Abstract
Many azole-resistant (AR) clinical isolates of Candida albicans display increased expression of the drug transporters CDR1 and CDR2. In this study, we evaluate the molecular mechanisms that contribute to the maintenance of constitutively high CDR1 transcript levels in two matched pairs of azole-susceptible (AS) and AR clinical isolates of C. albicans. To address this, we use reporter constructs of GFP and lacZ fused either to the CDR1 promoter (PCDR1-GFP/lacZ; transcriptional fusion) or to the CDR1 open reading frame (PCDR1-CDR1-GFP/lacZ; translational fusion) integrated at the native CDR1 locus. It is observed that expression of the two reporter genes as a transcriptional fusion in the AR isolates is higher than that in matched AS isolates. However, the difference in the reporter activity between the AS and AR isolates is even greater for the translational fusions, indicating that the sequences within the CDR1 coding region also contribute to its increased expression in AR isolates. Further analysis of these observations by transcription run-on assays demonstrated a ∼5- to 7-fold difference in the transcription initiation rates for the AR isolates from those for their respective matched AS isolates. Measurement of mRNA stability showed that the half-life of CDR1 mRNA in the AR isolates was threefold higher than that in the corresponding AS isolates. Our results demonstrate that both increased CDR1 transcription and enhanced CDR1 mRNA stability contribute to the overexpression of CDR1 in AR C. albicans isolates.
Resistance of the human fungal pathogen Candida albicans to azole antifungals is often caused by increased expression of genes encoding multidrug efflux pumps, the ATP-binding cassette transporter genes CDR1 and CDR2 and/or the major facilitator gene CaMDR1 (1, 7, 23, 29, 34, 36, 37, 38, 44, 45, 46). However, the molecular mechanisms leading to the constitutive overexpression of efflux pump-encoding genes in drug-resistant, clinical C. albicans isolates are only beginning to be understood. In particular, the regulation of CDR1 expression has been studied by many groups (2, 3, 4, 5, 10, 11, 17, 20, 32). Various unrelated stresses, including elevated temperature or the presence of drugs and steroids, induce a transient transcriptional activation of CDR1 in drug-susceptible C. albicans strains (20). Several cis elements in the CDR1 upstream region that affect basal as well as induced CDR1 expression have been identified. Puri et al. (32) identified four upstream activating sequences and four upstream repressing sequences in the 5′ noncoding region of CDR1. A basal regulatory element and a negative regulatory element, in the proximal region of the promoter, have also been characterized and implicated in the basal expression of CDR1 (10, 11). A specific steroid-responsive region in the distal part of the CDR1 promoter, consisting of two progesterone-responsive sequences and one β-estradiol-responsive sequence, has been shown to contribute exclusively to steroid responsiveness of CDR1 (17). Another basal expression element in the CDR1 upstream region and a drug response element (DRE), which is present in the upstream region of CDR1 and CDR2, have been identified by de Micheli et al. (5). The DRE was found to mediate both the transient upregulation of CDR1 and CDR2 by steroid hormones and drugs and their constitutive overexpression in a resistant strain (5).
Coste et al. identified a transcription factor, TAC1 (transcriptional activator of CDR genes), that binds to the DRE in the CDR1 and CDR2 promoters (3). Inactivation of TAC1 resulted in the loss of fluphenazine-induced upregulation of CDR1 and CDR2, with little impact on basal expression levels, and also abrogated the constitutive overexpression of these efflux pumps in a drug-resistant strain (3, 4, 5). CaNdt80p, a homolog of the meiosis-specific transcription factor Ndt80p of Saccharomyces cerevisiae, is another positive regulator of CDR1. Deletion of CaNDT80 abolished the induced expression of CDR1 and increased the sensitivity of C. albicans to antifungals (2). Interestingly, the global repressor CaTup1p acts as a negative regulator of CDR1 expression (26, 48).
Although transcriptional regulation is considered to be the key step accounting for complex basal and induced patterns of CDR1 expression, the possibility of posttranscriptional control of CDR1 expression, which could also affect drug resistance, still remains to be explored. The large amounts of Cdr1p, which correlate with high CDR1 mRNA levels, in azole-resistant (AR) C. albicans strains not only may be due to increased CDR1 transcription but could also be caused by increased stability of its mRNA and protein. It is therefore of interest to compare the following: (i) CDR1 transcription initiation rates, (ii) CDR1 mRNA stability, and (iii) Cdr1 protein stability in drug-susceptible and CDR1-overexpressing, drug-resistant C. albicans strains. In this study, we have addressed these issues by exploiting two pairs of matched azole-susceptible (AS) and CDR1-overexpressing, AR isolates. By using transcriptional and translational reporter gene fusions, transcriptional run-on (TRO), thiolutin, and cycloheximide chase assays, we demonstrate that CDR1 overexpression in C. albicans is caused by an increase in its transcriptional initiation rate and by increased mRNA stability.
MATERIALS AND METHODS
Materials.
Medium chemicals were obtained from HiMedia (Mumbai, India). Restriction endonuclease, DNA-modifying enzymes, ultra-pure deoxyribonucleotides (dATP, dGTP, dCTP, and dTTP) for PCR, and ribonucleotides (ATP, CTP, GTP, and UTP) for TRO were purchased from New England Biolabs (NEB, Inc.). Thiolutin (CP-4092) was a generous gift from Pfizer, Inc., Groton, CT. Radiolabeled [5,6-3H]uridine, [α-32P]dATP, and [α-32P]UTP were obtained from Amersham Biotech and Bhabha Atomic Research Center, India. Polyclonal anti-Cdr1p antibody was custom synthesized from Covance Research Products, Inc., PA. Oligonucleotides used were commercially synthesized from Sigma-Aldrich. All molecular biology-grade chemicals used in this study were obtained from Sigma Chemical Co. (St. Louis, MO), and routinely used chemicals (Tris, sodium chloride, glycine, potassium acetate, sodium carbonate, magnesium chloride, sodium hydroxide pellets, methanol, glacial acetic acid, urea, sodium dodecyl sulfate [SDS], formamide, and ethanol) were obtained from Qualigens and Merck, Mumbai, India.
Bacterial and yeast strains and growth media.
Escherichia coli DH5α was used as a host for plasmid constructions and propagation. C. albicans strains used in this study are listed in Table 1. C. albicans strains were maintained on yeast extract-peptone-dextrose (YEPD) medium. All strains were stored as frozen stocks with 15% glycerol at −80°C. Before each experiment, cells were freshly revived on YEPD plates from this stock.
TABLE 1.
C. albicans strains used
| Strain | Description | Reference |
|---|---|---|
| Gu4 | Fluconazole-susceptible clinical isolate | Franz et al. (6) |
| Gu4G1 | PCDR1-GFP integrated at CDR1 locus | This study |
| Gu4G2 | PCDR1-CDR1-GFP integrated at CDR1 locus | This study |
| Gu4L2 | PCDR1-lacZ integrated at CDR1 locus | This study |
| Gu4L3 | PCDR1-CDR1-lacZ integrated at CDR1 locus | This study |
| Gu5 | Fluconazole-resistant clinical isolate | Franz et al. (6) |
| Gu5G1 | PCDR1-GFP integrated at CDR1 locus | This study |
| Gu5G2 | PCDR1-CDR1-GFP integrated at CDR1 locus | This study |
| Gu5L2 | PCDR1-lacZ integrated at CDR1 locus | This study |
| Gu5L3 | PCDR1-CDR1-lacZ integrated at CDR1 locus | This study |
| DSY294 | Fluconazole-susceptible clinical isolate | Sanglard et al. (38) |
| DSY294G1 | PCDR1-GFP integrated at CDR1 locus | This study |
| DSY294G2 | PCDR1-CDR1-GFP integrated at CDR1 locus | This study |
| DSY294L2 | PCDR1-lacZ integrated at CDR1 locus | This study |
| DSY294L3 | PCDR1-CDR1-lacZ integrated at CDR1 locus | This study |
| DSY296 | Fluconazole-resistant clinical isolate | Sanglard et al. (38) |
| DSY296G1 | PCDR1-GFP integrated at CDR1 locus | This study |
| DSY296G2 | PCDR1-CDR1-GFP integrated at CDR1 locus | This study |
| DSY296L2 | PCDR1-lacZ integrated at CDR1 locus | This study |
| DSY296L3 | PCDR1-CDR1-lacZ integrated at CDR1 locus | This study |
Plasmid construction.
All primers and plasmids used in this study are listed in Table 2. Plasmids pCPL51 and pCPG3, harboring the PCDR1-lacZ and PCDR1-GFP transcriptional fusions, were constructed as follows. A CDR1 promoter fragment was first amplified from genomic DNA of isolate Gu5 with the primers CDR1F and CDR1R, digested at the introduced ApaI and XhoI sites, and substituted for the CDR1 promoter fragment in the previously described plasmid pCPL1 (11) to generate pCPL5. A CDR1 downstream fragment was then amplified from genomic DNA of strain SC5314 with the primers CDR29 and CDR30. The PCR product was digested at the introduced PstI and SacII sites and ligated with the PstI/SacII-digested plasmid vectors pCPL5 and pCPG1 (11) to generate pCPL51 and pCPG3, respectively. Notably, plasmid pCPG1 harbors the CDR1 promoter isolated from SC5314 genomic DNA, which has been used in our previous studies (11). The SC5314-derived CDR1 promoter showed sequence differences from the CDR1 promoters in the matched AS or AR isolates (at nucleotide positions −21, −150, −171, −215, −238, −315, −368, −381, −418, and −455 relative to the transcription start point) which have been used in this study (unpublished observation). These differences, however, did not affect the β-galactosidase reporter activity of integrated cassettes derived either from pCPL1 (11), which harbors the SC5314 CDR1 promoter, or from pCPL4 or pCPL5, which harbor the Gu4- or Gu5-derived CDR1 promoter, respectively (data not shown). Plasmid pCPG2, which contains the GFP reporter gene fused in frame with the last codon of the CDR1 open reading frame (ORF), was constructed as follows. The C-terminal region of CDR1 was amplified from SC5314 genomic DNA with the primers CDR32 and CDR31, digested at the introduced KpnI and BamHI sites, and ligated together with a BamHI-PstI fragment from pADH1G3 (14) containing GFP, the ACT1 transcription termination sequence, and the CaSAT1 marker into the KpnI/PstI-digested pCPL51. To generate pCPL52, in which the lacZ reporter gene instead of GFP is fused in frame to the CDR1 ORF, the C-terminal part of CDR1 was PCR amplified from SC5314 genomic DNA with the primers CDR32 and CT-CDR1-R-RML, digested at the introduced KpnI and XhoI sites, and ligated into the KpnI/XhoI-digested pCPL51. To ensure in-frame translational fusion of the CDR1 ORF with lacZ, pCPL62 was generated, in which an additional “A” in pCPL52 before the lacZ ATG was removed by site-directed mutagenesis employing the A-del pCPL52-F and A-del pCPL52-R primers. All constructs were confirmed by appropriate restriction digestion analysis and by sequencing. The flanking CDR1 sequences in all of these plasmids served for genomic integration of the reporter fusions at the native CDR1 locus of clinical C. albicans isolates, and the dominant CaSAT1 marker (33) was used to select nourseothricin-resistant (Nour) transformants. Briefly, for the electroporation, 5 μl (approximately 1 μg) of the linearized DNA fragments were mixed with 40 μl of electrocompetent cells (33) and electroporated using a Bio-Rad Genepulser XL system (0.2-cm cuvette, 1.5 kV). After electroporation, C. albicans transformants were washed with 1 ml of 1 M sorbitol, resuspended in 1 ml YEPD medium, incubated for 3 to 4 h with shaking at 30°C prior to plating on YEPD plates containing 200 μg/ml of nourseothricin, and grown at 30°C (33). Nour transformants were picked up after 24 h of growth, and single-copy integration of each construct at the desired locus was confirmed by Southern hybridization with gene-specific probes (data not shown).
TABLE 2.
Plasmids and oligonucleotides used
| Name | Description | Source (reference) or sequencea |
|---|---|---|
| Plasmids | ||
| pBS-KS(+) | Plasmid vector used for cloning purpose | Stratagene |
| pACT1 | Plasmid harboring ACT1 gene | Gift from Anand Bachhawat |
| pSAT1 | Plasmid harboring caSAT1 marker | Reuss et al. (33) |
| pLACZ6 | Plasmid harboring lacZ reporter gene | Gaur et al. (11) |
| pADH1G3 | Plasmid harboring GFP reporter gene and caSAT1 marker | Hiller et al. (14) |
| pCPG1 | Plasmid harboring PCDR1-GFP fusion for ACT1 locus integration | Gaur et al. (11) |
| pCPG3 | Plasmid harboring PCDR1-GFP fusion for CDR1 locus integration | This study |
| pCPG2 | Plasmid harboring CDR1-GFP fusion for CDR1 locus integration | Prasad et al. (30) |
| pCPL1 | Plasmid harboring PCDR1-lacZ fusion for ACT1 locus integration | Gaur et al. (11) |
| pCPL51 | Plasmid harboring PCDR1-lacZ fusion for CDR1 locus integration | This study |
| pCPL52 | Plasmid harboring CDR1-lacZ fusion for CDR1 locus integration | This study |
| pCPL62 | Plasmid harboring in frame CDR1-lacZ fusion for CDR1 locus integration | This study |
| Oligonucleotides | ||
| CDR1F | Forward primer for CDR1 promoter amplification | 5′-GATCGGGCCCTCGTTACTCAATAAGTAT-3′ |
| CDR1R | Reverse primer for CDR1 promoter amplification | 5′-AGCTCTCGAGTTCTTTTTGACCTTTTAAAG-3′ |
| CDR1-F-RML | Forward primer for CDR1 promoter amplification | 5′-GCTTCGCCTCAACTTCTTATAAAGTTTTGAAAG-3′ |
| CDR1-R-RML | Reverse primer for CDR1 promoter amplification | 5′-CGTGGTATTCATTAATGGAATGGTTTTCTGAAG-3′ |
| CDR29 | Forward primer for amplification of CDR1 downstream region | 5′-AATTCTGCAGTTTGTTTTTTGACATGGTGGTATC-3′ |
| CDR30 | Reverse primer for amplification of CDR1 downstream region | 5′-TCGTGCCGCGGAACACTTTTTCTTTCTAATTATAA-3′ |
| CDR32 | Forward primer for amplification of CDR1 C-terminal region | 5′-ATTTGGTACCATACATTAAATTTGCTGGTGGG-3′ |
| CDR31 | Reverse primer for amplification of CDR1 C-terminal region | 5′-GTTTGGATCCTTTCTTATTTTTTTTCTCTCTGTTACCC-3′ |
| CT-CDR1-R-RML | Reverse primer for amplification of CDR1 C-terminal region | 5′-GTTTCTCGAGTTTCTTATTTTTTTTCTCTCTGTTACCC-3′ |
| A-del pCPL52-F | Forward primer for in-frame translational fusion of CDR1-ORF with lacZ | 5′-GAGAGAAAAAAAATAAGAAACTCGAGATGAACATGACTGAAAAAATTCAAAC-3′ |
| A-del pCPL52-R | Reverse primer for in-frame translational fusion of CDR1-ORF with lacZ | 5′-GTTTGAATTTTTTCAGTCATGTTCATCTCGAGTTTCTTATTTTTTTTCTCTC-3′ |
| KM1 | Forward primer for amplification of CDR1 N-terminal region | 5′-CTTTTCCACTGGTAACTACT-3′ |
| KM2 | Reverse primer for amplification of CDR1 N-terminal region | 5′-CAATAAACCTGCTGACGAG-3′ |
Restriction sites introduced into the primers are underlined, while the flanking bases are written in italics.
Fluorescence microscopy and flow cytometry of the cells.
The strains were grown overnight in YEPD liquid medium, and aliquots were spotted on microscope slides. Fluorescence microscopy was performed with a Zeiss Axiolab microscope equipped for epifluorescence microscopy with a 50-W mercury high-pressure bulb and the Zeiss fluorescein-specific filter set 09. Imaging scans were acquired with an Argon laser of 488-nm wavelength and corresponding filter settings for green fluorescent protein (GFP) and parallel transmission images. For confocal microscopy, the cells were placed on glass slides and directly viewed with a Bio-Rad confocal microscope (Radiance 2100, AGR, 3Q/BLD; Bio-Rad, United Kingdom) under a 100× oil immersion objective (41). Fluorescence-activated cell sorter (FACS) analysis was performed using a FACSCalibur flow cytometer (Becton-Dickinson Immunocytometry Systems, San Jose, CA) equipped with an argon laser emitting at 488 nm. Fluorescence was measured on the FL1 fluorescence channel equipped with a 530-nm bandpass filter. A total of 20,000 events were counted at the low flow rate. Fluorescence data were collected using logarithmic amplifiers. Data analysis was performed using CellQuest software (Becton Dickinson Immunocytometry Systems). The mean fluorescence intensity was calculated using the histogram statistics program.
Immunodetection of GFP in AS and AR reporter strains.
Purified plasma membrane (PM) and crude-extract fractions of GFP reporter strains were prepared as described previously (13, 41). For Western blots, membranes were incubated with a 1:5,000 dilution of monoclonal anti-GFP antibody (JL-8) (BD Biosciences, Clontech) or a 1:1,000 dilution of polyclonal anti-Pma1p (plasma membrane ATPase) antibody. Immunoreactivity was detected using horseradish peroxidase-labeled secondary antibodies at a dilution of 1:5,000 (goat antimouse antibody for GFP and goat antirabbit antibody for Pma1p) using the enhanced chemiluminescence assay system (ECL kit; Amersham Biosciences, Arlington Heights, IL).
β-Galactosidase assays.
β-Galactosidase assays were performed using duplicate samples of cells from three independent experiments as described previously (11, 21, 42). β-Galactosidase activity was determined by the standard equation (11, 21, 42) and is expressed in Miller units per mg of protein (Miller units are arbitrary units): β-galactosidase activity (Miller units) = [OD420 × 1,000]/[OD600 × t × v], where t is time of reaction expressed in min, v is volume of culture used in the assay, expressed in ml, and OD420 and OD600 are optical densities at 420 and 600 nm, respectively.
TRO analysis.
TRO experiments were performed as described previously (8, 24) with the following modifications. Cells were grown at 30°C in YEPD with agitation until the culture reached an OD600 of 1.0. An aliquot of yeast cells (6 × 108 per ml) was used to perform TRO. The cells were centrifuged for 5 min at 4,000 × g and resuspended in 5 ml of cold TMN (10 mM Tris, 100 mM NaCl, 5 mM MgCl2; pH 7.4). The cells were again centrifuged for 5 min at 4,000 × g, and the cell pellet was resuspended in 900 μl of sterile cold diethyl pyrocarbonate (DEPC)-treated water. Next, the cell suspension was transferred to a fresh microcentrifuge tube containing 50 μl of 10% N-lauryl sarcosine sodium sulfate (sarkosyl) and was incubated for 20 min on ice. After the permeabilization step, cells were recovered by low-speed centrifugation at 6,000 rpm for 2 min at 4°C and the supernatant was removed. In vivo transcription was reinitiated by resuspending the permeabilized cell fraction in 120 μl of 2.5× transcription buffer (50 mM Tris-HCl [pH 7.7], 500 mM KCl, 80 mM MgCl2), 16 μl of AGC mix (10 mM each of ATP, GTP, and CTP), 6 μl of dithiothreitol (0.1 M), 1 U of RNase inhibitor per μl, 10 mM phosphocreatine, 1.2 μg of creatine phosphokinase per μl, and 15 μl of [α-32P]UTP (3,000 Ci/mmol, 10 μCi/μl). Cells were maintained on ice at all times. The final volume was adjusted to 300 μl with DEPC-treated water, and the mix was incubated for 15 min at 30°C to allow transcription elongation. The reaction was stopped by adding 1 ml of ice-cold DEPC-treated water to the mix. Cells were recovered by centrifugation to remove nonincorporated radioactive nucleotides. Total RNA was isolated using the Trizole reagent (Sigma) as per the manufacturer's specifications except that 200 μl of ice-cold acid-washed 0.4- to 0.6-mm-diameter glass beads (Sigma, St. Louis, MO) were also used for efficient and complete lysis of permeabilized cells. Isolated total labeled RNA was again precipitated by adding 2.5 M NH4 acetate and an equal volume of isopropanol. The mixture was stored overnight at −20°C. To pellet the RNA, tubes were centrifuged at 14,000 rpm for 15 min in the microcentrifuge. The isopropanol was removed, and the labeled RNA pellet was washed twice with 70% ethanol, dried, and resuspended in 100 μl of DEPC-treated water. This double precipitation of RNA was used to minimize DNA contamination. Total extracted RNA was spectrophotometrically quantified. An aliquot was used for specific radioactivity determination in a Tri-CARB 2900 TR liquid scintillation analyzer (Packard instrument Co., Inc.). All of the in vivo-labeled RNA of each isolate (∼2 × 106 to 2.5 × 106 cpm) was subsequently used for reverse Northern hybridization with a dot blotted Nylon membrane (Hybond-N+; Amersham Pharmacia Biotech) containing PCR-amplified gene-specific N-terminal CDR1 sequences (nucleotides −242 to +256 from the transcription start point), plasmid pACT1 (positive control), and pBlueScript-KS(+) (negative control) as probes, as per the manufacturer's recommendation. Northern blots were scanned with a phosphorimager scanner (FLA-5000 Fuji phosphorimager). Signal intensities of hybridized nuclear RNA were quantified and subsequently normalized to the actin intensities using densitometry scanning.
Thiolutin chase assay.
In order to measure the CDR1 mRNA half-life, a potent in vivo transcriptional inhibitor of C. albicans, thiolutin, was used (18, 40). AS and AR isolates of C. albicans were incubated with an optimized concentration (40 μg/ml) of thiolutin (data not shown). For this purpose, cultures were treated with 150 μM of the copper chelator bathocuprioinedisulphonic acid and incubated at 30°C for an additional 10 min at 200 rpm. Transcription was subsequently shut off by the addition of 150 nM of CuSO4 and 40 μg/ml of thiolutin. Addition of bathocuprioinedisulphonic acid and CuSO4 was found to enhance the response of the cells to thiolutin (40). Briefly, 100 ml of cells were grown to an OD600 of 1.0 at 30°C. Aliquots of cells were taken at the indicated times after transcriptional shutoff. Total RNA was isolated using Ambion's RiboPure-Yeast RNA isolation kit (catalog no. 1926) as per the manufacturer's instructions. For Northern blots, approximately 20 μg of total RNA from the above samples was hybridized with probes derived from gene-specific sequences of the CDR1 gene. Hybridization signal intensity was quantified with a phosphorimager and was normalized to the band intensity at time T0 and plotted as a line graph.
Cycloheximide chase assay.
Cycloheximide chase assays were performed as described earlier (9) with certain modifications that included the use of an optimized concentration of cycloheximide (16) (data not shown) and the alkaline extraction procedure used for the preparation of crude protein extract (13). Briefly, aliquots of mid-log-phase-grown cells were withdrawn for analysis after translational shutoff at the indicated times and lysed in solution containing 1.85 M NaOH and 7.5% β-mercaptoethanol. Crude proteins isolated were precipitated with 50% trichloroacetic acid and sedimented. The sediment was resuspended in loading buffer (40 mM Tris-HCl [pH 6.8], 8 M urea, 5% SDS, 0.1 M EDTA, 1% β-mercaptoethanol, and 0.1 mg/ml bromophenol blue) and incubated at 37°C for 10 min. Nonsolubilized material was cleared by a centrifugation step, and solubilized proteins (approximately 20 and 30 μg for AR and AS isolates, respectively) were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane for Western blotting. Immunodetected Cdr1p signal intensity was quantified with a phosphorimager and was normalized to the band intensity at time T0 and plotted as a line graph.
RESULTS
Expression of transcriptional and translational GFP reporter fusions in AS and AR isolates.
To analyze the molecular basis of CDR1 upregulation in azole-resistant, clinical C. albicans isolates in more detail, we employed two matched pairs of AS and AR isolates. The resistant isolates Gu5 and DSY296, which were obtained after fluconazole therapy for oropharyngeal candidiasis in two different human immunodeficiency virus-positive patients, were shown by DNA fingerprinting to be highly related to the susceptible isolates Gu4 and DSY294, respectively, which were recovered from earlier infection episodes in the same patients (6, 38). It was recently shown that a mutation in the transcription factor TAC1 is responsible for CDR1 and CDR2 upregulation in DSY296 (4). However, it has not yet been explored if other mechanisms contribute to the overexpression of the efflux pumps in these isolates.
We compared the expression of two different GFP reporter fusions in these isolates, one in which GFP was expressed from the CDR1 promoter (PCDR1-GFP) and another where GFP was fused in frame to the last codon of the CDR1 ORF and expressed from the CDR1 promoter (PCDR1-CDR1-GFP) (see Materials and Methods) (Fig. 1A). The reporter fusions were integrated at the native CDR1 locus, and two transformants of each of the four parental strains were used for further analysis. The reporter strains were designated Gu4G1 (PCDR1-GFP) and Gu4G2 (PCDR1-CDR1-GFP); Gu5G1 (PCDR1-GFP) and Gu5G2 (PCDR1-CDR1-GFP); DSY294G1 (PCDR1-GFP) and DSY294G2 (PCDR1-CDR1-GFP); and DSY296G1 (PCDR1-GFP) and DSY296G2 (PCDR1-CDR1-GFP).
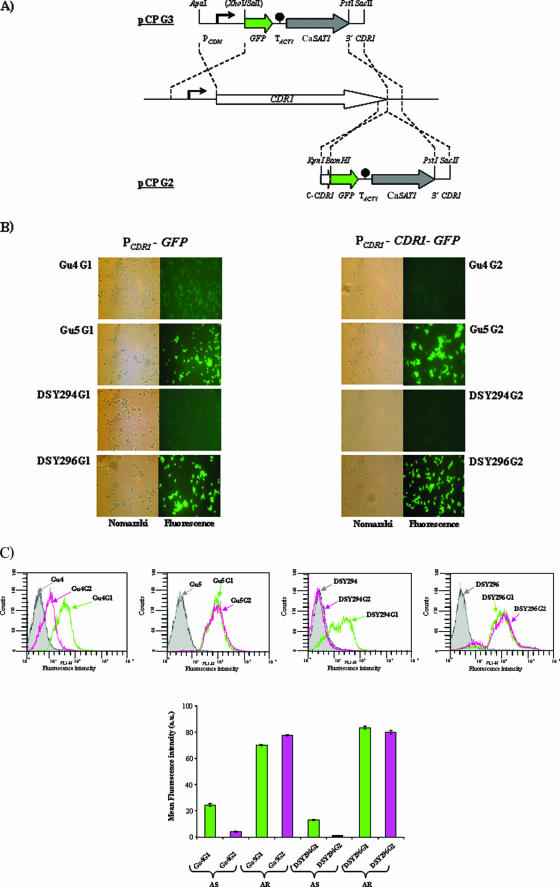
FIG. 1.
Schematic depiction of GFP reporter fusion integrants and their expression in AS and AR isolates. (A) Structure of the DNA cassettes which were used to integrate the transcriptional (PCDR1-GFP, top) and translational (PCDR1-CDR1-GFP, bottom) GFP reporter fusions into the CDR1 locus of the clinical C. albicans isolates (middle). The CDR1 and GFP coding regions are represented by white and green arrows, respectively, the CaSAT1 marker by the gray arrow, and the transcription termination sequence of the ACT1 gene (TACT1) by the filled circle. CDR1 upstream and downstream regions are represented by solid lines, and the CDR1 promoter (PCDR1) is symbolized by the bent arrow. Only relevant restriction sites are shown. (B) Nomarski and corresponding fluorescence micrographs of transformants containing the chromosomally integrated PCDR1-GFP (left) or PCDR1-CDR1-GFP (right) reporter fusion. (C) Cells of the reporter strains grown to exponential phase in YEPD medium were diluted to a density of 2 × 107 cells per ml in phosphate-buffered saline (pH 7.0), and a total of 20,000 events were analyzed by flow cytometry. The parental strains of the transformants, which do not contain GFP, were used as a negative control. The mean fluorescence intensity is shown for each population of cells (bottom panel) after normalization with values for their corresponding negative controls. Since the normalized mean fluorescence intensity of DSY294G2 was a negative value, we designated it “1.0′” for calculating the degree of change for this particular strain. a.u., arbitrary units.
Expression of the PCDR1-GFP transcriptional and PCDR1-CDR1-GFP translational fusions in cells grown to mid-exponential phase (OD600 of ∼1.0) was detected by epifluorescence microscopy (Fig. 1B) and quantified by FACS analysis (Fig. 1C). As expected, the fluorescence intensities of the AR reporter fusion strains were higher than those of the corresponding AS strains (2.5-fold for Gu5G1 versus Gu4G1, 19-fold for Gu5G2 versus Gu4G2; 6-fold for DSY296G1 versus DSY294G1; and 80-fold for DSY296G2 versus DSY294G2), confirming the previously reported increased CDR1 transcript and Cdr1p protein levels in the AR isolates (4, 6, 38). Interestingly, however, expression of the PCDR1-CDR1-GFP translational fusion resulted in much lower fluorescence than expression of the PCDR1-GFP transcriptional fusion in AS isolates (6-fold for Gu4G2 versus Gu4G1 and 13-fold for DSY294G2 versus DSY294G1), whereas the two types of reporter fusions produced comparable fluorescence in AR isolates. Notably, confocal microscopy confirmed that the Cdr1p-GFP fusion protein was correctly localized to the cell membrane in all reporter strains expressing the translational fusion (Fig. 2A). Immunoreactive bands of the expected sizes were observed in whole-cell extracts and plasma membrane preparations of the PCDR1-GFP and PCDR1-CDR1-GFP reporter strains, respectively, after Western immunoblotting with an anti-GFP antibody (Fig. 2B). Additionally, the tagging of PCDR1 and PCDR1-CDR1 with GFP did not alter the drug resistance profiles of AS and AR isolates, which ruled out that the GFP fusions caused any selective impact on Cdr1p functionality for either AS or AR isolates (data not shown).
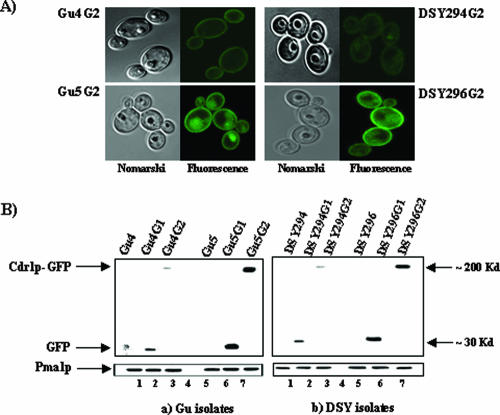
FIG. 2.
Localization of Cdr1p and immunodetection of GFP in reporter fusion transformants. (A) Nomarski (left) and corresponding confocal (right) pictures of the transformants harboring the chromosomally integrated PCDR1-CDR1-GFP (translational fusion) reporter construct are shown which indicate the proper plasma membrane localization of chimeric Cdr1p in clinical C. albicans isolates. The cells were viewed directly on a glass slide with a 100× oil immersion objective. (B) The Western blot analyses were done with an anti-GFP monoclonal antibody on both the transcriptional and translational fusion integrants. Equal loading of protein was assessed by using an anti-Pma1p polyclonal antibody.
Expression of transcriptional and translational lacZ reporter fusions in AS and AR isolates.
To rule out that the reduced expression of the PCDR1-CDR1-GFP translational fusion in AS isolates was an artifact intrinsic to the Cdr1p-GFP fusion protein, we used codon-optimized lacZ (42) as an alternative reporter gene. As for GFP, transcriptional (PCDR1-lacZ) and translational (PCDR1-CDR1-lacZ) lacZ reporter fusions were generated and integrated at the native CDR1 locus of the AS and AR isolates (see Materials and Methods) (Fig. 3A). The expression of Cdr1p in the PCDR1-CDR1-lacZ construct was unaffected by its fusion to lacZ as tested by Western blotting with an anti-Cdr1p antibody (data not shown). Two transformants of each parental strain containing a single copy of the reporter fusion were used for further analysis. The reporter strains were designated Gu4L2 (PCDR1-lacZ) and Gu4L3 (PCDR1-CDR1-lacZ); Gu5L2 (PCDR1-lacZ) and Gu5L3 (PCDR1-CDR1-lacZ); DSY294L2 (PCDR1-lacZ) and DSY294L3 (PCDR1-CDR1-lacZ); and DSY296L2 (PCDR1-lacZ) and DSY296L3 (PCDR1-CDR1-lacZ).
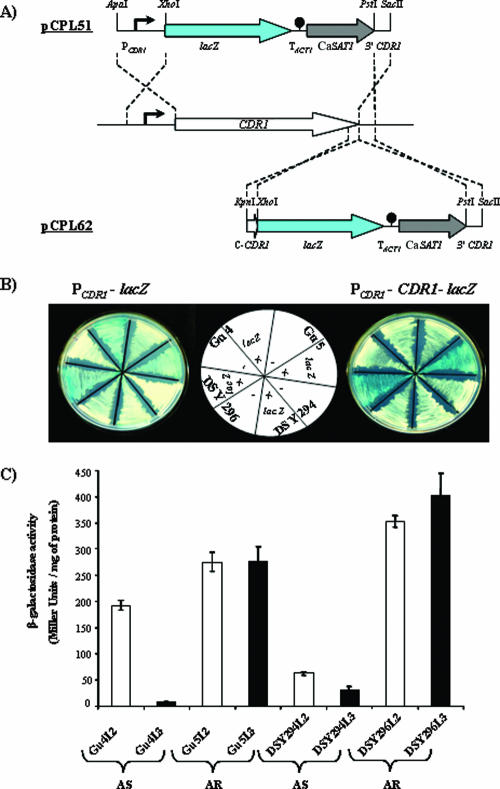
FIG. 3.
Schematic depiction of lacZ reporter fusion integrants and qualitative and quantitative assay of β-galactosidase activity in AS and AR isolates. (A) Structure of the DNA cassettes which were used to integrate the transcriptional (PCDR1-lacZ, top) and translational (PCDR1-CDR1-lacZ, bottom) lacZ reporter fusions into the CDR1 locus of the clinical C. albicans isolates (middle). The CDR1 and lacZ coding regions are represented by white and blue arrows, respectively, the CaSAT1 marker by the gray arrow, and the transcription termination sequence of the ACT1 gene (TACT1) by the filled circle. CDR1 upstream and downstream regions are represented by solid lines, and the CDR1 promoter (PCDR1) is symbolized by the bent arrow. Only relevant restriction sites are shown. (B) Transformants harboring chromosomally integrated PCDR1-lacZ (transcriptional fusion, left) and PCDR1-CDR1-lacZ (translational fusion, right) and their corresponding parental strain (without lacZ) were streaked on minimal medium plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and photographed after 3 days' growth at 30°C. The positions of the individual strains on the plates are shown in the scheme (middle). (C) β-Galactosidase quantitative reporter activities of each transformant were determined as described previously (11, 21, 42). The values are means ± standard deviations (indicated by the bars) of three independent experiments with duplicate measurements of two independent clones. Empty and filled bars indicate transcriptional (PCDR1-lacZ) and translational fusion (PCDR1-CDR1-lacZ) transformants in both AS and AR backgrounds.
Expression of the lacZ reporter gene in various strains was assessed by comparing the intensity of the blue color produced by cells grown on agar plates containing the indicator dye 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Fig. 3B) and was quantified by determining β-galactosidase activities in liquid assays (Fig. 3C). The lacZ reporter study confirmed the results obtained with GFP. Higher lacZ expression levels were observed in transformants of the AR isolates than in transformants of the AS isolates (1.4-fold for Gu5L2 versus Gu4L2, 37-fold for Gu5L3 versus Gu4L3, 5.6-fold for DSY296L2 versus DSY294L2, and 13-fold for DSY296L3 versus DSY294L3). In addition, while the transcriptional and translational fusions yielded comparable activities in the AR isolates, expression of the translational fusion was much lower than that of the transcriptional fusion in the AS isolates (26-fold for Gu4L3 versus Gu4L2 and 2-fold for DSY294L3 versus DSY294L2). Of note, the integration of PCDR1-lacZ reporter fusion constructs at the ectopic ACT1 locus resulted in β-galactosidase activity comparable to that of native CDR1 locus integrants (data not shown).
Growth phase versus β-galactosidase reporter activity.
To investigate whether the observed differences in the expression of transcriptional and translational reporter gene fusions in AS and AR isolates depended on the growth phase, we quantitatively monitored β-galactosidase activities in the lacZ reporter strains at various times during growth in batch cultures. As can be seen in Fig. 4, the low reporter expression levels of the translational PCDR1-CDR1-lacZ fusion compared with those of the transcriptional PCDR1-lacZ fusion in the AS isolates were observed at all growth stages (Fig. 4A and C). In contrast, both types of reporter fusion produced comparable β-galactosidase activities in the AR isolates throughout growth (Fig. 4B and D).
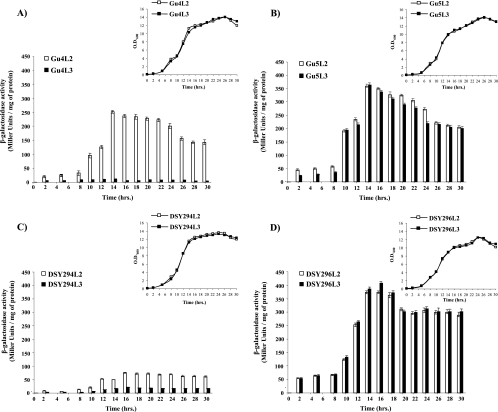
FIG. 4.
β-Galactosidase reporter activity of lacZ reporter fusion integrants of AS and AR isolates during growth phase. Transcriptional fusion (PCDR1-lacZ) and translational fusion (PCDR1-CDR1-lacZ) reporter transformants of each isolates were grown from an initial OD600 of 0.1 in YEPD broth and withdrawn at the indicated time points of growth for β-galactosidase reporter activity (Fig. 4A, B, C, and D). The inset depicts growth curves of the PCDR1-lacZ (□) and PCDR1-CDR1-lacZ (▪) reporter transformants in AS and AR isolates. The negative-control parental strain (without lacZ fusion constructs) reporter activity value was always below 0.5 Miller units, and it was subtracted from the reporter activity of each corresponding transcriptional and translational fusion transformant. The values are means ± standard deviations (indicated by the bars) for three independent experiments with duplicate measurements of two independent clones. Gu4 transformants (A), Gu5 transformants (B), DSY294 transformants (C), and DSY296 transformants (D) were analyzed. Empty and filled bars indicate transcriptional (PCDR1-lacZ) and translational fusion (PCDR1-CDR1-lacZ) transformants in both AS and AR backgrounds.
Taken together, exploitation of reporter fusions and their expression analysis indicated that an increase in CDR1 expression levels in the AR isolates compared to those in the corresponding AS isolates is contributed by affecting either CDR1 promoter activity, mRNA stability, translational efficiency, or protein stability. Therefore, we performed further experiments on native CDR1 (endogenous gene) to get a real insight into whether transcriptional/posttranscriptional control mechanisms are involved in the upregulation of CDR1 expression in AR isolates.
Transcriptional rate for CDR1 is increased in AR isolates.
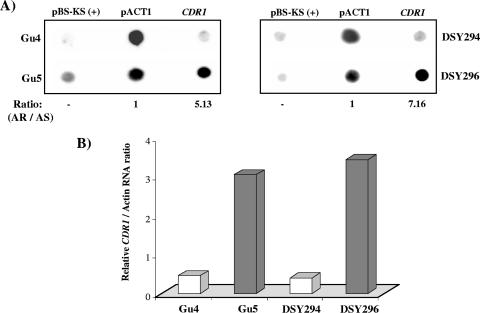
We first tested whether the transcription rate for CDR1 was elevated in the AR isolates. For this purpose, TRO assays were performed. Both AS and AR isolates were grown to an OD600 of ∼1.0, and the cells were permeabilized with the detergent N-lauryl sarcosine sodium sulfate (sarkosyl) for the isolation of intact nuclei (8, 24). The subsequent incubation of isolated nuclei with transcription buffer and radiolabeled [α-32P]UTP reinitiated the transcription (see Materials and Methods). The in vivo-labeled nascent RNAs were then used as probes in reverse Northern hybridizations with dot blotted CDR1-specific PCR-amplified DNA. As controls, pACT1 plasmid DNA, containing the constitutively expressed ACT1 gene, and the empty vector pBluescript were also dotted on the membranes. As shown in Fig. 5A and B, the AR isolates exhibited an increased rate of transcription of CDR1 compared with that for the AS isolates (fivefold for Gu5 versus Gu4 and sevenfold for DSY296 versus DSY294).
FIG. 5.
TRO analysis of AS and AR isolates. (A) Approximately 2 μg (each) of CDR1, pACT1, and empty vector pBlueScript-KS(+) DNA was blotted and immobilized on charged nylon membranes (Hybond-N+; Amersham Pharmacia Biotech) using a dot blot assembly apparatus. The blots were probed with total labeled nuclear run-on RNA as described in Materials and Methods. Hybridization signal intensities of nuclear RNA were quantified using densitometry scanning of phosphorimages. DNA from a pBlueScript-KS(+) plasmid was used as a negative control for nonspecific binding of nuclear RNA to a random DNA fragment. Signal intensities for each isolate were subtracted from the negative control values and subsequently normalized to the intensity corresponding to their AS isolate. The AR/AS ratio is the normalized nuclear RNA intensity between AR and AS isolates. (B) The relative intensity of CDR1 with respect to actin RNA of each isolate is plotted.
CDR1 mRNA stability is increased in AR isolates.
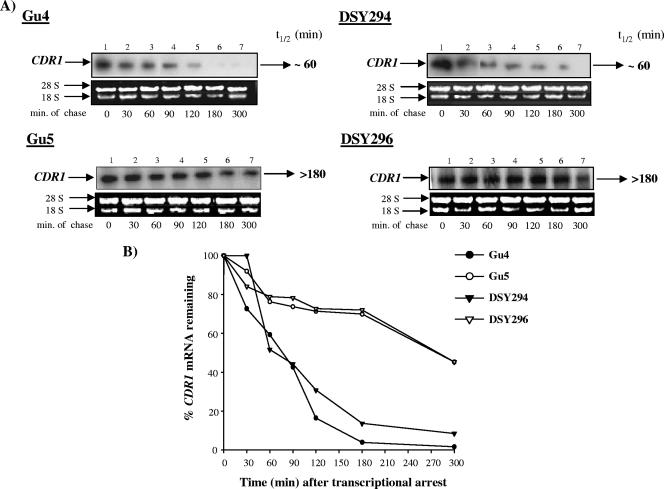
To investigate if in addition to the increased transcription rates posttranscriptional events also contribute to the higher level of CDR1 expression in drug-resistant strains, we determined CDR1 mRNA stability in the AS and AR isolates. To this end, we exploited an effective sulfur-containing purine analogue, thiolutin, as a potent inhibitor of de novo transcription to determine mRNA stability in C. albicans (18, 40). Thiolutin affected [3H]uridine incorporation into nascent RNAs in a concentration-dependent manner. About 40 μg/ml of thiolutin inhibited ∼95% of the [3H]uridine incorporation in total RNA (data not shown). Methylene blue staining revealed no decline in cell viability of AS and AR isolates treated with 40 μg/ml thiolutin, although growth was inhibited to a certain extent (data not shown). This optimized thiolutin concentration was subsequently used for the mRNA chase assays. Total RNA was isolated at different time points after transcriptional inhibition with thiolutin and analyzed by RNA gel blots (Fig. 6A). After probing the blots with a CDR1-specific probe, hybridization signals were quantified by densitometry scanning in a phosphorimager. Figure 6B depicts a typical CDR1 mRNA decay profile in the AS and AR isolates over a 300-min period from one of these experiments. CDR1 mRNA could be detected in both AS isolates Gu4 and DSY294 at time T0, and the signal intensity diminished progressively with time (mRNA half-life was approximately 60 min). The turnover of the CDR1 transcript occurred much more slowly in the AR isolates Gu5 and DSY296, with a half-life of >180 min. These results demonstrated that CDR1 mRNA stability was increased in the AR isolates over that in the AS isolates.
FIG. 6.
CDR1 mRNA decay assay. Exponentially growing cultures of C. albicans were incubated with the optimized thiolutin concentration (40 μg/ml) to inhibit ongoing in vivo transcription. Total RNA was isolated at the indicated times thereafter and fractionated on a 1% (wt/vol) agarose-2.2 M formaldehyde denaturing gel. (A) The gel was stained with ethidium bromide before blotting to monitor equal loading of the RNA and subsequently blotted onto a charged nylon membrane. The blot was hybridized with a CDR1-specific probe. Time points in minutes are indicated below each phosphorimage. (B) The hybridization signals were quantified using densitometry scanning in a phosphorimager. The signal intensity at each time point was normalized to that of time T0 (expressed as a percentage) and plotted as described in Materials and Methods. t1/2, half-life.
Cdr1 protein stability does not differ in AS and AR isolates.
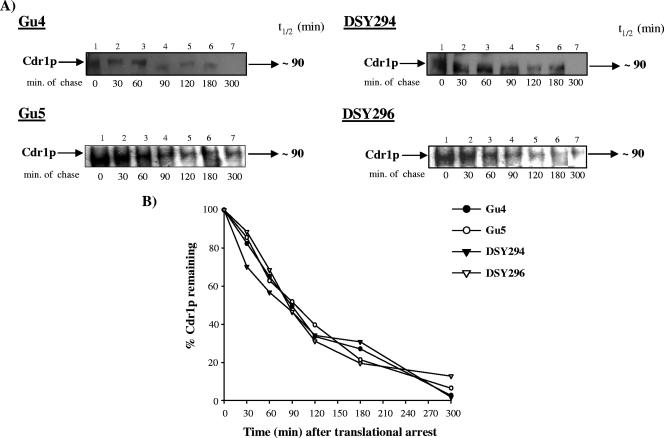
To test whether increased protein stability might also contribute to the high Cdr1p levels in AR isolates, cycloheximide chase assays were performed. Total crude protein extracts were isolated at different times after treatment of the cells with an optimized concentration (75 mM) of cycloheximide (16) and analyzed by Western immunoblotting with a rabbit polyclonal anti-Cdr1p antibody. Figure 7A shows the Western blot of the decay experiment, while Fig. 7B shows the quantitative decay profile. The half-life of Cdr1p was similar in AS and AR isolates and was calculated to be approximately 90 min.
FIG. 7.
Cdr1p decay assay. (A) Exponentially grown cultures of C. albicans were translationally halted at 30°C by addition of 75 mM of cycloheximide for 1 h. Whole-cell extracts were prepared at the indicated times after cycloheximide treatment. For AR isolates, ∼20 μg, and for AS isolates, ∼30 μg (because of relatively low expression of Cdr1p) of crude extract for each time was loaded and separated by SDS-polyacrylamide gel electrophoresis. Equal loading of protein was assessed using a Coomassie-stained gel (data not shown). Cdr1p was detected using a polyclonal anti-Cdr1p antibody. The Cdr1p-specific bands were subsequently quantified by densitometry scanning in a phosphorimager. (B) Band intensities (represented as percentages of the value at T0) for each isolate were plotted against the chased time. t1/2, half-life.
DISCUSSION
In this study, we used two pairs of matched AS and AR C. albicans clinical isolates to study the mechanisms of CDR1 overexpression in AR isolates. Our results demonstrate that both increased transcriptional activation and enhanced mRNA stability contribute to increased CDR1 expression in these drug-resistant isolates. Interestingly, we found that in the AS isolates reporter fusions with the CDR1 coding region were expressed at lower levels than fusions in which the reporter genes were directly fused to the CDR1 promoter, whereas in the AR isolates the two types of reporter fusions were expressed at comparable levels. This would mean that sequences in the CDR1 coding region can also contribute to the increased CDR1 expression in AR isolates.
It has been shown previously that CDR1 overexpression in C. albicans is caused by an increased CDR1 transcription rate in AR isolates compared with that in AS isolates (24). Our TRO experiments confirmed that the transcriptional initiation rate from the CDR1 promoter was five- to sevenfold higher in the AR isolates than in the AS isolates used in the present study (Fig. 5). The CDR1 upstream region contains many sequence elements which are involved in the regulation of CDR1 expression (5, 10, 11, 17, 32); however, no sequence differences were found in the CDR1 upstream region of these matched pairs of AS and AR isolates (5, 11; also unpublished observations). In line with this, it has recently been shown that a gain-of-function mutation in the transcription factor TAC1, which controls CDR1 expression, causes CDR1 upregulation in the AR isolate DSY296 (4).
In order to evaluate if, in addition to transcriptional activation of CDR1, differential mRNA and protein stability also contribute to the enhanced Cdr1p levels in AR isolates, we performed thiolutin and cycloheximide chase assays and observed that the up-regulation of CDR1 mRNA in AR isolates was due to an increase in the mRNA half-life (>180 min), which was approximately threefold greater than that in AS isolates (Fig. 6). In contrast, no difference in Cdr1p protein stability was observed between AS and AR isolates (Fig. 7). There are examples in other organisms where overexpression of efflux pumps can be caused by increased mRNA stability. An increase in the mRNA half-life of MDR1 (a CDR1 homologue in humans) has been shown to contribute to doxorubicin and colchicine resistance in the myelogenous leukemic cell line K562 (47). An enhanced mRNA stability of bmr3, encoding a multidrug transporter, also leads to a multidrug-resistant (MDR) phenotype in Bacillus subtilis (28). In addition, the reported MDR phenotype of Entamoeba histolytica trophozoites is also caused by transcriptional activation (27), as well as an increase in mRNA stability of the EhPgp5 gene (22).
Notably, though, cis determinants located in the 3′ untranslated region (UTR) regulate the degradation of mRNA (35). Among these cis elements, adenylate-uridylate-rich-element motifs of the 3′ UTR involved in destabilization of their corresponding mRNAs are of prime importance (22, 31, 35). Several reports have also suggested a relationship between the relative affinity of a given RNA for RNA-binding protein(s) and the stability of an mRNA containing these sequences (31, 35). Our preliminary results reveal that the CDR1 3′ UTR is ∼78% AU rich and also possesses several putative consensus binding sequences for a regulatory RNA-binding protein(s). Therefore, any contribution of CDR1 3′ UTR cis elements and of the mutation or alteration in trans-acting regulatory factor(s) corresponding to these conserved elements in determining mRNA stability between AS and AR isolates requires an in-depth analysis.
Our results with the reporter fusion transformants also suggest that sequences in the CDR1 coding region could also be an important contributor for increased CDR1 expression in AR isolates. In this context, it should be mentioned that synonymous and nonsynonymous nucleotide polymorphisms have been observed in the CDR1 coding region, but so far none of these has been linked to CDR1 overexpression (12, 15). Our present study did not consider the role of these allelic differences in sustained overexpression of CDR1 in AR isolates. However, a recent study has reported that a silent polymorphism does not influence human P-gp/MDR1 mRNA and protein expression but affects posttranslational events in terms of timing of cotranslational folding and membrane insertion (19, 43).
In conclusion, our results demonstrate for the first time that CDR1 is regulated by both transcriptional and posttranscriptional events. Our finding that the acquisition of azole resistance involves transcriptional activation as well as decreased mRNA turnover opens up new possibilities for treatment regimes to circumvent MDR in C. albicans. In this context, it is worth mentioning that the intervention of overexpressing MDR1 in MDR cell lines by verapamil (25) and ecteinascidin 743 (39) has been reported to be due to the transcriptional down-regulation of the gene.
Acknowledgments
We thank Dominique Sanglard for providing the C. albicans isolates DSY294 and DSY296. Thiolutin (CP-4092) was a generous gift from Pfizer, Inc. (Groton, CT). We also acknowledge R. Serrano for the PM-ATPase antibody. We especially thank K. Natarajan for his valuable suggestions in improving the manuscript.
The work presented in this paper has been supported in part by grants to R.P. from the Department of Biotechnology, India [BT/PR3825/MED/14/488(a)/2003 and BT/PR4862/BRB/10/360/2004], the Council of Scientific and Industrial Research [38(1122)/06/EMR-II], Department of Science Technology (SR/SO/BB-12/2004), Indo-French (IFC/A/3403-2/2006). Work in J.M.'s lab was supported by the Deutsche Forschungsgemeinschaft (SFB 630). S.L.P acknowledges a grant from the Department of Science and Technology (SR/FT/L-26/2006), India. R.M. thanks the Council of Scientific and Industrial Research (C.S.I.R.) for the award of junior and senior research fellowships.
Footnotes
Published ahead of print on 11 February 2008.
REFERENCES
- 1.Akins, R. A. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43:285-318. [DOI] [PubMed] [Google Scholar]
- 2.Chen, C. G., Y. L. Yang, H. I. Shih, C. L. Su, and H. J. Lo. 2004. CaNdt80 is involved in drug resistance in Candida albicans by regulating CDR1. Antimicrob. Agents Chemother. 48:4505-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coste, A., V. Turner, F. Ischer, J. Morschhäuser, A. Forche, A. Selmecki, J. Berman, J. Bille, and D. Sanglard. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 6.Franz, R., M. Ruhnke, and J. Morschhäuser. 1999. Molecular aspects of fluconazole resistance development in Candida albicans. Mycoses 42:453-458. [DOI] [PubMed] [Google Scholar]
- 7.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhäuser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Martinez, J., A. Aranda, and J. E. Perez-Ortin. 2004. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell 15:303-313. [DOI] [PubMed] [Google Scholar]
- 9.Gardner, R. G., G. M. Swarbrick, N. W. Bays, S. R. Cronin, S. Wilhovsky, L. Seelig, C. Kim, and R. Y. Hampton. 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling: transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151:69-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaur, N. A., N. Puri, N. Karnani, G. Mukhopadhyay, S. K. Goswami, and R. Prasad. 2004. Identification of a negative regulatory element which regulates basal transcription of a multidrug resistance gene CDR1 of Candida albicans. FEMS Yeast Res. 4:389-399. [DOI] [PubMed] [Google Scholar]
- 11.Gaur, N. A., R. Manoharlal, P. Saini, T. Prasad, G. Mukhopadhyay, M. Hoefer, J. Morschhäuser, and R. Prasad. 2005. Expression of the CDR1 efflux pump in clinical Candida albicans isolates is controlled by a negative regulatory element. Biochem. Biophys. Res. Commun. 332:206-214. [DOI] [PubMed] [Google Scholar]
- 12.Haque, A., V. Rai, B. S. Bahal, S. Shukla, A. A. Lattif, G. Mukhopadhyay, and R. Prasad. 2007. Allelic variants of ABC drug transporter Cdr1p in clinical isolates of Candida albicans. Biochem. Biophys. Res. Commun. 352:491-497. [DOI] [PubMed] [Google Scholar]
- 13.Hiller, D., D. Sanglard, and J. Morschhäuser. 2006. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob. Agents Chemother. 50:1365-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiller, D., S. Stahl, and J. Morschhäuser. 2006. Multiple cis-acting sequences mediate upregulation of the MDR1 efflux pump in a fluconazole-resistant clinical Candida albicans isolate. Antimicrob. Agents Chemother. 50:2300-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, A. R., S. Tsao, S. W. Ong, E. Lamping, K. Niimi, B. C. Monk, M. Niimi, A. Kaneko, B. R. Holland, J. Schmid, and R. D. Cannon. 2006. Heterozygosity and functional allelic variation in the Candida albicans efflux pump genes CDR1 and CDR2. Mol. Microbiol. 62:170-186. [DOI] [PubMed] [Google Scholar]
- 16.Imanishi, Y., K. Yokoyama, and K. Nishimura. 2004. Inductions of germ tube and hyphal formations are controlled by mRNA synthesis inhibitor in Candida albicans. Nippon Ishinkin Gakkai Zasshi 45:113-119. [DOI] [PubMed] [Google Scholar]
- 17.Karnani, N., N. A. Gaur, S. Jha, N. Puri, S. Krishnamurthy, S. K. Goswami, G. Mukhopadhyay, and R. Prasad. 2004. SRE1 and SRE2 are two specific steroid-responsive modules of Candida drug resistance gene 1 (CDR1) promoter. Yeast 21:219-239. [DOI] [PubMed] [Google Scholar]
- 18.Kebaara, B. W., L. E. Nielsen, K. W. Nickerson, and A. L. Atkin. 2006. Determination of mRNA half-lives in Candida albicans using thiolutin as a transcription inhibitor. Genome 49:894-899. [DOI] [PubMed] [Google Scholar]
- 19.Kimchi-Sarfaty, C., J. M. Oh, I. W. Kim, Z. E. Sauna, A. M. Calcagno, S. V. Ambudkar, and M. M. Gottesman. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525-528. [DOI] [PubMed] [Google Scholar]
- 20.Krishnamurthy, S., V. Gupta, R. Prasad, S. L. Panwar, and R. Prasad. 1998. Expression of CDR1, a multidrug resistance gene of Candida albicans: in vitro transcriptional activation by heat shock, drugs and human steroid hormones. FEMS Microbiol. Lett. 160:191-197. [DOI] [PubMed] [Google Scholar]
- 21.Li, D., V. Gurkovska, M. Sheridan, R. Calderone, and N. Chauhan. 2004. Studies on the regulation of the two-component histidine kinase gene CHK1 in Candida albicans using the heterologous lacZ reporter gene. Microbiology 150:3305-3313. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Camarillo, C., J. P. Luna-Arias, L. A. Marchat, and E. Orozco. 2003. EhPgp5 mRNA stability is a regulatory event in the Entamoeba histolytica multidrug resistance phenotype. J. Biol. Chem. 278:11273-11280. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White, D. Sanglard, and T. F. Patterson. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42:2932-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons, C. N., and T. C. White. 2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob Agents Chemother. 44:2296-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller, C., F. Goubin, E. Ferrandis, I. Cornil-Scharwtz, J. D. Bailly, C. Bordier, J. Benard, B. I. Sikic, and G. Laurent. 1995. Evidence for transcriptional control of human mdr1 gene expression by verapamil in multidrug-resistant leukemic cells. Mol. Pharmacol. 47:51-56. [PubMed] [Google Scholar]
- 26.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 27.Nieto, A., D. Guillermo Perez, E. Orozco, F. Paz, and C. Gomez. 2005. Entamoeba histolytica EhPgp5 transcriptional activation depends on putative emetine response elements. Exp. Parasitol. 110:233-237. [DOI] [PubMed] [Google Scholar]
- 28.Ohki, R., and K. Tateno. 2004. Increased stability of bmr3 mRNA results in a multidrug-resistant phenotype in Bacillus subtilis. J. Bacteriol. 186:7450-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasad, R., P. D. Worgifosse, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 30.Prasad, T., P. Saini, N. A. Gaur, R. A. Vishwakarma, L. A. Khan, Q. M. Haq, and R. Prasad. 2005. Functional analysis of CaIPT1, a sphingolipid biosynthetic gene involved in multidrug resistance and morphogenesis of Candida albicans. Antimicrob Agents Chemother. 49:3442-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prokipcak, R. D., A. Raouf, and C. Lee. 1999. The AU-rich 3′ untranslated region of human MDR1 mRNA is an inefficient mRNA destabilizer. Biochem. Biophys. Res. Commun. 261:627-634. [DOI] [PubMed] [Google Scholar]
- 32.Puri, N., S. Krishnamurthy, S. Habib, S. E. Hasnain, S. K. Goswami, and R. Prasad. 1999. CDR1, a multidrug resistance gene from Candida albicans, contains multiple regulatory domains in its promoter and the distal AP-1 element mediates its induction by miconazole. FEMS Microbiol. Lett. 180:213-219. [DOI] [PubMed] [Google Scholar]
- 33.Reuss, O., A. Vik, R. Kolter, and J. Morschhäuser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 34.Riggle, P. J., and C. A. Kumamoto. 2006. Transcriptional regulation of MDR1, encoding a drug efflux determinant, in fluconazole-resistant Candida albicans strains through an Mcm1p binding site. Eukaryot. Cell 5:1957-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 37.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 38.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scotto, K. W., and R. A. Johnson. 2001. Transcription of the multidrug resistance gene MDR1: a therapeutic target. Mol. Interv. 1:117-125. [PubMed] [Google Scholar]
- 40.Setiadi, E. R., T. Doedt, F. Cottier, C. Noffz, and J. F. Ernst. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J. Mol. Biol. 361:399-411. [DOI] [PubMed] [Google Scholar]
- 41.Shukla, S., P. Saini, Smriti, S. Jha, S. V. Ambudkar, and R. Prasad. 2003. Functional characterization of Candida albicans ABC transporter Cdr1p. Eukaryot. Cell 2:1361-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhl, M. A., and A. D. Johnson. 2001. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology 147:1189-1195. [DOI] [PubMed] [Google Scholar]
- 43.Wang, D., A. D. Johnson, A. C. Papp, D. L. Kroetz, and W. Sadee. 2005. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet. Genomics 15:693-704. [PubMed] [Google Scholar]
- 44.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increased azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wirsching, S., S. Michel, G. Kohler, and J. Morschhäuser. 2000. Activation of the multidrug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J. Bacteriol. 182:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yague, E., A. L. Armesilla, G. Harrison, J. Elliott, A. Sardini, C. F. Higgins, and S. Raguz. 2003. P-glycoprotein (MDR1) expression in leukemic cells is regulated at two distinct steps, mRNA stabilization and translational initiation. J. Biol. Chem. 278:10344-10352. [DOI] [PubMed] [Google Scholar]
- 48.Yang, Y. L., Y. H. Lin, M. Y. Tsao, C. G. Chen, H. I. Shih, J. C. Fan, J. S. Wang, and H. L. Lo. 2006. Serum repressing efflux pump CDR1 in Candida albicans. BMC Mol. Biol. 7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]