Abstract
This study investigated the genetic organization of erm(B)-carrying transposons of Streptococcus pneumoniae and their distribution in tetracycline-resistant clinical isolates. By comparatively analyzing reference pneumococci carrying erm(B)/tet(M) transposon Tn1545, Tn6003, Tn6002, or Tn3872, we demonstrated a substantial correspondence between Tn1545 and Tn6003, which have the same resistance gene combination [tet(M) (tetracycline), erm(B) (erythromycin), and aphA-3 (kanamycin)]; share the macrolide-aminoglycoside-streptothricin element, containing a second erm(B); and only differ by a ca. 1.2-kb insertion (containing a putative IS1239 insertion sequence) detected in Tn1545 from S. pneumoniae reference strain BM4200. These results enabled elucidation of the structure of Tn1545, the first erm(B)-carrying transposon described in S. pneumoniae. A collection of 83 erythromycin- and tetracycline-resistant clinical pneumococci, representative of recent Italian isolates carrying erm(B) as the sole erythromycin resistance gene, was used to investigate the distribution of the different transposons. All 83 organisms were positive for tet(M) and bore an erm(B)/tet(M) transposon that could be characterized by using a specific set of primer pairs; Tn3872 was detected in 18 isolates, Tn6002 in 59 isolates, and Tn6003 in 6 (the sole kanamycin-resistant) isolates. The genetic organization of transposon Tn1545, with its specific insertion, was not detected in any of the isolates tested. The erm(B)-carrying elements of tetracycline-resistant pneumococci substantially corresponded to those [bearing a silent tet(M) gene] recently detected in tetracycline-susceptible pneumococci. Overall, in erm(B)-positive pneumococci, Tn6003 was the least common erm(B)-carrying Tn916-related element and Tn6002 the most common.
A posttranscriptional methylase-mediated modification of the 23S rRNA encoded by the erm(B) gene, which can be expressed either constitutively or inducibly, is the most common mechanism of resistance to macrolide, lincosamide, and streptogramin B antibiotics in Streptococcus pneumoniae (21). In Italy, this mechanism accounts for approximately two-thirds of the erythromycin-resistant pneumococci (24, 25), approximately 90% of which are cross-resistant to tetracycline (24). Different erm(B)-carrying Tn916-related elements (Tn1545, Tn3872, Tn6002, and Tn6003) have been detected in S. pneumoniae isolates with erm(B)-mediated erythromycin resistance.
Tn1545, the first of these elements described, originally detected in multiply resistant S. pneumoniae strain BM4200, and reported to have a size of 25.3 kb (12, 13), is a transposon carrying the resistance genes tet(M) (tetracycline), erm(B) (macrolide, lincosamide, and streptogramin B antibiotics), and aphA-3 (kanamycin and structurally related aminoglycosides). Its genetic organization has been analyzed essentially by restriction mapping (3); only specific portions have been sequenced (4, 5, 22, 28, 34).
Tn3872 (reported size, 23.3 kb) is a composite element resulting from the insertion of the erm(B)-containing Tn917 transposon (33) into Tn916 (23).
Tn6002 is a 20.9-kb element resulting from the insertion of a ca. 2.8-kb erm(B)-containing DNA fragment [erm(B) element] into Tn916 (2, 10). Tn6002 was originally detected in Streptococcus cristatus and designated Tn916Erm, but it has not been published even though its complete sequence has been available in GenBank since 2005 (accession no. AY898750). In S. pneumoniae isolates, Tn6002 may also be part of composite transposon Tn2010 (26.3 kb) (14).
Tn6003 (25.1 kb)—unlike Tn3872 and Tn6002 and similar to Tn1545—also carries, besides the tetracycline [tet(M)] and erythromycin [erm(B)] resistance determinants, the kanamycin resistance gene aphA-3. In Tn6003, aphA-3 has been shown to be contained in a 4.2-kb insertion called the MAS (macrolide-aminoglycoside-streptothricin) element, also containing a second erm(B) gene (10).
All of these erm(B)-carrying elements are derivatives of Tn916, a ubiquitous 18.0-kb transposon originally detected in Enterococcus faecalis (17), that confers tetracycline resistance via the tet(M) gene on a wide range of clinically important (especially gram-positive) bacteria (8, 29). As we recently showed, a minority of both S. pneumoniae (10) and Streptococcus pyogenes (2) isolates with erm(B)-mediated erythromycin resistance are tetracycline susceptible because the tet(M) gene is silent.
The present study, by comparatively analyzing the genetic organization of erm(B)-carrying transposons in S. pneumoniae reference strains and investigating their distribution in tetracycline-resistant clinical isolates, shows a substantial correspondence between Tn1545 and Tn6003, while Tn6002 appears to be the most common Tn916-related transposon in pneumococci.
MATERIALS AND METHODS
Bacterial strains.
The following four S. pneumoniae reference strains carrying specific erm(B)/tet(M) transposons were used and compared in PCR- and sequencing-based experiments: BM4200, carrying Tn1545; Ar4, carrying Tn6003; Pg1, carrying Tn6002; and Vt2, carrying Tn3872. Strain BM4200 was from the Pasteur Institute Collection, Paris, France; the other three reference strains (10) and the reference pneumococcus (An51) carrying Tn916 were from our Institute's collection.
A collection of 83 erythromycin- and tetracycline-resistant nonreplicate clinical isolates of S. pneumoniae, all recovered in different areas of Italy in 2005 to 2006 and carrying erm(B) as the sole erythromycin resistance gene, was used to investigate the distribution of the different transposons.
Antibiotics and susceptibility tests.
Erythromycin, tetracycline, and kanamycin were purchased from Sigma Chemical Co., St. Louis, MO. Erythromycin and tetracycline MICs were determined by agar dilution as recommended by the Clinical and Laboratory Standards Institute (9). Kanamycin susceptibility was determined by agar diffusion using the disk drug content (1,000 μg) and breakpoints (susceptible, ≥14 mm; resistant, <10 mm) recommended by the French Society for Microbiology (http://www.sfm.asso.fr).
Serotyping.
All isolates were serotyped by the capsular-swelling test with specific antisera (Statens Seruminstitut, Copenhagen, Denmark). Serotypes were indicated with conventional capsular type designations.
PFGE.
Macrorestriction with SmaI endonuclease (New England BioLabs, Beverly, MA) and pulsed-field gel electrophoresis (PFGE) analysis were performed and the relevant patterns were examined and compared as described elsewhere (30). For clusters with at least two isolates, types were designated with lowercase letters in order of size.
Amplification experiments.
The principal primer pairs used in this study are shown in Table 1. Other primer pairs used to detect particular genes or to analyze transposon structures have been reported elsewhere (10, 11). DNA preparation and amplification and electrophoresis of PCR products were carried out by established procedures as described previously (11). DNAs from the reference strains carrying specific transposons were used as specific controls.
TABLE 1.
Principal oligonucleotide primer pairs used in this studya
| Gene | Primer designation | Sequence (5′-3′) | Reference | Product size(s) (bp) |
|---|---|---|---|---|
| Primer pairs used to discriminate between Tn916 family erm(B)/tet(M)-carrying pneumococcal transposons | ||||
| orf20b | J12 | CCCATTGAAGACGCAGAAGT | 10 | 801, 3,649, 7,874c |
| IR18-19d | J11 | AAAAATCCCTACCGCACT | 10 | |
| tet(M) | TETM2 | GAACTCGAACAAGAGGAAAGC | 26 | 9,102, 3,835e |
| xis | xis-rev | GCGTCCAATGTATCTATAA | 1 | |
| erm(B) | ERMB1 | GAAAAGGTACTCAACCAAATA | 32 | 11,944f |
| tet(M) | TETM3 | ATGGAAGCCCAGAAAGGAT | 26 | |
| tet(M) | TETM2 | GAACTCGAACAAGAGGAAAGC | 26 | 3,203g |
| erm(B) | ERMB2 | AGTAACGGTACTTAAATTGTTTAC | 32 | |
| orf13b | 06 | GGTACTTGAAAAGAACGGGAG | 7 | 2,434, 3,589h |
| tet(M) | TETM2dg | CTATCTCCTCCTTTACAC | 15 | |
| Primer pairs used to analyze and compare Tn916 family transposons from reference pneumococci | ||||
| orf24b | TN6-rev | CCATCAAACATTCATTCAGC | 10 | 3,358, 3,356e |
| orf20b | J13 | GGTTTTGTGGTTAGTTTT | 10 | |
| IR18-19d | J11-rev | TTTTTAGGGATGGCGTGA | 10 | 4,059 |
| orf15b | J15-rev | AAAGGAAGCCGATAGGATAAA | 10 | |
| orf15b | J15 | TTTATCCTATCGGCTTCCTTT | 10 | 3,955, 3,956i |
| tet(M) | TETM3 | ATGGAAGCCCAGAAAGGAT | 26 | |
| tet(M) | TETM2 | GAACTCGAACAAGAGGAAAGC | 26 | 10,438, 5,171i |
| int | int-rev | GACGCTCCTGTTGCTTCT | 16 | |
| Primer pair used for the detection of kanamycin resistance determinant | ||||
| aphA-3 | APHA1 | GCCGATGTGGATTGCGAAAA | 20 | 292 |
| aphA-3 | APHA2 | GCTTGATCCCCAGTAAGTCA | 20 |
Other primer pairs used to detect particular genes or to analyze transposon structures are reported elsewhere (10, 11).
Numbered according to the reported sequence of Tn916 (accession no. U09422).
The first product size was expected according to the reported sequence of Tn916 and the reported organization of Tn3872, the second according to the reported sequence of Tn6002 (accession no. AY898750), and the third according to the reported sequences of Tn6003 (accession no. AM410044) and Tn1545 (accession no. AM903082).
IR18-19, intergenic region between orf19 and orf18 (numbered as specified in footnote b).
The first product size was expected according to the reported organization of Tn3872, the second according to the reported sequence of Tn6002 or Tn6003 and the reported organization of Tn1545.
Expected according to the reported sequence of Tn6002 or Tn6003.
The first product size was expected according to the reported sequence of Tn916, Tn6002, or Tn6003 and the reported organization of Tn3872, the second according to the reported sequence of Tn1545 (accession no. AM889142).
The first product size was expected according to the reported sequence of Tn916 and the reported organization of Tn3872, the second according to the reported sequences of Tn6002 and Tn6003.
The xis gene was taken as the marker of Tn916 family transposons. On the basis of previous data (10) and the present findings, PCR-based discrimination among erm(B)/tet(M)-carrying transposons Tn3872, Tn6002, Tn6003, and Tn1545 was performed with a set of primer pairs (Table 1) that enabled their particular genetic organization to be recognized.
Nucleotide sequence accession numbers.
Two regions of transposon Tn1545 newly sequenced from S. pneumoniae reference strain BM4200 were submitted to the GenBank/EMBL sequence database and assigned accession no. AM889142 (the O6/TETM2dg amplicon) and AM903082 (the J12/J11 amplicon). The sequence of transposon Tn6003, previously determined only for the region corresponding to the J12/J11 amplicon (10), is now entirely available under the same accession number (AM410044).
RESULTS AND DISCUSSION
Comparative analysis of reference strains.
PCR and sequencing experiments were performed in order to compare the four reference pneumococci carrying specific erm(B)/tet(M) transposons (BM4200 [Tn1545], Ar4 [Tn6003], Pg1 [Tn6002], and Vt2 [Tn3872]) and the reference pneumococcus (An51) carrying Tn916.
The main results of PCR assays are summarized in Table 2, where the Tn916 type structure is subdivided into five regions from the left end to the right end [(i) orf24 to orf20, (ii) orf20 to orf19, (iii) orf18 to orf15, (iv) orf15 to tet(M), and (v) tet(M) to int], analyzed, and compared with appropriate primers. (i) No differences were found in the DNA regions of the five reference transposons between the left end (orf24) and orf20. (ii) Between orf20 and orf19, only Tn3872 exhibited the same PCR-based structure as Tn916. Tn6002 showed the insertion of the 2.8-kb erm(B) element (accession no. AY898750; 10, 14), whereas an insertion of ca. 7.1 kb, corresponding in Tn6003 to the 4.2-kb MAS element, in turn inserted into the erm(B) fragment (10), was likewise detected in Tn1545. (iii) No differences were found in the DNA regions of the five reference transposons between orf18 and orf15. (iv) Between orf15 and tet(M), only Tn1545 exhibited a PCR-based structure different from that of Tn916, due to a ca. 1.2-kb insertion that additional PCR experiments (not shown) indicated to be situated between orf13 and tet(M). (v) Between tet(M) and the right end (int) of the five reference transposons, only Tn3872 differed from Tn916, due to the insertion of the Tn917 transposon (ca. 5.3 kb) (23).
TABLE 2.
Comparison of the amplicons obtained from reference pneumococci carrying different Tn916 family transposons with specific primer pairs
| Reference strain (transposon carried) | Sizea (kb) of amplicon obtained from ORFs (with primer pair indicated):
|
||||
|---|---|---|---|---|---|
| orf24/orf20 (TN6-rev/J13) | orf20/orf19 (J12/J11) | orf18/orf15 (J11-rev/ J15-rev) | orf15/tet(M) (J15/ TETM3) | tet(M)/int (TETM2/ int-rev) | |
| An51 (Tn916) | 3.4 | 0.8 | 4.1 | 4.0 | 5.2 |
| BM4200 (Tn1545) | 3.4 | 7.9 | 4.1 | 5.2 | 5.2 |
| Ar4 (Tn6003) | 3.4 | 7.9 | 4.1 | 4.0 | 5.2 |
| Pg1 (Tn6002) | 3.4 | 3.6 | 4.1 | 4.0 | 5.2 |
| Vt2 (Tn3872) | 3.4 | 0.8 | 4.1 | 4.0 | 10.4 |
Five genetic regions in the sequence of Tn916 family transposons are considered. The relevant primer pairs indicated are described in Table 1.
New sequencing experiments and comparative analysis of previously determined sequences were performed in order to clarify the structural relationships of Tn1545 to Tn6003 and Tn6002. Homology between formerly sequenced segments of Tn1545 and the corresponding segments of Tn6003 and Tn6002 (accession no. AM410044 and AY898750, respectively) ranged from 98.2% to 100%. In particular, 100% homology to Tn6003 was detected for the aphA-3 gene promoter (596 bp; accession number X05577), a region that is absent in Tn6002. Homologies to other individual regions of Tn6003 and Tn6002, respectively, were as follows: 99.8% and 99.1% for the erm(B) gene (1,196 bp; accession no. X52632), 99.5% and 98.2% for a region (2,094 bp, sequenced in E. faecalis BM4127; accession no. X04388) including the tet(M) gene and its leader peptide and ribosomal binding site, and 98.7% and 99.9% for the segment containing the int and xis genes (2,058 bp; accession no. X61025).
Two new regions of Tn1545 were sequenced from reference strain BM4200, namely, the J12/J11 amplicon (spanning from orf20 to the intergenic region between orf19 and orf18; accession no. AM903082) and the O6/TETM2dg amplicon [spanning from orf13 to tet(M); accession no. AM889142]. The J12/J11 amplicon (7,874 bp) was virtually identical (99.7% homology) to the one sequenced in Tn6003 from reference strain Ar4 (accession no. AM410044) (10); in particular, its right portion contained the erm(B) element (99.6% and 99.4% homology to Tn6003 and Tn6002, respectively), and its left portion contained the MAS element (99.7% homology to Tn6003). Remarkably, the second erm(B) gene of the MAS element lacked the stop codon, exactly like that found in Tn6003 (10). The O6/TETM2dg amplicon displayed a 1,245-bp insertion (accession no. AM889142) between orf13 and orf12, containing a putative IS1239 insertion sequence with a transposase-encoding open reading frame (ORF) of 951 bp flanked by direct repeats. This insertion, found in Tn1545 but not in Tn6003, was the sole dissimilarity between these two closely related transposons. The right portion of the amplicon contained the tet(M) gene with its regulatory region, whose homologies to the corresponding segment were as follows: 100% for Tn1545 from E. faecalis BM4127, 99.5% for Tn6003 from reference strain Ar4 (tetracycline susceptible), and 99.8% for Tn6003 from a randomly selected strain from the six tetracycline-resistant pneumococci of this study, which yielded a 7.9-kb amplicon with primer pair J12/J11. Neither Tn6003 nor Tn6002 had an EcoRI endonuclease restriction site, as demonstrated for Tn1545 since the earliest studies (12).
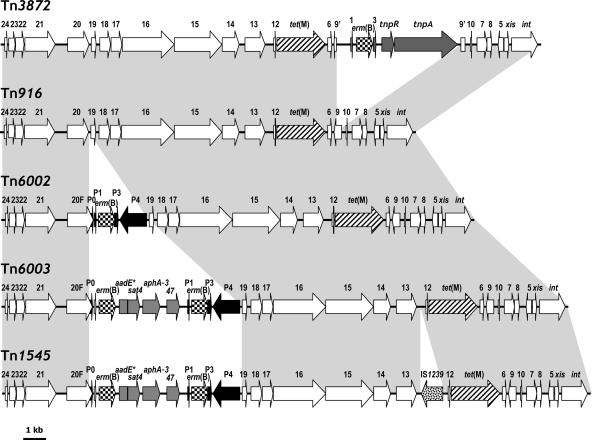
The maps of Tn916 and Tn916 family erm(B)-carrying transposons Tn3872, Tn6002, Tn6003, and Tn1545 are represented and compared in Fig. 1.
FIG. 1.
Comparative representation of the ORF maps of Tn916 and Tn916 family erm(B)/tet(M)-carrying transposons Tn3872, Tn6002, Tn6003, and Tn1545. White arrows indicate Tn916 ORFs other than tet(M) (striped). Dark gray arrows indicate ORFs from Tn917 other than erm(B) (checkered). Black arrows indicate ORFs from the ca. 2.8-kb erm(B) element, except for erm(B) (checkered). Light gray arrows indicate ORFs from the ca. 4.2-kb MAS element (10), except for erm(B) (checkered). A spotted arrow indicates the ORF encoding a putative IS1239 transposase detected in Tn1545 of the reference pneumococcus BM4200. Gray areas between ORF maps indicate areas of homology to Tn916. It should be noted that only the sizes of Tn916 (18,032 bp), Tn6002 (20,880 bp), and Tn6003 (25,101 bp) are the result of sequence analysis and are therefore accurate. In fact, the actual size of Tn1545 is likely to be approximately 1 kb greater than the 25.3 kb reported originally (3, 12, 13), the sole difference from Tn6003 appearing to be a 1,245-bp insertion.
Characterization of 83 erm(B)-positive and tetracycline-resistant clinical S. pneumoniae isolates.
Eighty-three isolates representative of recent erm(B)-positive tetracycline-resistant pneumococci circulating in Italy were analyzed for a number of phenotypic and molecular characteristics, summarized in Table 3.
TABLE 3.
Phenotypic and molecular characteristics of the 83 tested pneumococcal isolates, grouped according to the erm(B)/tet(M) genetic element carried
| erm(B)/tet(M) genetic element carried (n) | Presencea of gene:
|
Susceptibility to kanamycinb | Serotype (n)c | PFGE type (n)c,d | ||
|---|---|---|---|---|---|---|
| tnpR | tnpA | aphA-3 | ||||
| Tn3872 (18) | + | + | − | S | 19A (3), 9V (2), 19F (2), 23F (2), 3, 6A, 6B, 7F, 9N, 10A, 14, 15A, 36 | ost (9), b (2), c (2), n (2), a, e, k |
| Tn6002 (59) | − | − | − | S | 3 (12), 23F (12), 6B (9), 19F (6), 19A (5), 6A (2), 11A (2), 7F, 9A, 9L, 9N, 9V, 10A, 14, 15A, 17F, 23A, 33F | ost (29), a (4), b (3), d (3), f (3), c (2), g (2), h (2), i (2), j (2), l (2), o (2), e, k, m |
| Tn6003 (6) | − | − | + | R | 14 (3), 1, 9A, 23F | A (2), b, d, e, m |
+, present; −, absent. All tested isolates were positive for tet(M) and xis and negative for tet(O) and the putative IS1239 insertion sequence.
S, susceptible; R, resistant.
The number of isolates is indicated in parentheses for types represented by more than one.
ost, one-strain type.
Erythromycin MICs were uniformly >128 μg/ml. All isolates were positive for tet(M) and negative for tet(O), with tetracycline MICs ranging between 32 and >128 μg/ml. Six isolates were kanamycin resistant and bore the aphA-3 gene.
Twenty different serotypes (14 represented by at least two isolates) were detected. The most common was 23F (n = 15), followed by 3 (n = 13), 6B (n = 10), 19A (n = 8), 19F (n = 8), and 14 (n = 5). Fifty-seven PFGE types (15 represented by at least two isolates) were detected. Type a, the most common, included seven isolates.
An erm(B)/tet(M) linkage was detected in all 83 isolates, which were all positive for the xis gene. The genetic organization of transposon Tn3872, confirmed by positive PCRs for tnpR and tnpA, was detected in 18 isolates; that of Tn6002 in 59 isolates; and that of Tn6003 in the 6 kanamycin-resistant isolates. Interestingly, different transposons were often carried by isolates with the same PFGE profile. The genetic organization of transposon Tn1545, with its specific insertion, was detected in none of the isolates. It is worth noting that the putative IS1239 insertion sequence was found neither in the Tn6003 and Tn6002 elements of the tetracycline-resistant isolates of this study nor in those of the tetracycline-susceptible isolates investigated previously (10).
Conclusions.
This study elucidates the complete organization and structure of Tn1545—the first erm(B)-carrying transposon to be described in S. pneumoniae—and shows that the erm(B)-carrying elements [bearing an expressed tet(M) gene] of tetracycline-resistant pneumococci substantially correspond to those [bearing a silent tet(M) gene] recently detected in tetracycline-susceptible pneumococci (10). Such transposons, though often presenting a number of variants and minor differences, substantially fit into one of two groups (probably also denoting different origins or evolutions), depending on the erm(B)-carrying fragment. In one group (Tn3872), erm(B) is contained in the Tn917 transposon, inserted into orf9 of Tn916 at base 14525 of its published sequence (accession no. U09422); this group is typically associated with positive PCRs for tnpR and tnpA, the transposase genes of Tn917. In the other group (Tn6002, Tn6003, and Tn1545), erm(B) is contained in the erm(B) element, inserted at base 3847 of the Tn916 sequence. This insertion distinguishes Tn6002, whereas the further insertion of the MAS element [containing a second erm(B) gene] at base 4110 of the published sequence of Tn6002 (accession no. AY898750) distinguishes Tn6003/Tn1545. It should be noted that a PCR with primer pair J12/J11 is sufficient to distinguish among Tn3872, Tn6002, and Tn6003/Tn1545, which yield amplicons of 0.8 kb, 3.7 kb, and 7.9 kb, respectively (Table 1). Tn6003 and Tn1545 can be distinguished with primer pair O6/TETM2dg (amplicons of 2.4 kb and 3.6 kb, respectively).
Curiously, although its prevalence may obviously vary from area to area and over time, the first-described erm(B)-carrying transposon of the Tn916 family, i.e., Tn1545, was never detected in our isolates, and its variant lacking the IS1239 insertion sequence, i.e., Tn6003, was the least common of the pneumococcal erm(B)-carrying Tn916-related elements. Indeed, in terms of frequency, Tn6002 seems to rank first (roughly from half to two-thirds) among both tetracycline-susceptible (10) and tetracycline-resistant pneumococci, followed by Tn3872, while Tn6003 consistently ranks last. Kanamycin resistance, or the presence of the aphA-3 gene, has usually been ignored in most epidemiological surveys of erythromycin-resistant pneumococci; however, the few studies addressing the point confirm that kanamycin resistance is associated with a tiny minority of erm(B)-carrying isolates (6, 18, 19, 31).
Acknowledgments
We are grateful to Eleonora Giovanetti, Andrea Brenciani, and Alessandro Bacciaglia for helpful discussions.
This work was partly supported by the Italian Ministry of University and Research.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Amezaga, M. R., P. E. Carter, P. Cash, and H. McKenzie. 2002. Molecular epidemiology of erythromycin resistance in Streptococcus pneumoniae isolates from blood and noninvasive sites. J. Clin. Microbiol. 40:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenciani, A., A. Bacciaglia, M. Vecchi, L. A. Vitali, P. E. Varaldo, and E. Giovanetti. 2007. Genetic elements carrying erm(B) in Streptococcus pyogenes, and association with tet(M) tetracycline resistance gene. Antimicrob. Agents Chemother. 51:1209-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caillaud, F., C. Carlier, and P. Courvalin. 1987. Physical analysis of the conjugative shuttle transposon Tn1545. Plasmid 17:58-60. [DOI] [PubMed] [Google Scholar]
- 4.Caillaud, F., and P. Courvalin. 1987. Nucleotide sequence of the ends of the conjugative shuttle transposon Tn1545. Mol. Gen. Genet. 209:110-115. [DOI] [PubMed] [Google Scholar]
- 5.Caillaud, F., P. Trieu-Cuot, C. Carlier, and P. Courvalin. 1987. Nucleotide sequence of the kanamycin resistance determinant of the pneumococcal transposon Tn1545: evolutionary relationships and transcriptional analysis of aphA-3 genes. Mol. Gen. Genet. 207:509-513. [DOI] [PubMed] [Google Scholar]
- 6.Calatayud, L., C. Ardanuy, E. Cercenado, A. Fenoll, E. Bouza, R. Pallares, R. Martin, and J. Liñares. 2007. Erythromycin-resistant Streptococcus pneumoniae isolated in Spain: serotypes, clones, and mechanisms of resistance. Antimicrob. Agents Chemother. 51:3240-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. [DOI] [PubMed] [Google Scholar]
- 8.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Cochetti, I., E. Tili, M. Vecchi, A. Manzin, M. Mingoia, P. E. Varaldo, and M. P. Montanari. 2007. New Tn916-related elements causing erm(B)-mediated erythromycin resistance in tetracycline-susceptible pneumococci. J. Antimicrob. Chemother. 60:127-131. [DOI] [PubMed] [Google Scholar]
- 11.Cochetti, I., M. Vecchi, M. Mingoia, E. Tili, M. R. Catania, A. Manzin, P. E. Varaldo, and M. P. Montanari. 2005. Molecular characterization of pneumococci with efflux-mediated erythromycin resistance and identification of a novel mef gene subclass, mef(I). Antimicrob. Agents Chemother. 49:4999-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courvalin, P., and C. Carlier. 1986. Transposable multiple antibiotic resistance in Streptococcus pneumoniae. Mol. Gen. Genet. 205:291-297. [DOI] [PubMed] [Google Scholar]
- 13.Courvalin, P., and C. Carlier. 1987. Tn1545: a conjugative shuttle transposon. Mol. Gen. Genet. 206:259-264. [DOI] [PubMed] [Google Scholar]
- 14.Del Grosso, M., R. Camilli, F. Iannelli, G. Pozzi, and A. Pantosti. 2006. The mef(E)-carrying genetic element (mega) of Streptococcus pneumoniae: insertion sites and association with other genetic elements. Antimicrob. Agents Chemother. 50:3361-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Grosso, M., A. Scotto d'Abusco, F. Iannelli, G. Pozzi, and A. Pantosti. 2004. Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gherardi, G., M. Del Grosso, A. Scotto d'Abusco, F. D'Ambrosio, G. Dicuonzo, and A Pantosti. 2003. Phenotypic and genotypic characterization of two penicillin-susceptible serotype 6B Streptococcus pneumoniae clones circulating in Italy. J. Clin. Microbiol. 41:2855-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izdebski, R., E. Sadowy, J. Fiett, P. Grzesiowski, M. Gniadkowski, and W. Hryniewicz. 2007. Clonal diversity and resistance mechanisms in tetracycline-nonsusceptible Streptococcus pneumoniae isolates in Poland. Antimicrob. Agents Chemother. 51:1155-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jos, A. M., D. van Klundert, and J. S. Vliegenthart. 1993. PCR detection of genes coding for aminoglycoside-modifying enzymes, p. 547-552. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular biology: principles and applications. ASM Press, Washington, DC.
- 21.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, P., P. Trieu-Cuot, and P. Courvalin. 1986. Nucleotide sequence of the tetM tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 14:7047-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDougal, L. K., F. Tenover, L. N. Lee, J. K. Rasheed, J. E. Patterson, J. H. Jorgensen, and D. J. Leblanc. 1998. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2312-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monaco, M., R. Camilli, F. D'Ambrosio, M. Del Grosso, and A. Pantosti. 2005. Evolution of erythromycin resistance in Streptococcus pneumoniae in Italy. J. Antimicrob. Chemother. 55:256-259. [DOI] [PubMed] [Google Scholar]
- 25.Montanari, M. P., M. Mingoia, I. Cochetti, and P. E. Varaldo. 2003. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J. Clin. Microbiol. 41:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsvik, B., I. Olsen, and F. C. Tenover. 1995. Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol. Immunol. 10:87-92. [DOI] [PubMed] [Google Scholar]
- 27.Poyart, C., G. Quesne, P. Acar, P. Berche, and P. Trieu-Cuot. 2000. Characterization of the Tn916-like transposon Tn3872 in a strain of Abiotrophia defectiva (Streptococcus defectivus) causing sequential episodes of endocarditis in a child. Antimicrob. Agents Chemother. 44:790-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poyart-Salmeron, C., P. Trieu-Cuot, C. Carlier, and P. Courvalin. 1989. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 8:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripa, S., C. Zampaloni, L. A. Vitali, E. Giovanetti, M. P. Montanari, M. Prenna, and P. E. Varaldo. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 7:65-71. [DOI] [PubMed] [Google Scholar]
- 31.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. Garca, and R. Gómez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 32.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomich, P. K., F. Y. An, and D. B. Clewell. 1980. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 141:1366-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]