacetylH4) was used to compare histone acetylation in untreated (−) or NaB-treated (+) BCBL-1 cells at the ORF50 promoter (left panel) or the GAPDH coding sequence (right panel). Control IgG ChIP and input DNA are indicated in the lower panels. (B) The ORF50 promoter was compared to the latency-associated ORF72 and ORF73 promoters by ChIP assay with antibodies to acetylated histone H3 and H4. (C) 293 cells transfected with ORF50-Luc plasmid were assayed by ChIP for association of acetylated histones. Quantification of ethidium-stained DNA in agarose gels is indicated by the bar graphs to the right.
Ectopic expression of CBP stimulates ORF50 transcription.
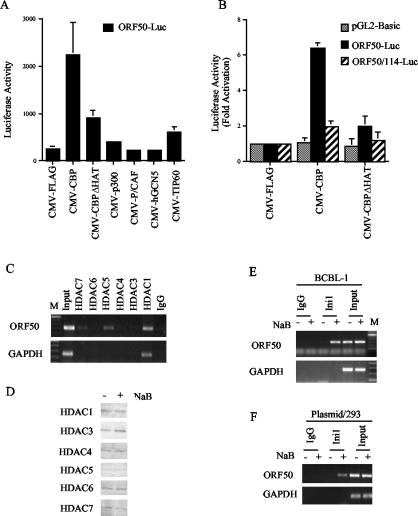
One prediction of these results is that overexpression of a histone acetylase should reverse the transcriptional repression mediated by HDACs. To test this, we cotransfected JSC-1 cells with the ORF50 luciferase reporter and expression vector for various HAT-containing coactivator proteins (Fig. 7A). All of these expression vectors have been shown to express functional proteins that can stimulate transcription of specific genes (data not shown). In these experiments, we found that CBP stimulated the ORF50 promoter by ∼10-fold while p300, P/CAF, hGCN5, and TIP60 had no significant effect. Importantly, we found that expression of a CBP deletion mutant lacking HAT activity was compromised for activation of ORF50 transcription. CBP activation was shown to be relatively specific for ORF50, since CBP did not activate the pGL3 control promoter or the ORF50 promoter containing a substitution mutation in the Sp1/Sp3 binding site (Fig. 7B). These results indicate that overexpression of a specific HAT, CBP, can activate transcription of the ORF50 promoter in a HAT- and GC-box-dependent manner.
FIG. 7.
Recruitment of chromatin-modifying activities at ORF50. (A) JSC1 cells were transfected with ORF50 (−298/+1) luciferase and CMV-driven expression vectors for CBP, CBP-ΔHAT, p300, P/CAF, hGCN5, or TIP60. Luciferase activity is indicated in relative light units. (B) ORF50-Luc (−298/+1), the GC-box mutant (ORF50/−114), and pGL2-Basic were compared for their responsiveness to CMV-FLAG, CMV-CBP, or CMV-CBPΔHAT. (C) Antibodies to HDACs 7, 6, 5, 4, 3, and 1 were used in a ChIP assay to determine association with the ORF50 promoter (top panel) in BCBL-1 untreated cells. The GAPDH coding sequence is indicated in the lower panel. IgG control ChIP, input DNA, and marker DNA (M) are indicated. (D) Western blotting with HDAC antibodies was used to measure protein levels in total cell extracts from untreated (−) or NaB-treated (+) BCBL1 cells. (E) ChIP assay with antibodies specific for the Ini1/Snf5 protein in untreated (−) or NaB-treated (+) BCBL-1 cells comparing ORF50 (top panel) and GAPDH (lower panel) DNA amplification. Control IgG ChIP and input DNA are indicated. (F) 293 cells transfected with ORF50-Luc were assayed by ChIP for association with Ini1/Snf5 in the absence (−) and presence (+) of NaB.
Association of HDACs with ORF50 during latent infection.
Histone deacetylation has been correlated with transcriptional repression and the stable positioning of repressive nucleosomes (reviewed in reference 48). Moreover, Sp1 and Sp3 have been shown by others to interact with several HDACs, including HDAC1 and HDAC2 (65, 71). To determine whether HDACs were associated with ORF50, we used several HDAC antibodies in the ChIP assay (Fig. 7C). We found that HDAC7, HDAC5, and HDAC1 could be detected at ORF50, although HDAC1 was also found at GAPDH. We did not find HDAC7 or HDAC5 at GAPDH, indicating that these HDACs were specific for the ORF50 promoter. We did not detect specific association of HDAC6, 4, 3, or 2 (Fig. 7C and data not shown) in these assays. All of the HDAC antibodies reacted with protein species of the predicted molecular weight in Western blot analysis (Fig. 7D). We also did not find any significant change in the association of HDACs when cells were treated with NaB (data not shown). Together, these results suggest that HDACs are stably associated with ORF50 and that they contribute to the stable repression and binding of the promoter-specific nucleosome.
NaB-dependent recruitment of Snf5/Ini1 chromatin remodeling complex to ORF50.
Work by others has shown that histone acetylation can lead to the recruitment of bromodomain-containing chromatin remodeling complexes (31). One of the major ATP-dependent chromatin remodeling complexes involved in transcription activation is the BRG1 (human homolog of Swi/Snf) complex. Antibody specific for the Ini1/Snf5 subunit of the BRG1 complex was assayed by ChIP for its association with ORF50 (Fig. 7E). We found that treatment with NaB led to a robust increase in Ini1/Snf5 association with ORF50. This association could be detected as early as 30 min after NaB treatment, suggesting that these are early events in the transcription activation process. We did not detect Ini1/Snf5 at GAPDH in the absence or presence of NaB, indicating that the association with ORF50 was specific. We also found that Ini1/Snf5 associated with transfected plasmid-based ORF50 after treatment with NaB (Fig. 7F). These results suggest that NaB-induced histone acetylation leads to the recruitment of the BRG1 chromatin remodeling complex to ORF50 and that this complex is responsible for the derepression of the promoter-associated nucleosome.
DISCUSSION
Several levels of control regulate the switch from latent to lytic infection of gammaherpesviruses. Expression of the KSHV ORF50 protein Rta is sufficient to initiate lytic cycle gene expression and complete productive replication in latently infected PEL cells (28, 39, 66). The mechanisms regulating the expression of ORF50 have not been well characterized. Several chemical reagents stimulate ORF50 gene expression and consequently lead to viral lytic reactivation. In this work, we investigated the mechanism of one potent chemical activator of the KSHV lytic cycle and found that NaB can act directly and specifically on the ORF50 core promoter elements. In particular, we show that NaB activates the ORF50 promoter in the context of the viral genome and on transfected plasmids (Fig. 1A and B). We also found that NaB activation of ORF50 was robust and specific relative to other gammaherpesvirus promoters and independent of other viral gene products, including autoactivation by Rta (Fig. 1C and D). We were able to identify a GC-rich sequence motif near the ORF50 transcriptional initiation sites that conferred responsiveness to NaB (Fig. 2). This GC box was shown to bind Sp1/Sp3 proteins in vitro by DNA affinity chromatography and in vivo by ChIP assay (Fig. 3). Mnase studies were used to map a nucleosome positioned over the Sp1/3 and transcriptional initiation sites (Fig. 4 and 5). NaB treatment altered the Mnase-hypersensitive sites near the downstream end of the nucleosome and increased the endonuclease accessibility of nucleosome-protected DNA (Fig. 4 and 5). We demonstrated that NaB induced histone H3 and H4 hyperacetylation at ORF50 (Fig. 6), that ectopic expression of CBP activated the plasmid-based ORF50 promoter in a HAT-dependent manner (Fig. 7A), and that HDACs 1, 5, and 7 associated with the ORF50 promoter in latently infected cells (Fig. 7C). Finally, we show that Ini1/Snf5, a component of the Brg chromatin remodeling complex, was recruited to the ORF50 promoter soon after NaB treatment (Fig. 7D). Taken together, these data suggest that histone modification and chromatin remodeling of a nucleosome positioned over the transcriptional initiation site are early events in the transcription of ORF50 promoter and correlate with control mechanisms that regulate KSHV reactivation from latency.
The NaB-induced transcriptional initiation site of ORF50 has been mapped to position 71518 by 5′ rapid amplification of cDNA ends (74) and to position 71489 by RNase1 protection (22). Our data revealed that the position 71518 initiation site corresponds to an Sp1/Sp3 binding site and is absolutely required for basal and activated transcription of ORF50 (Fig. 2 and 3 and data not shown). We did not find any evidence that ORF50 has a functional TATA element, since mutagenesis of the ORF50 TAAATA element at position 71452 (−33 and −59 nucleotides upstream of the transcriptional initiation sites) did not inhibit basal or activated transcription. Although several TA-rich sequences in the minimal promoter exist and may also serve as a binding site for the TATA-binding protein, our data suggests that ORF50 is a TATA-less promoter that utilizes a GC box as an initiator element. Sp1 and Sp3 have been shown to have complex behavior at the GC boxes of many promoters, and mutations in Sp1 binding sites often have profound effects on transcription levels. The GC box in ORF50 is the dominant element that establishes the transcription initiation site and supports high-level transcription when cells are treated with NaB or TSA.
Several NaB response elements have been mapped and found to correspond in some cases to GC-rich Sp1/3 binding sites (19, 71). Sp1 has been shown to bind to HDAC1 and Sp3 has been shown to bind to HDAC2 by IP assay (71). We did not find strong evidence that HDAC1 and HDAC2 could bind to ORF50-bound Sp1/3 in vitro by DNA affinity chromatography (data not shown). This may be a result of too-stringent wash conditions that eliminate weakly associated HDACs with Sp1-bound promoter DNA. Alternatively, HDACs 1, 5, and 7 were detected by the more sensitive ChIP assay (Fig. 7C), suggesting that these HDACs do associate with ORF50 promoter in vivo. One possible explanation to resolve the discrepancy between in vivo and in vitro findings for HDACs is that nucleosome assembly is required for stable HDAC binding to ORF50 promoter. It is also interesting that CBP was a specific and efficient activator of ORF50 when ectopically expressed in transient transfection assays (Fig. 7A and B). CBP has been isolated in a complex with Sp1 and has been shown to coactivate Sp1 in transcription assays (47, 51). At present, we do not know which promoter-specific binding factors recruit CBP or HDACs to ORF50. Several other sequence elements of ORF50 contributed to NaB inducibility (Fig. 2), and it is likely that additional sequence-specific factors contribute to the regulation of ORF50, both in response to NaB and in response to endogenous stimulatory signals.
The most significant finding from these studies is our demonstration that the region from position 71462 to 71605 is associated with a nucleosome in latently infected PEL cells (Fig. 4 and 5). This region covers both transcriptional initiation sites (positions 71489 and 71518) as well as the GC-box binding site for Sp1 (positions 71484 to 71493). Upon NaB stimulation, we observed significant changes in the accessibility of this region to Mnase and to HincII restriction endonuclease. We did not detect a translational sliding of the nucleosome, but we cannot rule out the possibility that the nucleosome does slide to a position outside the detection of our primer sets used in primer extension assays. However, an equally likely explanation for these findings is that the nucleosome is remodeled in response to NaB treatment to increase the accessibility of nucleases and general transcription factors. Perhaps the most interesting finding is that Sp1, which falls within the boundary of the nucleosome and overlaps the transcription initiation site, was stably associated with ORF50 in latent and NaB-treated PEL cells. This indicates that Sp1 binds to the nucleosomal DNA at a site near the internucleosomal linker region. One important question in gene regulation is how the nucleosomes are positioned. We speculate that Sp1 is important for positioning the nucleosome at ORF50 and that this positioning is maintained by HDAC activity. Future experiments will be required to determine what additional factors are important for the positioning of a nucleosome over the ORF50 initiation site. Our data also suggest that Sp1 binds constitutively to ORF50 during latency and reactivation, and it is not clear what cellular conditions or factors convert Sp1 from a chromatin-associated repressor into a potent transcriptional activator of ORF50.
The activation of the KSHV lytic cycle can be initiated by HDAC inhibitors (42, 43), as we have shown here, as well as by phorbol esters (14, 55) and by DNA methylation inhibitors (5′ azacytidine) (18). ORF50 can also be autoactivated by Rta, but the precise sequence requirements have not been fully characterized (22). Interestingly, the EBV homologue of Rta can also autoactivate its own promoter BRLF1, and the response element in this EBV immediate early promoter has been mapped to an Sp1 site (53). Sp1/3 has also been implicated in the regulation of the EBV BZLF1 immediate early promoter, although at BZLF1, the HDACs have been shown to be recruited by MEF2D proteins that bind adjacent sequences (63). Because of the abundant and generic nature of Sp1, it is unlikely that it can account for all of the complex regulation of ORF50, and it will be important to identify additional factors that work in cooperation with Sp1/3 to regulate this promoter, especially in response to natural activation signals.
In addition to HDAC inhibition, our results indicated that ectopic expression of CBP stimulated plasmid-based ORF50 transcription. CBP stimulation of ORF50 was dependent upon the HAT domain, and no other HAT coactivator proteins could be substituted, suggesting that CBP has specificity for some factors associated with the ORF50 promoter. Acetylation by CBP has been shown to be important for recruitment of the BRG1 remodeling complex (2, 3, 31). Using the ChIP assay, we were able to find that the BRG1 subunit Ini1/Snf5 could associate with ORF50 promoter after treatment with HDAC inhibitors (Fig. 7C). These results suggest that histone acetylation regulates the recruitment of BRG1 complex and that BRG1-dependent remodeling of the positioned nucleosome permits transcription initiation of ORF50. These results also suggest that HDACs 1, 5, and 7 are constitutively associated with ORF50 and that their inhibition allows a constitutively associated HAT, perhaps CBP, to tip the balance towards the acetylation of the nucleosome at ORF50 initiation site. It is interesting that the EBV EBNA2 protein has been shown to recruit Ini1/Snf5, and this has been implicated in the transcription activation of EBV-associated growth-transforming genes in latently infected lymphocytes (72).
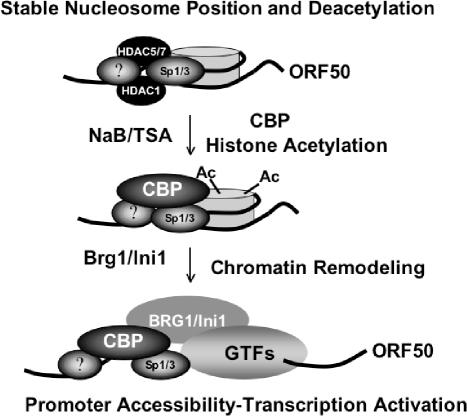
Chromatin structure and nucleosome positioning have been shown to have profound effects on gene regulation in eukaryotes. Our findings are consistent with this general role of chromatin and suggest that chromatin plays a critical role in transcriptional repression and maintenance of the latent state of gammaherpesviruses. The tendency to establish latent infection is largely dictated by the transcriptional repression of the gammaherpesvirus immediate early genes during initial infection. It remains unclear whether the transcriptional inactivity of gammaherpesvirus immediate early genes results from active repression or from a deficiency of positively acting transcription factors. Results from our study indicate that Sp1, which can be a potent activator of some genes, is inactive at ORF50 in the absence of a costimulatory signal, like NaB treatment. It seems likely that HDACs associated with the ORF50 promoter maintain a deacetylated and transcriptionally repressive nucleosome over the initiation site. Only upon inhibition of HDACs, or recruitment of CBP, does histone acetylation relieve nucleosome position and repression (Fig. 8). While we do not know if this is a general rule for other gammaherpesvirus immediate early promoters, it is likely that nucleosome acetylation and chromatin remodeling play a similar role in regulating the transcription from latency to lytic infection. Future studies will try to identify factors that position the nucleosome over the ORF50 initiation site, establish a stable latent state, and constitute the molecular switch from latent to lytic gene expression for KSHV.
FIG. 8.
Nucleosome modification and chromatin remodeling can regulate the ORF50 promoter during reactivation signals. Sp1 binds ORF50 near the transcription initiation site and within the boundary of the nucleosome. Histone acetylation caused by NaB, TSA, or ectopic expression of CBP leads to nucleosome remodeling and transcription activation. Contributions from unknown additional factors are represented by question marks.
Acknowledgments
We thank Steve McMahon, Gerd Blobel, and Shelley Berger for plasmids and critical comments and Latasha Day for technical support.
This work was supported by a pilot grant from the PENN Center for AIDS Research (to P.M.L.) and by NIH grants CA85678 (to P.M.L.) and CA86839 (to Y.Y.).
REFERENCES
- 1.Ablashi, D. V., L. G. Chatlynne, J. E. Whitman, Jr., and E. Cesarman. 2002. Spectrum of Kaposi's sarcoma-associated herpesvirus, or human herpesvirus 8, diseases. Clin. Microbiol. Rev. 15:439-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 3.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad, K., and S. Henikoff. 2002. Epigenetic consequences of nucleosome dynamics. Cell 111:281-284. [DOI] [PubMed] [Google Scholar]
- 5.Ambroziak, J. A., D. J. Blackbourn, B. G. Herndier, R. G. Glogau, J. H. Gullett, A. R. McDonald, E. T. Lennette, and J. A. Levy. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582-583. [DOI] [PubMed] [Google Scholar]
- 6.Bais, C., B. Santomasso, O. Coso, L. Arvanitakis, E. G. Raaka, J. S. Gutkind, A. S. Asch, E. Cesarman, M. C. Gerhengorn, and E. A. Mesri. 1998. G-protein coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86-89. [DOI] [PubMed] [Google Scholar]
- 7.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff, C., Y. Endo, P. D. Collins, Y. Takeuchi, J. D. Reeves, V. L. Schweickart, M. A. Siani, T. Sasaki, T. J. Williams, P. W. Gray, M. A. Moore, J. Chang, and R. A. Weiss. 1997. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science 278:290-294. [DOI] [PubMed] [Google Scholar]
- 9.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 10.Burysek, L., and P. M. Pitha. 2001. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J. Virol. 75:2345-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cesarman, E. 2002. The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Recent Results Cancer Res. 159:27-37. [DOI] [PubMed] [Google Scholar]
- 13.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 14.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 15.Chang, J., R. Renne, D. Dittmer, and D. Ganem. 2000. Inflammatory cytokines and the reactivation of Kaposi's sarcoma-associated herpesvirus lytic replication. Virology 266:17-25. [DOI] [PubMed] [Google Scholar]
- 16.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 17.Chen, C.-J., Z. Deng, A. Y. Kim, G. A. Blobel, and P. M. Lieberman. 2001. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol. 21:476-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, C. Parravicini, M. Corbellino, and K. Yamanishi. 2001. Activation of latent Kaposi's sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc. Natl. Acad. Sci. USA 98:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi, H. S., J. H. Lee, J. G. Park, and Y. I. Lee. 2002. Trichostatin A, a histone deacetylase inhibitor, activates the IGFBP-3 promoter by upregulating Sp1 activity in hepatoma cells: alteration of the Sp1/Sp3/HDAC1 multiprotein complex. Biochem. Biophys. Res. Commun. 296:1005-1012. [DOI] [PubMed] [Google Scholar]
- 20.Cotter, M. A., II, and E. S. Robertson. 2002. Molecular biology of Kaposi's sarcoma-associated herpesvirus. Front. Biosci. 7:d358-d375. [DOI] [PubMed] [Google Scholar]
- 21.Creaven, M., F. Hans, V. Mutskov, E. Col, C. Caron, S. Dimitrov, and S. Khochbin. 1999. Control of the histone-acetyltransferase activity of Tip60 by the HIV-1 transactivator protein, Tat. Biochemistry 38:8826-8830. [DOI] [PubMed] [Google Scholar]
- 22.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 23.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 24.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ensoli, B., C. Sgadari, G. Barillari, M. C. Sirianni, M. Sturzl, and P. Monini. 2001. Biology of Kaposi's sarcoma. Eur. J. Cancer 37:1251-1269. [DOI] [PubMed] [Google Scholar]
- 26.Flore, O., S. Rafii, S. Ely, J. J. O'Leary, E. M. Hyjek, and E. Cesarman. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature 394:588-592. [DOI] [PubMed] [Google Scholar]
- 27.Foreman, K. E., J. Friborg, Jr., W. P. Kong, C. Woffendin, P. J. Polverini, B. J. Nickoloff, and G. J. Nabel. 1997. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N. Engl. J. Med. 336:163-171. [DOI] [PubMed] [Google Scholar]
- 28.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo, H. G., M. Sadowska, W. Reid, E. Tschachler, G. Hayward, and M. Reitz. 2003. Kaposi's sarcoma-like tumors in a human herpesvirus 8 ORF74 transgenic mouse. J. Virol. 77:2631-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 32.Jenner, R. G., and C. Boshoff. 2002. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim. Biophys. Acta 1602:1-22. [DOI] [PubMed] [Google Scholar]
- 33.Katano, H., Y. Sato, H. Itoh, and T. Sata. 2001. Expression of human herpesvirus 8 (HHV-8)-encoded immediate early protein, open reading frame 50, in HHV-8-associated diseases. J. Hum. Virol. 4:96-102. [PubMed] [Google Scholar]
- 34.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komatsu, T., M. E. Ballestas, A. J. Barbera, and K. M. Kaye. 2002. The KSHV latency-associated nuclear antigen: a multifunctional protein. Front. Biosci. 7:d726-d730. [DOI] [PubMed] [Google Scholar]
- 36.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jk (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 38.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 40.Martin, D. F., B. D. Kuppermann, R. A. Wolitz, A. G. Palestine, H. Li, and C. A. Robinson. 1999. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. N. Engl. J. Med. 340:1063-1070. [DOI] [PubMed] [Google Scholar]
- 41.McMahon, S. B., M. A. Wood, and M. D. Cole. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, G., M. O. Rigsby, L. Heston, E. Grogan, R. Sun, C. Metroka, J. A. Levy, S. J. Gao, Y. Chang, and P. Moore. 1996. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 334:1292-1297. [DOI] [PubMed] [Google Scholar]
- 43.Miller, G. L., E. Heston, L. Grogan, M. Gradoville, R. Rigsby, D. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma-associated herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 45.Morfeldt, L., and J. Torssander. 1994. Long-term remission of Kaposi's sarcoma following foscarnet treatment in HIV-infected patients. Scand. J. Infect. Dis. 26:749-752. [DOI] [PubMed] [Google Scholar]
- 46.Muralidhar, S., A. M. Pumfery, M. Hassani, M. R. Sadaie, M. Kishishita, J. N. Brady, J. Doniger, P. Medveczky, and L. J. Rosenthal. 1998. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J. Virol. 72:4980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naar, A. M., P. A. Beaurang, K. M. Robinson, J. D. Oliner, D. Avizonis, S. Scheek, J. Zwicker, J. T. Kadonaga, and R. Tjian. 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng, H. H., and A. Bird. 2000. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25:121-126. [DOI] [PubMed] [Google Scholar]
- 49.Nicholas, J., V. R. Ruvolo, W. H. Burns, G. Sandford, X. Wan, D. Ciufo, S. B. Hendrickson, H. G. Guo, G. S. Hayward, and M. S. Reitz. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of marophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287-292. [DOI] [PubMed] [Google Scholar]
- 50.Niepel, F., J. C. Albrecht, A. Ennser, Y. Q. Huang, J. J. Li, A. E. Friedman-Kien, and B. Fleckenstein. 1997. Human herpesvirus 8 encodes a homolog of interleukin-6. J. Virol. 71:839-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Owen, G. I., J. K. Richer, L. Tung, G. Takimoto, and K. B. Horwitz. 1998. Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J. Biol. Chem. 273:10696-10701. [DOI] [PubMed] [Google Scholar]
- 52.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 53.Ragoczy, T., and G. Miller. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 75:5240-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renne, R., W. Zhong, B. G. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 56.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadler, R., L. Wu, B. Forghani, R. Renne, W. Zhong, B. Herndier, and D. Ganem. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saveliev, A., F. Zhu, and Y. Yuan. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299:301-314. [DOI] [PubMed] [Google Scholar]
- 60.Schulz, T. F., J. Sheldon, and J. Greensill. 2002. Kaposi's sarcoma associated herpesvirus (KSHV) or human herpesvirus 8 (HHV8). Virus Res. 82:115-126. [DOI] [PubMed] [Google Scholar]
- 61.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 63.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 64.Staskus, K., W. Zhong, K. Gebhardt, B. G. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun, J. M., H. Y. Chen, M. Moniwa, D. W. Litchfield, E. Seto, and J. R. Davie. 2002. The transcriptional repressor Sp3 is associated with CK2-phosphorylated histone deacetylase 2. J. Biol. Chem. 277:35783-35786. [DOI] [PubMed] [Google Scholar]
- 66.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Shu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 68.Varthakavi, V., P. J. Browning, and P. Spearman. 1999. Human immunodeficiency virus replication in a primary effusion lymphoma cell line stimulates lytic-phase replication of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:10329-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Varthakavi, V., R. M. Smith, H. Deng, R. Sun, and P. Spearman. 2002. Human immunodeficiency virus type-1 activates lytic cycle replication of Kaposi's sarcoma-associated herpesvirus through induction of KSHV Rta. Virology 297:270-280. [DOI] [PubMed] [Google Scholar]
- 70.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, A. S. Denton, et al. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 71.Won, J., J. Yim, and T. K. Kim. 2002. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem. 277:38230-38238. [DOI] [PubMed] [Google Scholar]
- 72.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]