Abstract
A remarkable feature of the prototype foamy virus (PFV) replication pathway has been reported to consist of the ability to retrotranspose intracellularly with high efficiency (M. Heinkelein, T. Pietschmann, G. Jármy, M. Dressler, H. Imrich, J. Thurow, D. Lindemann, M. Bock, A. Moebes, J. Roy, O. Herchenröder, and A. Rethwilm, EMBO J. 19:3436-3345, 2000). PFV intracellular retrotransposition (IRT) was reported to be enhanced by coexpression of fusion-defective envelope protein. To investigate the possibility of cell-to-cell transfer of PFV genomes, which could mimic IRT, we performed cocultivation experiments with cells transfected with an IRT-competent and marker gene-expressing PFV vector together with cells expressing a different marker and measured cells positive for both markers. The findings corroborated the initial report on IRT of Env-deficient PFV. Furthermore, they indicated that viral cores that have incorporated fusion-deficient Env can be transferred from cell to cell in a cell type-specific manor. One possible explanation consists of a minor alternative cleavage site in Env that can be used to expose the fusion peptide of the Env transmembrane protein, which appears to be required for virus uptake.
It has been reported previously that a marker gene-tagged vector that was derived from prototype foamy virus (PFV) is able to recycle the genome following reverse transcription into the nucleus and correctly integrate into the cellular genome with high efficiency (7). PFV intracellular retrotransposition (IRT) was found to require capsid (Gag) and polymerase (Pol) protein expression and active reverse transcriptase and integrase (IN) and to be sensitive to the reverse transcriptase-inhibiting drug zidovudine (7). Results of coexpression experiments with PFV envelope (Env) that was mutated at the surface (SU)-transmembrane (TM) protein cleavage site and therefore able to allow PFV capsid export but unable to perform vector transduction via the cell-free supernatant (2, 15) revealed a significant increase in IRT, suggesting that PFV IRT is an ongoing process in virus-infected cells (7). However, an alternative explanation for the finding of supposed PFV IRT and its observed enhancement by coexpression of functionally disabled Env could consist of a cell-to-cell transfer of naked or enveloped PFV cores, which are defective in spread through the cell-free supernatant. To corroborate the initial report on PFV IRT and to address the role of Env, we performed the following experiments.
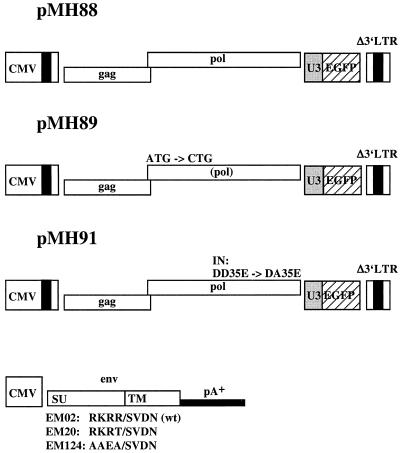
293T cells were transfected with the murine leukemia virus vector pLEN conferring G418 resistance (1), the murine leukemia virus capsid (Gag) and polymerase (Pol) protein-expressing plasmid pHIT60 (18), and the vesicular stomatitis virus G Env protein-expressing plasmid pcVG-wt (14) by calcium phosphate coprecipitation as described previously (8, 9). HeLa and 293 cells were transduced with the vector supernatant and selected with G418 (1 mg/ml) for 2 weeks. Bulk G418-resistant HeLa/neo and 293/neo cells were used in consecutive experiments. The PFV vector and Env expression plasmids used this study are shown in Fig. 1. The PFV vector pMH88 is, with respect to the viral sequences, identical to previously described plasmid pMH92 (7) except that it harbors an internal enhanced green fluorescent protein (EGFP) marker gene cassette instead of an EGFPneo cassette; similarly, pMH89 is an EGFP-expressing derivative of the pol gene ATG-to-CTG mutant plasmid pMH96, which expresses the marker EGFPneo (7); pMH91, the corresponding derivative of the IN active site mutant plasmid pMH97 (7); the Env expression vector EM02, which encodes the wild-type PFV Env protein (11); EM02-derived EM20, which has an Arg571 → Thr571 mutation at the Env SU-TM cleavage site (7); and EM124, in which the basic residues preceding the SU-TM cleavage site were converted from Arg568-Lys-Arg-Arg to Ala568-Ala-Glu-Ala. To create EM124, a recombinant PCR (10) on a 276-bp Eco147I/Bsp1407I fragment with EM02 template DNA and oligonucleotide primers incorporating the desired sequence exchanges was performed. The amplimer was sequenced to exclude unwanted mutations. The PFV vectors and Env expression constructs are in a Neo− backbone (pcDNAzeo from Invitrogen) and are shown in Fig. 1.
FIG. 1.
Schematic representation of the vectors and expression constructs used in this study. All plasmids harbor the human cytomegalovirus (CMV) immediate-early gene promoter to direct transient expression. In pMH88, this is followed by the PFV start of transcription and the leader region (with R-U5 of the 5′ LTR), the wild-type gag and pol genes, an internal expression cassette consisting of the constitutively active spleen focus-forming virus U3 promoter and the gene for EGFP, and the 3′ LTR which was partially deleted in the U3 region (7). pMH89 is structurally identical to pMH88 except that it harbors the pol gene ATG-to-CTG mutation disabling the expression of a functional polymerase protein (4). pMH91 contains a mutation in the active center of IN that changes the conserved DD35E motif to DA35E (5). The env expression constructs consist of EM02 with the wild-type (wt) sequence and the two SU-TM cleavage site mutants EM20 and EM124, which bear one or four alterations, respectively, of the conserved basic amino acids preceding the cleavage site. The poly(A)+ signal was derived from bovine growth hormone of the vector backbone (pcDNAzeo). All vectors are in a Neo− background.
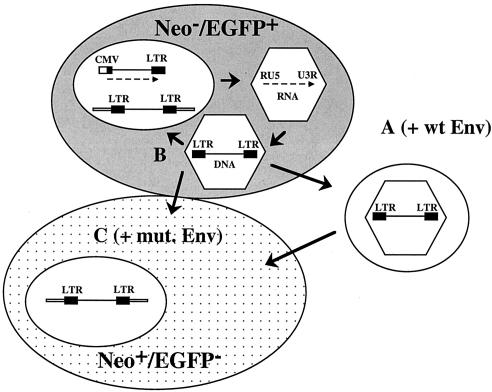
The experimental outline is schematically illustrated in Fig. 2. HeLa (0.8 × 106) or 293 (1.6 × 106) cells were transfected with a total of 4 (HeLa cells) or 20 (293 cells) μg of plasmid DNA by PolyFect (Qiagen) or calcium phosphate coprecipitation, respectively. Three milliliters of supernatant (0.45-μm-pore-size filtrate), harvested at 2 days posttransfection (dpt), was used for the inoculation of 104 target HeLa/neo or 293/neo cells to determine the vector transmission rate via the cell-free medium (pathway A in Fig. 2 and leftmost column of data in Table 1). These cells were analyzed by flow cytometry 3 to 4 days later for marker gene transfer mediated by viral particles in the supernatant. From cultures transfected in parallel, the cells were detached from the plastic support and split into three parts at 1 dpt. One-third was used for flow cytometry (fluorescence-activated cell sorter analysis) as previously described (8, 9) to determine the initial transfection efficiency. One-third was further cultivated and analyzed on a weekly basis by flow cytometry to determine the persistence of the marker gene expression. Persistence of marker gene expression in the absence of marker transfer from cell to cell or via extracellular virus particles is indicative of IRT (pathway B in Fig. 2 and center column of data in Table 1). To analyze marker gene transfer from cell to cell, one-third of the initially transfected cells was mixed with an equal number of HeLa/neo or 293/neo cells, respectively (pathway C in Fig. 2 and rightmost column of data in Table 1). These cells were cocultured for 1 week, selected with G418 (1 mg/ml) for an additional 2 weeks, and then subjected to fluorescence-activated cell sorter analysis to determine the percentage of EGFP-positive and G418-resistant cells. Control experiments revealed that after 2 weeks of selection in G418, no viable naive cells were left (data not shown).
FIG. 2.
Design of the key experiment. Normal HeLa or 293 cells (grey background) were transfected with wild-type (pMH88) or control (pMH89 and pMH91) vectors in the absence or presence of wild-type (EM02) or fusion-defective (EM20 and EM124) Env expression constructs. The transfected cells were cocultivated with Neor derivatives (dotted background), and it was determined whether they had taken up the EGFP marker 3 weeks later. Pathway A describes the generation of vector particles that have wild-type (wt) Env and are infectious through the cell-free supernatant, pathway B illustrates IRT, and pathway C describes the cell-to-cell transfer of vector particles, which were pseudotyped with mutated (mut.) fusion-deficient Env. While the donor cells were Neo− EGFP+, the recipient cells were initially Neo+ EGFP− and were then analyzed for a Neo+ EGFP+ phenotype.
TABLE 1.
Percentages of marker-positive cells after transfection of HeLa and 293 cells with the indicated plasmids as determined by flow cytometrya
| Cells and plasmidsb | % of EGFP-positive recipient cells after transfer of cell-free supernatant | % of stable EGFP-positive cells (21 dpt) | % of EGFP-positive and G418-resistant cells after cocultivation (22 dpt) |
|---|---|---|---|
| HeLa | |||
| pMH91 + pcDNAzeo | <0.1 | 0.6 | <0.1 |
| pMH89 + pcDNAzeo | <0.1 | 0.5 | <0.1 |
| pMH88 + pcDNAzeo | <0.1 | 7.2 ± 1.9 | <0.1 |
| pMH88 + EM02 | 60.0 ± 26.0 | 186.7 ± 17.2 | 121.8 ± 30.8 |
| pMH88 + EM20 | 0.1 | 31.8 ± 10.6 | 28.2 ± 19.3 |
| pMH88 + EM124 | 0.3 | 25.6 ± 6.0 | 30.8 ± 8.8 |
| 293 | |||
| pMH91 + pcDNAzeo | <0.1 | 1.0 | <0.1 |
| pMH88 + pcDNAzeo | <0.1 | 11.3 ± 8.5 | 0.1 |
| pMH88 + EM02 | 43.1 ± 30.4 | 180.8 ± 12.7 | 58.5 ± 17.2 |
| pMH88 + EM20 | <0.1 | 21.9 ± 6.4 | 1.9 ± 1.0 |
The cells were transfected with the indicated amounts of vector DNAs (as shown in Fig. 1), the transient transfection efficiency was determined 1 dpt, and these reference values were arbitrarily set to 100%. The absolute values were in the range of 25 to 30 and 35 to 40% for HeLa and 293 cells, respectively. The data shown address the different pathways by which PFV particles can be transmitted (as shown in Fig. 2). In the leftmost column the cell-free supernatant of primary transfected cells was transferred at 2 dpt to recipient cells, which were monitored for the EGFP marker 3 to 4 days after the transfer. In the center column, the primary transfected cells were analyzed 3 weeks after transfection for stable marker gene expression. The rightmost column addresses the question of cell-to-cell transfer of PFV cores. Primary transfected cells were cocultured with G418-resistant sister cells, and double-marker-positive cells were monitored 3 weeks later. The values correspond to the mean ± the standard error of the mean three independent experiments.
HeLa cells received 2 μg of each, and 293 cells received 10 μg of each.
We speculated that by demonstrating cells positive for both markers (EGFP expression and G418 resistance), we can measure cell-to-cell transfer of either naked viral cores (pMH88) or cores enveloped with env mutants deficient in cell-free transfer (pMH88 plus EM20 or pMH88 plus EM124, respectively). Such cell-to-cell transfer could mimic PFV IRT.
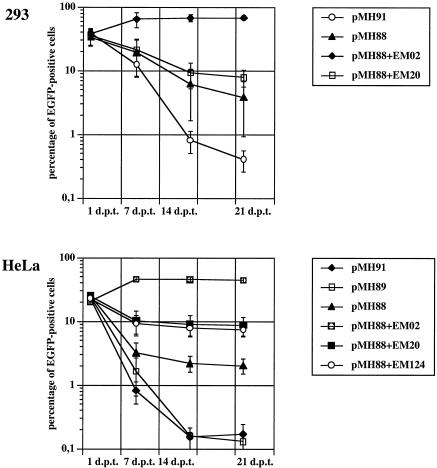
As shown in Fig. 3, the part B cells behaved as previously reported (7). Compared to the negative control vectors (pMH89 and pMH91), transfection of cells with wild-type vector pMH88 led to a certain stable amount of marker-expressing cells, which is indicative of PFV IRT (7). Cotransfection of cells with env mutant EM20 or EM124 resulted in a significant increase in cells with stable marker gene expression. The results obtained with part C cells and the vector transduction rates with cell-free supernatant (part A) are summarized in Table 1. In contrast to wild-type EM02, neither of the two env cleavage site mutants (EM20 or EM124) led to detectable vector transfer via the cell-free supernatant (Table 1, leftmost column of data). Interestingly, the coculture experiment (Table 1, rightmost column of data) produced a different result. Cotransfection of HeLa cells with pMH88 together with either of the two env mutants and consecutive cocultivation with HeLa/neo cells, followed by G418 resistance selection, resulted in similar percentages of double-marker-positive cells (Table 1, rightmost column of data) compared to the nonselected singly positive cells cultivated in parallel (Table 1, center column of data). This indicated cell-to-cell transfer of the vector-bearing capsid pseudotyped with supposedly fusion-deficient Env. When HeLa cells were transfected by the calcium phosphate coprecipitation method, the outcome of the experiment was basically the same as that shown in Fig. 3 and Table 1, although the absolute values were lower than those obtained by PolyFect transfection (data not shown). In 293 cells, the situation was somewhat less clear, since the amount of double-positive cells was far less than that of single-marker-positive cells (compare the two rightmost columns of Table 1). Thus, cell-to-cell transfer of apparently fusion-defective PFV was most prominent in HeLa cells.
FIG. 3.
Normal HeLa and 293T cells were transfected with the indicated plasmids and monitored by flow cytometry for EGFP marker expression on a weekly basis. Cotransfection of cells with fusion-defective Env constructs (EM20 and EM124) resulted in a higher percentage of stably EGFP-positive cells than the sole transfection of cells with the IRT-competent vector pMH88.
Importantly, we were unable to demonstrate double-positive cells upon cocultivation of either HeLa or 293 cells with their G418-resistant derivatives after transfection with pMH88 only. Since stably EGFP-positive cells can be easily obtained following transfection with pMH88 (Fig. 3), this result corroborates the previous report on IRT of an env-deficient PFV vector (7).
PFV IRT is reminiscent of the transposition of classical long terminal repeat (LTR) retrotransposons (3). Although the different experimental systems cannot be exactly compared, an extraordinarily high frequency of PFV IRT has been calculated (7). PFV IRT also resembles the nuclear reshuttling of hepatitis B virus genomes in infected cells (6). However, in contrast to hepatitis B virus, PFV integrates obligatorily into the host cell genome (5, 12, 16) and it is unresolved whether integrants due to IRT can be harmful to the host cell. To address the questions of the relationship to other movable elements, a quantifiable system for PFV IRT is required, which is currently not available. Such a system should also allow the determination of how PFV IRT is regulated.
In addition, it is not known whether IRT occurs upon natural foamy virus infection, for example, in monkeys, and what, if any, function this may have. Furthermore, IRT does not seem to be a common feature of foamy viruses, since it could not be demonstrated for the related virus of feline origin (17). Thus, we cannot exclude the possibility that IRT is an epiphenomenon of PFV vectors in which env is deleted.
The discovery of Env mutants that are obviously fusion deficient and disabled in virus infection through the cell-free supernatant but competent in allowing virus infection by cell-to-cell transfer is surprising. One possible explanation for this finding is the cleavage of an alternative site in Env by a cellular protease that is active only under the condition of direct cell-to-cell contact. Such hypothesized alternative cleavage could enable the fusion of a TM-like protein with endosomal membranes, because it has been shown recently that PFV fusion is low pH dependent (13). This hypothesized alternative cleavage site is obviously minor, since it was not detected upon protein analysis of EM20 by radioimmunoprecipitation assay (15). Furthermore, the data presented here suggest that this cleavage is cell specific, since efficient cell-to-cell transfer was observed only in HeLa cells and not in 293 cells.
Acknowledgments
We are indebted to Carolyn Rinke and Maxine Linial for communicating results prior to publication and Ottmar Herchenröder for critical reading of the manuscript.
This work was supported by grants from the DFG (Re627/6-2, Li621/2-3), the BMBF (01KV9817/0 and BEO 0312191), the Bayerische Forschungsstiftung, the Sächsisches Staatsministerium für Umwelt und Landwirtschaft (13-8811.61/142), the EU (BMH4-CT97-2010), and Bayer AG.
REFERENCES
- 1.Adam, M. A., and A. D. Miller. 1991. Internal initiation of translation in retroviral vectors carrying picornavirus 5′ nontranslated regions. J. Virol. 65:4985-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bansal, A., K. L. Shaw, B. H. Edwards, P. A. Goepfert, and M. J. Mulligan. 2000. Characterization of the R527T point mutant of a putative cleavage site in human foamy virus Env. J. Virol. 74:2949-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman, F. 2002. Lateral DNA transfer. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Enssle, J., I. Jordan, B. Mauer, and A. Rethwilm. 1996. Foamy virus reverse transcriptase is expressed independently from the gag protein. Proc. Natl. Acad. Sci. USA 93:4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enssle, J., A. Moebes, M. Heinkelein, M. Panhuysen, B. Mauer, M. Schweizer, D. Neumann-Haefelin, and A. Rethwilm. 1999. An active human foamy virus integrase is required for viral replication. J. Gen. Virol. 80:1445-1452. [DOI] [PubMed] [Google Scholar]
- 6.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 7.Heinkelein, M., T. Pietschmann, G. Jármy, M. Dressler, H. Imrich, J. Thurow, D. Lindemann, M. Bock, A. Moebes, J. Roy, O. Herchenröder, and A. Rethwilm. 2000. Efficient intra-cellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 19:3436-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinkelein, M., M. Schmidt, N. Fischer, A. Moebes, D. Lindemann, J. Enssle, and A. Rethwilm. 1998. Characterization of a cis-acting sequence in the pol region required to transfer human foamy virus vectors. J. Virol. 72:6307-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinkelein, M., J. Thurow, M. Dressler, H. Imrich, D. Neumann-Haefelin, M. O. McClure, and A. Rethwilm. 2000. Complex effects of deletions in the 5′ untranslated region of primate foamy virus on viral gene expression and RNA packaging. J. Virol. 74:3141-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, and T. J. White (ed.), PCR protocols, a guide to methods and applications. Academic Press, San Diego, Calif.
- 11.Lindemann, D., and A. Rethwilm. 1998. Characterization of a human foamy virus 170 kilodalton Env-Bet fusion protein generated by alternative splicing. J. Virol. 72:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meiering, C. D., K. E. Comstock, and M. L. Linial. 2000. Multiple integrations of human foamy virus in persistently infected human erythroleukemia cells. J. Virol. 74:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picard-Maureau, M., G. Jármy, A. Berg, A. Rethwilm, and D. Lindemann. 2003. Foamy virus envelope glycoprotein mediated entry involves a pH-dependent fusion process. J. Virol. 77:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietschmann, T., M. Heinkelein, M. A. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pietschmann, T., H. Zentgraf, A. Rethwilm, and D. Lindemann. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J. Virol. 74:4474-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rethwilm, A. 2003. Foamy virus replication strategy. Curr. Top. Microbiol. Immunol. 277:1-26. [DOI] [PubMed] [Google Scholar]
- 17.Roy, J., W. Rudolph, T. Juretzek, K. Gärtner, M. Bock, O. Herchenröder, D. Lindemann, M. Heinkelein, and A. Rethwilm. 2003. Feline foamy virus genome and replication strategy. J. Virol. 77:11324-11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffith, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]