Abstract
Endothelial cells are permissive to dengue virus (DV) infection in vitro, although their importance as targets of DV infection in vivo remains a subject of debate. To analyze the virus-host interaction, we studied the effect of DV infection on gene expression in human umbilical vein endothelial cells (HUVECs) by using differential display reverse transcription-PCR (DD-RTPCR), quantitative RT-PCR, and Affymetrix oligonucleotide microarrays. DD identified eight differentially expressed cDNAs, including inhibitor of apoptosis-1, 2′-5′ oligoadenylate synthetase (OAS), a 2′-5′ OAS-like (OASL) gene, galectin-9, myxovirus protein A (MxA), regulator of G-protein signaling, endothelial and smooth muscle cell-derived neuropilin-like protein, and phospholipid scramblase 1. Microarray analysis of 22,000 human genes confirmed these findings and identified an additional 269 genes that were induced and 126 that were repressed more than fourfold after DV infection. Broad functional responses that were activated included the stress, defense, immune, cell adhesion, wounding, inflammatory, and antiviral pathways. These changes in gene expression were seen after infection of HUVECs with either laboratory-adapted virus or with virus isolated directly from plasma of DV-infected patients. Tumor necrosis factor alpha, OASL, and MxA and h-IAP1 genes were induced within the first 8 to 12 h after infection, suggesting a direct effect of DV infection. These global analyses of DV effects on cellular gene expression identify potentially novel mechanisms involved in dengue disease manifestations such as hemostatic disturbance.
Dengue virus (DV) is a Flavivirus; the Flaviviridae family includes >60 known human pathogens, such as those causing yellow fever, Japanese encephalitis, tick-borne encephalitis, Saint Louis encephalitis, and West Nile encephalitis. DVs are classified into four different serotypes: 1, 2, 3, and 4. Infection with any of these four serotypes can result in dengue fever (DF) or dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS). DHF/DSS is particularly a concern during heterologous secondary DV infections in patients with preexisting cross-reactive antibodies and memory T lymphocytes from the previous (primary) DV infection (44). It is not fully understood how the same virus can produce a mild or severe outcome in a secondary infection. Halstead and O'Rourke (14) proposed that the pathogenesis of DHF/DSS might involve an immunopathologic process as a result of immune sensitization from the prior DV infection, such as antibody-dependent enhancement of infection.
Because the endothelium forms the primary barrier of the circulatory system, dysfunction of endothelial cells during acute disease could broadly affect immune cell function (by regulating cytokines, chemokines, and cell receptors), vascular permeability, and plasma leakage (by regulating cell adhesion molecules and modifying cell-cell interactions). DV-induced plasma leakage may involve a complex interplay between host and viral factors, including the production of inflammatory cytokines (44). Virus-infected monocytes have been shown to produce vasoactive factors that affect endothelial cells in culture (2) and induce permeability (8, 14) Also, other immune cells, such as lymphocytes, may interact with the endothelium to cause plasma leakage indirectly (23, 43).
Due to the lack of a good animal model, in vitro cellular responses to DV have been used to elucidate virus interactions with host cells such as endothelial cells. Although there have been previous data on in vitro infection of endothelial cells (3, 5, 11, 21) by DV, in vivo results have been divided between those that indicate little evidence of endothelial cell damage (3) and those that indicate endothelial damage (27, 29, 30, 37). The effects of in vitro DV infection on human umbilical vein endothelial cells (HUVECs) reported to date include increased cell death, induction of cytokine production, NF-κB activation, and complement activation (3, 5). For other diseases, direct damage of the endothelium has been recognized. Among these agents are hantaviruses (13), Lassa fever virus (32), cytomegalovirus and herpesvirus (52), and Rickettsia conorii (12).
We used differential display reverse transcription-PCR (DD-RTPCR) (28) and oligonuceotide microarrays (Affymetrix HG-U133A) to identify candidate genes differentially expressed during a dengue 2 virus (D2V) infection. DD identified human inhibitor of apoptosis-1 (h-IAP1), 2′-5′ oligoadenylate synthase (OAS), 2′-5′ OAS-like (OASL), myxovirus protein A (MxA), phospholipid scramblase 1 (PLSCR1), galectin-9 (Gal-9), and regulator of G protein signaling 2 (RGS2) genes as upregulated and the endothelial and smooth muscle cell-derived neuropilin-like protein (ESDN) gene as downregulated in D2V-infected HUVECs. Microarray analysis detected the upregulation of 269 genes and the downregulation of 126 genes from an oligonucleotide library of 22,283 known human genes. The majority of the differentially expressed genes belonged to stress, defense, and immune responses. The two techniques provided complementary information regarding the proinflammatory alpha/beta interferon (IFN-α/β) response, as well as tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β) receptor signaling, as a result of viral infection. DD results were confirmed by quantitative RT-PCR and with microarray DNA chip data.
MATERIALS AND METHODS
Cell culture.
HUVECs from a second passage (Clonetics Corp., San Diego, Calif.) were maintained at 37°C in fresh cell culture media (EGM bullet kit; Clonetics) supplemented with hydrocortisone (1 mg/ml), gentamicin sulfate (50 mg/ml), amphotericin-B (GA-1000), 2% fetal calf serum (FCS), 10 μg of recombinant human epidermal growth factor/ml, and 3 mg of bovine brain extract/ml. Cell culture took place in a controlled environment of 100% humidity and 5% CO2.
Infection of endothelial cells.
Monolayers of HUVECs were resuspended in fresh cell culture medium. D2V New Guinea C (NGC) grown in C6/36 cell monolayers was added to confluent monolayers of cells at a multiplicity of infection (MOI) of 1 based on titration in Vero cells. The culture supernatant was then removed, and fresh growth medium containing 10% fetal calf serum was added to each well. After 48 h of infection, cells were harvested, centrifuged at 700 × g for 7 min at 4°C, washed with phosphate-buffered saline (PBS), pelleted again at 700 × g for 7 min at 4 C, and stored at −70°C for future experiments. For earlier time points of 8, 12, and 24 h, the MOI was 1 and the percentage of final infected cells was determined by flow cytometry.
Flow cytometry analysis.
Infected HUVECs were analyzed by using indirect intracellular labeling (Cytofix/Cytoperm Plus kit; Pharmingen/Becton Dickinson, San Diego, Calif.) with mouse polyclonal D2V antibodies (American Type Culture Collection) diluted 1:500 in PBS and a secondary antibody against mouse immunoglobulin G coupled with fluorescein isothiocyanate (Sigma).
Primer sequences.
Synthetic oligonucleotides (Table 1) were obtained from Qiagen.
TABLE 1.
Primer sequences used in RT-PCR semiquantitative analysis
| Gene | Primer sequence (5′ to 3′)
|
|
|---|---|---|
| Forward | Reverse | |
| h-IAP1 | cagaagacacagacgtcttta | cgaactgtacccttgattgta |
| 2′-5′ OAS | actttaaaaaccccattattgaaa | ggagaggggcagggatgaat |
| 2′-5′ OASL | catcgtgaacccttatgagc | ctgctggtcttcaatctgc |
| Gal-9 | cctgcccagtgctcagaggtt | gcacatgggtcagctggatga |
| ESDN | ctacctttaaaccagaagaa | ccccatcccttcctgctc |
| RGS2 | cggaattctgtgaagaaaatat | gcatgaggctctgtggtgattt |
| MxA | gccaggaccaggtatacag | gcctgcgtcagccgtgc |
| PLSCR1 | cgagcgccagcgcgggaac | ctctggctgccgctgtttcc |
| IL-8 | aagagagctctgtctggacc | gatattctcttggcccttgg |
| L35A | cttctcttaccgccatcttc | tccttgaggggtacagcatc |
| IL-1β | aagcttggtgatgtctgg | tgagaggtgctgatgtacca |
| TNF-α | ccgggcgtggtggtgag | tctgccttttgggtcttgtgaata |
Total cellular RNA preparation.
Total cellular RNA was extracted from 2 × 107 uninfected or D2V-infected HUVECs by using an RNA extraction kit (Qiagen, Hilden, Germany). Typical yields were 10 μg of total RNA per million cells. Total RNA was digested with DNase I to eliminate residual DNA by using the Message Clean Kit (GenHunter, Nashville, Tenn.). DNase I treatment was used to avoid false positives in the DD technique (33). The RNA was resuspended in diethyl pyrocarbonate-treated 0.01% distilled H2O (Ambion, Austin, Tex.) and quantified by spectrophotometry (Bio Photometer, Eppendorf, Germany) at 260 nm.
RT for Differential Display.
RT was performed by using SensiScript reverse transcriptase (Qiagen). cDNA was synthesized in 20-μl reactions containing 200 ng of mRNA, RT buffer (25 mM Tris-Cl [pH 8.3], 37.2 mM KCl, 1.5 mM MgCl2, 5 mM dithiothreitol [DTT]), a 0.5 mM concentration of deoxynucleoside triphosphate (dNTP), 10 μmol of H-T11C anchor primer (5′-AAGCTTTTTTTTTTTC-3′)/μl, and RNase inhibitor (10 U/μl). Reactions were incubated at 37°C for 60 min and then at 95°C for 5 min.
DD.
DD was performed as described previously (6) with minor modifications as follows: cDNA was amplified by PCR according to the RNAimage protocol (GenHunter) on 10% of the RT reaction product by using Taq DNA polymerase (Promega, Madison, Wis.) or TitaniumTaq polymerase (Clontech, Palo Alto, Calif.). The PCR contained 200 nM concentrations of each primer (H-T11C anchor primer with either of the random primers; GenHunter), 2 μl of 10× PCR buffer (final concentrations of 10 μM Tris-Cl [pH 8.4], 50 mM KCl, 1.5 mM MgCl2, and 5 mM DTT), 2 μl of a 25 μM dNTP mix, 0.5 μl of 3,000 of Ci [α-33P]dCTP/mmol, and 5 U of Taq polymerase (Promega)/μl in a total volume of 20 μl. Reactions with Taq polymerase were cycled at 94°C for 30 s, 40°C for 2 min, and 72°C for 30 s for 40 cycles, followed by 72°C for 10 min. Reactions with Titanium Taq were run at 95°C for 1 min, followed by 40 cycles at 93°C for 30 s, 40°C for 2 min, and 68°C for 30 s, followed finally by one cycle at 68°C for 10 min. Primers H-AP 1, 3, 4, 6, 8, 12, 15, 16, 19, 21, 22, 23, 41, or 42 were used for each cDNA synthesis.
PCRs were analyzed by extended-format denaturing 6% polyacrylamide gels by using the programmable Genomyx LR DNA sequencer (Beckman Coulter, Columbia, Md.).
The RNAimage (GenHunter) protocol was followed to reamplify candidate bands observed on the DD gels. The bands were extracted from the denaturing polyacrylamide gel by soaking in 100 μl of 2× PCR buffer (final concentrations of 10 μM Tris-Cl [pH 8.4], 50 mM KCl, 1.5 mM MgCl2, and 5 mM DTT) for 10 min at room temperature. The supernatant was treated with 3 M sodium acetate (10 μl), 5 μl of glycogen (10 mg/ml), and 450 μl of 100% ethanol. It was then stored at −70°C for 30 min and pelleted by centrifugation at 15,000 rpm in an Eppendorf centrifuge. The pellet was rinsed with 200 μl of ethanol, air dried, and dissolved in 10 μl of distilled H2O. The extracted DNA was amplified by using the RNAimage protocol. The 40-μl PCR included 4 μl of DNA, 4 μl of 10× PCR buffer (final concentrations of 10 μM Tris-Cl [pH 8.4], 50 mM KCl, 1.5 mM MgCl2, and 5 mM DTT), 3.2 μl of dNTP (250 μM), 200 nM concentrations of arbitrary primers, a 200 nM concentration of H-T11C, and 2.5 U of Taq polymerase (Promega). Reactions were subjected to 40 rounds of PCR amplification at 94°C for 30 s, 40°C for 2 min, and 72°C for 30 s, followed by 72°C for 10 min. The amplified PCR products were analyzed in 1.5% agarose gels (Life Technologies, Grand Island, N.Y.) containing 0.5 μg of ethidium bromide/ml for staining. The results were documented with Quantity One Software version 4.2.3 (Bio-Rad).
QIAquick PCR purification kit protocol (Qiagen) was used to extract DNA from PCR samples. DNA samples were eluted with distilled H2O and sequenced in the UMASS Medical School sequencing core facility using the anchor primer H-T11C.
Semiquantitative RT-PCR.
A total of 0.2 μg of total cellular RNA obtained from infected and uninfected cells cultures was reverse transcribed by using Omniscript/SensiScript reverse transcriptase (Qiagen) in the presence of 1 μM anchor primer and a 500 μM dNTP concentration in a volume of 20 μl. RT was performed at 37°C for 60 min, followed by 94°C for 15 min. PCR was performed with 0.5, 1, or 2 μl of cDNA. The 50-μl PCR included 2.5 U of Taq DNA polymerase (Clontech), 100 μM dNTP, and 100 μM concentrations of primers. A PCR was run at 94°C for 1 min, followed by various cycles of 58°C for 1 min, 72°C for 45 s, and 94°C for 30 s, followed by 58°C for 1 min, and finally by 72°C for 10 min.
Real-time SYBR Green RT-PCR.
RT-PCR was performed in two steps. cDNA was obtained as described for semiquantitative analysis. For PCR, we used a 50-μl reaction, including 1 to 4 μl of cDNA and 15.75 μl of PCR master mix containing 5 μl of 10× SYBR PCR buffer, 6 μl of 25 mM MgCl2, 4 μl of dNTP blend (2.5 mM dATP, 2.5 mM dCTP, 2.5 mM dGTP, 5 mM dUTP), 0.25 μl of AmpliTaq Gold (1.25 U), and 0.50 μl of AmpErase UNG (0.5 U) in deionized water. A calibration curve containing five points ranging from 100 fg to 1 ng was used as a standard for the experiment, and L35A was used as an internal control for the unknown samples. Reactions were cycled at 50°C for 2 min and then 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min in the PCR signal detection system 5700 (Perkin-Elmer).
Multiplex RT-PCR for Toll-like receptors (TLRs).
For the multiplex PCR, cDNA template was obtained from the semiquantitative RT step. The CytoXpress multiplex RT-PCR kit protocol (BioSource International) was performed as follows: a 50-μl reaction contained 4 μl of 3.12 mM dNTP, 5 μl of Taq DNA polymerase (2.5 U), 5 μl of 10× multiplex PCR (MPCR) primers, 5 μl of 10× MPCR buffer, 5 μl of 10× MPCR enhancer, and 5 μl of cDNA (200 ng/μl) in distilled H2O. After an initial denaturation step at 96°C for 1 min, the PCR was cycled twice at 96°C for 1 min and 60°C for 4 min, followed by 30 cycles of 94°C for 1 min and 60°C for 2.5 min. The final step was incubation at 70°C for 10 min. Then, 20 μl of PCR product was analyzed by agarose gel electrophoresis with a 1.5% agarose gel (Life Technologies) containing 0.5 μg of ethidium bromide/ml.
Microarray hybridization and analysis.
For the Affymetrix data, HUVECs were pooled from two to five umbilical cords and established as primary cultures in M199 containing 20% FCS. HUVECs were passaged at a 1:3 or 1:4 split ratio in M199, 10% FCS, 1 mM glutamine, endothelial cell growth stimulant, porcine intestinal heparin, and antibiotics. RNA isolated from D2V-infected and uninfected HUVECs was biotin-labeled and hybridized to human oligonucleotide microarrays (Affymetrix) by using standard methods as follows. First-strand cDNA was prepared by using a T7-(dT)24 primer and SuperScript II reverse transcriptase (Invitrogen/Life Technologies) from 8 μg of total cellular RNA. Second strand synthesis was performed by using the Superscript Choice system (Invitrogen/Life Technologies), and product was purified by using a Qiagen PCR purification kit. Synthesis of biotin-labeled cRNA was carried out by using a BioArray high-yield transcript labeling kit (Enzo), and cRNA product was purified by using RNAeasy kit (Qiagen). cRNA was fragmented and spiked with bacterial control genes (bioB, bioC, bioD, and cre) according to the Affymetrix protocol. Hybridization to Affymetrix HG-U133A oligonucleotide microarrays (22,283 genes) was carried out overnight with 10 μg of biotin-labeled cRNA product. Microarrays were washed by using the GeneChip Fluidics Station (Affymetrix) according to the manufacturer's protocol. Staining with R-phycoerythrin-streptavidin (Molecular Probes) was followed by antibody amplification with a biotinylated streptavidin antibody (Vector Laboratories). Scanning was carried out by using the GeneArray scanner (Hewlett-Packard). The data were collected with software provided by Affymetrix (MAS 5) and analyzed by using the Statistical Expression Algorithm to give signal values. Gene expression signals from 22,283 genes for each of the five arrays were collected in Microsoft Excel. Arrays were median normalized, genes with low signals (defined as genes with signals within 20-fold of the minimum array signal on two or more of the arrays, unless the signal was low on only both D2V-infected arrays) were omitted (ca. 10% of genes), and remaining signals were log2 transformed. Genes were normalized to the mean of the controls. The data were then filtered to retain only differentially expressed genes (at least fourfold differential on at least two arrays). Hierarchical cluster analysis and identification of functional local clusters were performed on the remaining 395 genes by using dChip (public software developed by the Wong Lab, Department of Biostatistics, Harvard School of Public Health).
The two D2V infection array experiments (i.e., the experiments with D2V and D2N) represent repeated experiments with the same RNA sample. A comparison of these experiments showed that 95% of the values were reproducible within ±3.4-fold. Control (C) experiments were performed with three different RNA samples isolated from two different pools of HUVECs (each pool represented two or three different human donors pooled together for final cell isolation). An additional three controls from independent lots produced comparable results (data not shown).
PEG virus precipitation from human serum.
Precipitation and concentration of D3V from serum obtained from two Venezuelan patients was performed by using the protocol of Terajima and Ennis (52a). Briefly, polyethylene glycol (PEG) 2000 was previously prepared in sterile PBS at 12%. Tubes containing 120 μl of serum and 28 μl of 12% PEG were incubated at 4°C overnight with constant mixing. The samples were centrifuged at 4°C for 25 min at 5,000 rpm in a Eppendorf centrifuge, and all of the pellets were pooled and resuspended under sterile conditions with 220 μl of PBS. Then, 100 μl of the precipitated viral isolate was added to 5 × 106 HUVECs in M199 medium supplemented with growth factors plus 2% FCS (MOI ∼0.1 based on the viremia levels in serum). After 2 h, medium with 10% FCS was added, and the cells were incubated for an additional 72 h. The supernatant was discarded, and the cell monolayer was washed three times before cells were collected by centrifugation at 600 × g, the cell pellets were washed three times again with PBS, and RNA was extracted by using the Qiagen RNeasy kit. The RNA was subjected to TaqMan RT-PCR with forward primer D3VF (5′-AAAAGCCACGGTTTGAGCAA-3′), reverse primer D3VR (5′-CCCAGGCTTTACGTCCCC-3′), and probe (5′-CGTGCTGCCTGTAGCTCCGTCGTA-3′). The quantities of DV RNA (femtograms per million cells) were reported by normalizing the cellular RNA with β-actin levels. The percentage of cells infected with the clinical DV isolates was also determined by using indirect inmunofluorescence flow cytometry with an anti-D3V antibody (American Type Culture Collection).
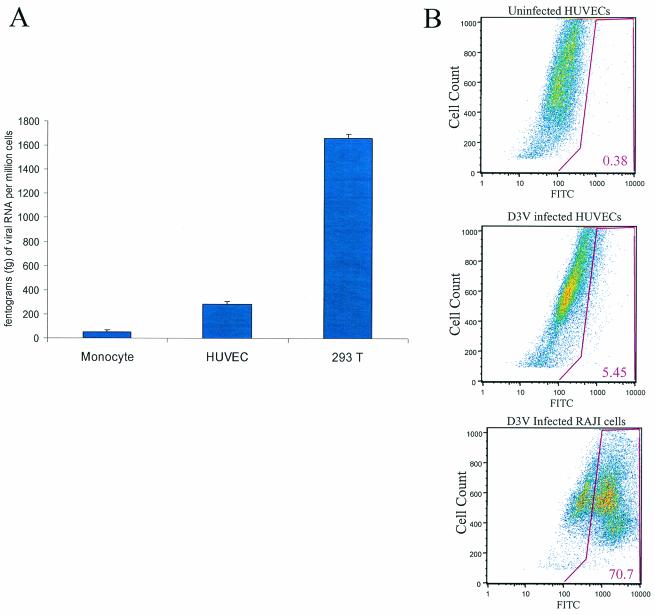
We measured viral RNA levels in plasma by quantitative fluorogenic RT-PCR in two subjects with documented D3V viremia. Viral RNA levels in plasma during the febrile period ranged up to >8.5-log RNA copies/ml and were undetectable by day 5 after the onset of illness. PEG precipitation was used to make complement-free viral inoculum from day 3 of febrile illness. The PEG-precipitated virus was added in equal parts to three different cell types: primary monocytes isolated from healthy donors, HUVECs, and epithelial human cell line 293T, which has been previously reported to be very competent for DV infection was used as a positive control for infection. Cell cultures were infected at an MOI of ∼0.1 for 48 h, and cellular RNA isolated from cellular pellets that had been washed times. The amount of cellular RNA was normalized by actin mRNA levels. The amounts of D3V RNA (in femtograms)/million cells detected by using D3V TaqMan primers and probes in the sequence detection system (see Materials and Methods) were plotted in the three cell types to compare efficiencies of infection. The measurements were done in triplicate, and the standard errors are shown for each cell type. The results obtained with a representative patient are shown.
RESULTS
Infection of HUVECs by D2V NGC and clinical D3V isolates.
To determine whether HUVECs support DV infection in the absence of antibody enhancement, we performed infection in HUVECs by using the laboratory virus strain D2V NGC and confirmed previous results of other researchers showing in vitro infection of endothelial cells (3). Infection rates were assessed by using both quantitative PCR and immunofluorescence flow cytometry. In one representative experiment, after 48 h at an MOI of 1 10% of HUVECs were infected with D2V NGC as determined by flow cytometry (data not shown). By using quantitative PCR, intracellular RNA levels of virus were compared to those of the housekeeping gene β-actin. From 0 to 48 h after inoculation of DV, the ratio of D2V RNA to β-actin mRNA generally increased by fourfold. The D2V levels were up to 10-fold higher in highly permissive 293T cells than for monocytes and HUVECs or for chronically infected Raji cells (data not shown). Infection of HUVECs by primary D3V isolated directly from serum by both quantitative RT-PCR and flow cytometry is shown in Fig. 1A and B, respectively. The relative amounts of D3V RNA in three different cell types is shown in Fig. 1A. Higher levels of intracellular D3V RNA were detected in HUVECs versus primary monocytes. The percentage of infected HUVECs was 5.5% as determined by flow cytometry. These data demonstrate the ability of clinical isolates of D3V to infect HUVECs.
FIG. 1.
Primary isolate of DV infects HUVECs in vitro. (A) Quantitative Taqman PCR. We measured DV RNA levels in plasma by quantitative fluorogenic RT-PCR in two subjects with documented D3V viremia. PEG precipitation was used to isolate DV from 120 μl of sera obtained on day 3 of illness (see Materials and Methods). The PEG-precipitated virus was added to three different cell types: primary monocytes isolated from healthy donors, HUVECs, and epithelial human cell line 293T. Cell cultures were infected at an MOI of ∼0.1 for 48 h, and cellular RNA was isolated. The amount of cellular RNA was normalized by actin mRNA levels. Particles of D3V RNA/million cells detected by D3V TaqMan primers and probes (Materials and Methods) were plotted in the three cell types to compare the efficiencies of infection. The measurements were done in triplicate, and the standard error is shown for each cell type. The results from a representative patient are shown. (B) Indirect immunofluorescence. The percentage of infection of HUVECs with clinical isolates of D3V was detemined by flow cytometry. HUVECs were infected with PEG-precipitated D3V for 48 h, and the cells were fixed by using 1% formaldehyde. The cells were stained with mouse anti-D3V antibody.
DD analysis of DV-infected HUVECs.
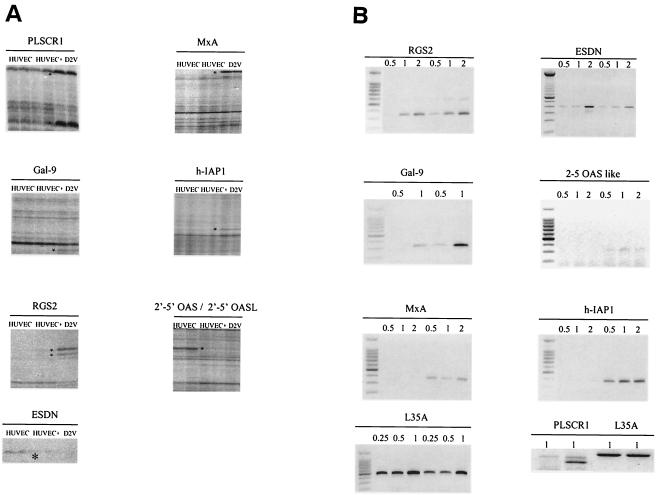
DD was performed with combinations of 16 different forward primers and the HT11C reverse primer. Fourteen differential bands were detected, excised from the gel, amplified, and prepared for DNA sequence analysis. Of these, eight cDNAs (Fig. 1B) were identified by a basic local alignment search tool (BLAST) search. Five cDNAs could not be sequenced, probably due to low template DNA concentrations (data not shown). Seven genes were overexpressed, while one gene was underexpressed.
GenBank accession numbers and functions of the DD-identified genes are listed (Table 2). Among the overexpressed genes PLSCR1, 2′-5′ OAS, 2′-5′ OASL, MxA, and Gal-9 genes are induced by the IFN-α/β pathway, h-IAP1 is induced by the TNF-α pathway, while Gal-9 is also activated by the IL-1β pathway. RGS2 (overexpressed) and ESDN (underexpressed) genes have not been linked to any known viral activation pathway, nor is it known whether they are induced or repressed by IFN.
TABLE 2.
GenBank accession numbers and functions of the DD-identified genes
| Gene | GenBank accession no. | Function(s) |
|---|---|---|
| h-IAP1 | AF070674 | Antiapoptotic, inhibitor of caspases |
| ESDN | AF387547 | Regulation of vascular cell growth |
| 2′-5′ OAS | XM_056785 | Activates RNase L |
| 2′-5′ OASL | NM_003733 | Binds dsRNA but function unknown |
| MxA | XM_009773 | Antiviral IFN-α/β response |
| RGS2 | XM_029884 | Regulates G protein signaling and T-cell activation |
| Gal-2 | NM_002308 | Eosinophil chemoattractant, cell adhesion, apoptosis, and cell proliferation |
Multiple alignments of the complete family of oligonucleotide adenylate synthases showed that the genetic region of the 2′-5′ OAS gene detected by DD was homologous to the 2′-5′ OASL gene (data not shown).
We incorporated a novel approach of spiking the uninfected cellular RNA samples with viral RNA at the time of RT to avoid the isolation of viral genome cDNAs. At least five virus-specific DD bands were detected by using the spiked control. These bands represented DV genes as confirmed by sequence analysis (data not shown). The spiking control avoided the high number of false-positive differentially expressed genes reported by others (45, 61).
RT-PCR confirmation of differential expression.
The results of semiquantitative RT-PCR analyses performed to confirm differential expression of the genes identified are shown in Table 2. The L35A gene, a housekeeping gene, was used as a control. We confirmed that eight genes were overexpressed and one gene was underexpressed in HUVECs during D2V infection (Fig. 2B).
FIG. 2.
(A) Differentially displayed genes in HUVEC 48 h after D2V infection. HUVEC RNA was used to prepare cDNA with SensiScript reverse transcriptase and HT11C primer. Asterisks indicate differentially displayed bands identified in DV-infected cultures (D2V) compared to uninfected cells (HUVEC). +, addition of exogenous viral RNA to uninfected cells. (B) Semiquantitative RT-PCR analysis. One of three semiquantitative RT-PCR analyses is shown for seven of the eight genes identified by DD; uninfected (left) or virus-infected cultures (right) at 48 h postinfection are included in each panel. L35A was used as a control gene. We used 0.5, 1, and 2 μl of cDNA of either uninfected and infected samples in the PCR for all reactions except for L35A, in which 0.25, 0.5, and 1 μl of cDNA were used. The PLSCR1 gene analysis was performed independently. For the PLSCR1 RT-PCR, results obtained with 1 μl of cDNA are shown. Panel B, left side, uninfected cells; right side, infected cells. The PLSCR1 panel has uninfected cells in lanes 1 and 3 and infected cells in lanes 2 and 4.
To accurately measure the fold change of expression of the DD-identified genes in HUVECs 48 h after D2V infection, we performed quantitative real-time RT-PCR with SYBR Green and the primers listed in Table 1. The values obtained agreed with the findings of semiquantitative RT-PCR (Table 3). In Table 3 the data extracted from the Affymetrix analysis correlated with those obtained by PCR amplification techniques. In most cases the fold induction was replicated in either of the techniques. Of the overexpressed genes, four were highly differentially expressed (>70-fold increased): 2′-5′ OAS, 2′-5′ OASL, MxA, and PLSCR1. The values of overexpression obtained by chip analysis were lower in some instances, probably due to saturation of the fluorescence signal in the array (7). Additional results on the temporal expression are available online at http://biotools.umassmed.edu/ibosch/.
TABLE 3.
Comparison of semiquantitative RT-PCR and quantitative and nonamplified mRNA detection on microarrays as indicated by the induction (n-fold) of gene expression
| Gene | Fold inductiona 48 h postinfection as determined by:
|
||
|---|---|---|---|
| SYBR Green analysis | RT-PCR analysis | Affymetrix analysis | |
| h-IAP1 | 8.1 | 7.79 | 11 |
| 2′-5′ OAS | 72.8 | >10 | 67 |
| 2′-5′ OASL | >100 | >10 | 16 |
| RGS2 | 2.8 | 3.9 | <4 |
| Gal-9 | 4.59 | 4.7 | 6.5 |
| MxA | >100 | >11 | 48.5 |
| ESDN | ND | 0.45 | <4 |
| TNF-α | 3.76 | 3.82 | <4 |
| IL-1β | 40.77 | 10.23 | 9.4 |
| TLR3 | ND | ND | 14 |
| PLSCR1 | ND | >10 | 7.9 |
ND, not determined.
Oligonucleotide microarray analysis.
Oligonucleotide microarray analysis (Affymetrix HG-U133A) was performed with RNAs isolated from one sample of D2V-infected HUVECs (analysis performed in duplicate) and three samples of uninfected HUVECs. The data were collected for 22,283 human genes and filtered to remove ca. 2,000 genes expressed at very low levels. Cluster analysis was performed for the 395 genes that were differentially expressed by >4-fold. The results are shown (Fig. 2). The experimental reproducibility assessed in replicate Affymetrix chip analyses showed that 95% of the values were reproducible within 3.4-fold. Hence, we were confident that differential gene expression of at least fourfold from control levels represented truly up- or downregulated genes.
D2V infection had a marked effect on gene expression of HUVECs. Two groups of differentially expressed genes were seen, including a large upregulated (cluster I) and a smaller downregulated group (cluster II). The upregulated cluster included all of the DD-identified genes that were differentially expressed by >4-fold. A full cluster diagram, including the identities of all 395 differentially expressed genes, is shown at the website under the title of Fig. 3.
FIG. 3.
Affymetrix H133A analysis. Hierarchical cluster analysis of 22,283 human genes measured by using Affymetrix gene chips. The results were filtered to retain only the 395 genes differentially expressed by >4-fold. Cluster I (269 genes) was upregulated, and cluster II (126 genes) was downregulated by infection. The key shows the range of relative differential expression of the 395 genes. Displayed expression levels were standardized. A full cluster diagram with all genes identities is available at our website at http://biotools.umassmed.edu/ibosch/.
dChip software was used to identify regions of the cluster diagram where specific functional classes of genes were significantly enriched. Functional classes that were most significantly enriched (lowest P values) in the D2V upregulated group (cluster I) included the categories of response to biotic stimulus and defense response (Fig. 3; see also Table 5). Functional classes that were most significantly enriched in the downregulated group (cluster II) included categories related to cytoskeletal and membrane proteins.
TABLE 5.
Categories of genes whose expression was altered by DV infection of HUVECsa
| Gene cluster and function | P |
|---|---|
| DV-induced genes (cluster I) | |
| Stress response | 0.0012 |
| Defense response | 0.000020 |
| Immune response | 0.0012 |
| Response to external stimulus | 0.0036 |
| Response to biotic stimulus | 0.000007 |
| Response to pest, pathogen, or parasite | 0.00097 |
| Defense or immunity protein | 0.0014 |
| Response to wounding | 0.00112 |
| Inflammatory response | 0.0014 |
| Growth factor | 0.00077 |
| Soluble fraction | 0.0014 |
| Cell communication | 0.0045 |
| Pathogenesis | 0.0036 |
| Ligand | 0.0012 |
| Extracellular space | 0.0049 |
| Cell-cell signaling | 0.0035 |
| Cell fraction | 0.00029 |
| Extracellular | 0.00087 |
| Extracellular space | 0.00038 |
| Proteolysis and peptidolysis | 0.0020 |
| Macromolecule catabolism | 0.0020 |
| Protein degradation | 0.0020 |
| Antiviral response | 0.0043 |
| Receptor signaling protein | 0.00038 |
| DV-repressed genes (cluster II) | |
| Cytoskeleton | 0.0033 |
| Radiation response | 0.0037 |
| Perception of abiotic stimulus | 0.0032 |
| Response to abiotic stimulus | 0.0025 |
| Membrane | 0.0049 |
| Integral membrane protein | 0.0026 |
| Plasma membrane | 0.0040 |
| Integral plasma membrane protein | 0.0047 |
dChip was used to identify functional local clusters in the genes altered by DV infection. After we performed hierarchical clustering, we used dChip to determine whether genes with a specific function were enriched in a particular cluster relative to the remainder of the genes in the analysis. Functional clusters with P values smaller than 0.00005 were considered highly significant and are shown. The P value reflects the probability that the observed degree of functional group enrichment would be found in a random distribution. (Individual gene identities of the functional classifications of clusters I and II in HUVECs infected with D2V NGC can be found in the online Table 5 at www.umassmed.edu/Labs/CIDVR/Supplements.)
Expression of IL-1β, TNF-α, and TLR in HUVECs.
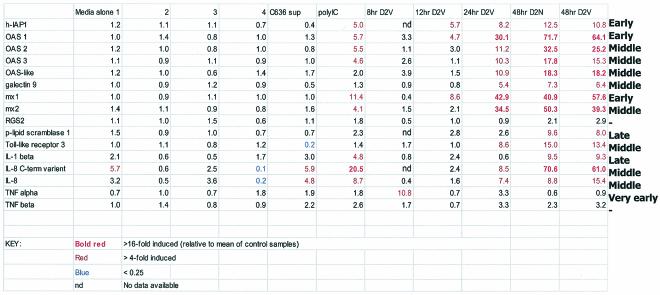
Based on the previous finding that h-IAP1 expression was induced by stimulation with TNF-α or IL-1β (10, 50), we analyzed the expression of TNF-α and IL-1β in D2V-infected and uninfected HUVECs. Both semiquantitative RT-PCR and microarray analysis showed approximately 10-fold induction for IL-1β at 48 h postinfection (Table 3). TNF-α was indeed expressed very early in infection, but h-IAP1 was induced before IL-1β (Table 4).
TABLE 4.
Time course study of D2V infection in HUVECs showing expression of DD genes obtained using Affymetrix Genechip analysisa
n = 4 for control, n = 2 for 48-h DV infection, and n = 1 for all other conditions. Mx1-MxA and Mx2-MxB in the Affymetrix gene bank.
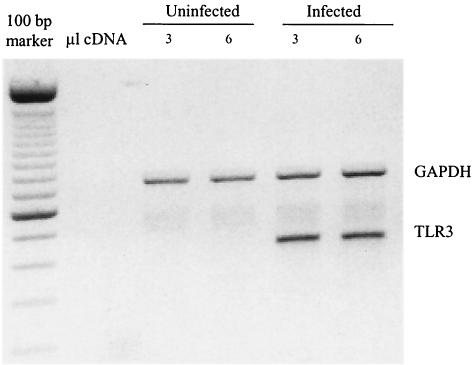
TLR3 has been shown to recognize dsRNA and activate the transcription factor NF-κB (1). Since TLR3 gene expression is induced by IFN-α/β and IFN-γ (35), we wanted to determine whether TLR3 expression was regulated during D2V infection in HUVECs. It is interesting that TLR3 did not show induction by poly(dIC), whereas it showed specific induction in infected cells. Multiplex PCR of TLR1 to TLR5 showed a fourfold increase of TLR3 only (whereas microarrays showed a ninefold increase by 24 h after D2V infection) (Fig. 4). The predominance of TLR3 has been previously reported for endothelial and epithelial cells (35). The role of viral infections (DV infections in particular) and IFNs in the overexpression of TLR3 indicates its potential role in innate immunity.
FIG. 4.
Multiplex PCR for the expression of TLR1 to -5 in uninfected and D2V-infected HUVECs. cDNA template was obtained from an RT step performed on total RNA isolated from control and D2V-infected cultures 48 h postinfection. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is given as a control for equal loading. In the PCR step, two different quantities of cDNA (3 and 6 μl) were used. GAPDH is shown at 680 bp.
DV infection time course in HUVECs.
Previous studies have reported conflicting data on whether altered gene expression in infected cells is due to a direct or an indirect effect on the virus. To help distinguish between these effects, we analyzed gene expression at early time points after DV infection. Early changes are more likely to represent direct effects of DV infection, whereas later changes may be caused by early induced genes acting on uninfected cells (bystander effects). We have used Affymetrix microarray analysis to compare gene expression at 8, 12, 24, and 48 h postinfection. Controls included poly(dI-dC) and the C6/36 insect cell supernatant used for production of D2V (mock infection). The results are tabulated (Table 4).
We observed very early and transient induction of TNF-α expression. Induction of h-IAP1, OAS1, and MxA expression was observed starting at 12 h postinfection. The 8- and 12-h time points precede viral spread to neighboring cells, and hence gene induction at these time points is more likely to be the direct result of viral infection of HUVECs. We cannot distinguish whether the changes were due to the late effects of direct infection or to bystander effects. For Gal-9, TLR3, and OASL genes, DV had a specific effect since these genes were not induced when poly(dI-dC) was used to stimulate HUVECs.
DISCUSSION
The endothelium forms the primary barrier of the circulatory system and dysfunction of endothelial cells during acute dengue disease could broadly affect immune cell function and contribute to dengue pathology. We show in vitro that the laboratory strain D2V NGC and primary DV isolates obtained from patient serum infected HUVECs. The rate of HUVEC infection in both cases was similar to that of primary monocytes, which are widely considered to be an important site of DV infection in vivo. Although it remains debated whether DV infects endothelial cells during the acute phase of disease in vivo, the proximity of these cells to the blood and their involvement in disease pathology makes them a target for investigation.
We used DD, RT-PCR, and Affymetrix oligonuceotide microarray analyses to study gene expression changes in HUVECs during infection with DV. Using DD, we identified several genes, some of which are involved in the innate antiviral response and some of which were not known to be involved in the response to DV infection. We confirmed these results by RT-PCR. Using microarrays, we identified genomewide expression changes, characterized functional categories of genes that were differentially expressed in response to the virus, and confirmed our DD results. Both techniques demonstrated comparable expression patterns for the DD-identified genes.
Using 16 different arbitrary primers in DD, we found eight genes to be differentially expressed in HUVECs during D2V infection. Since each primer pair detected approximately 50 mRNAs and a given cell expresses approximately 30,000 genes, we estimate that we screened by DD approximately 3% of the HUVEC transcriptome. In agreement with this, microarray analysis identified 395 genes to be differentially regulated in HUVECs during D2V infection. The corresponding abundance of the eight genes detected by DD were in the same range of Affymetrix Genechip assay for sensitivity. Therefore, DD offers detection of mRNA in the medium to high range (www.umassmed.edu/Labs/CIDVR/Supplements).
The genes identified as differentially expressed by microarray analysis comprised two groups (Table 5). Cluster I included 269 genes that were upregulated, and cluster II included 126 genes that were downregulated. Cluster I included genes that play a role in broad functional categories of stress, defense, immune responses, wounding, inflammatory, and antiviral responses.
The genes downregulated by DV infection (cluster II) included a significant number of genes broadly characterized as membrane and cytoskeletal genes. The functional categories were ordered by statistical significance (see Table 6 in the additional online data at www.umassmed.edu/Labs/CIDVR/Supplements). The list of specific genes in these categories of dengue infection since provides further information about the activation pathways being utilized in DV-infected cells.
The identification of several specific differentially expressed genes also contributes to our understanding of DV mechanisms. DD, RT-PCR, and microarrays consistently identified the apoptotic inhibitor, h-IAP1, as overexpressed during dengue infection. Because DV promotes cell death in HUVECs (3), we postulate that h-IAP1 counterbalances apoptosis, reducing cell death so that the virus can achieve replication and transmission. The hypothesis that the function of h-IAP1 is to prevent virus-induced apoptosis is further supported by the early time point of upregulation of h-IAP1 at 12 h after D2V infection in HUVECs (Table 4). Similar modulation of apoptosis is seen in other viral systems (42, 56, 58).
Activation of the myxovirus pathway during D2V infection in endothelial cells may be an important antiviral response. The MxA gene was highly induced by 12 h in D2V infected HUVECs (Table 4). Myxovirus antiviral proteins belong to the family of GTPases and are induced preferentially by IFN-α/β (18). These proteins have been shown to impair influenza virus and vesicular stomatitis virus growth at the transcriptional level (62). They may also block the intracellular trafficking of nucleocapsids during viral assembly (17, 19, 26, 36).
The 2′-5′ OASL protein is highly homologous to 2′-5′ OAS. Like 2′-5′ OAS protein, it binds to DNA and dsRNA (15). However, unlike 2′-5′ OAS protein,, it cannot activate RNase L to cause viral RNA degradation (16, 20, 41). The results from DD and microarrays showed that OASL protein was highly upregulated by 24 h (Table 4) after D2V infection. We postulate that 2′-5′ OASL protein overexpression may play a role in the control of DV infection.
An additional DV-induced gene found in HUVECs was PLSCR1. It is a plasma membrane protein (57) that has been implicated in the increased synthesis and rapid translocation of phospholipids, phosphatidylserine, and phosphatidylethanolamine to the cell surface in response to cell activation, cell injury, or apoptotic stimuli. Phosphatidylserine translocation promotes the activation of cell binding (40), plasma complement and coagulation proteases, accelerates clearance of activated platelets and apoptotic cells by the reticuloendothelial system, and interacts with epidermal growth factor (48, 51, 60). PLSCR1 contributes to cytokine-regulated cell proliferation and differentiation and is highly induced by IFN in HUVECs (47, 51). PLSCR1 expression was upregulated by 48 h after D2V infection in HUVECs (Table 4).
A previously unknown response that we found in DV-infected HUVECs is the overexpression of the RGS2. RGS2 is a member of a family of proteins that regulate G-protein linked signaling pathways by enhancing the hydrolysis of GTP and also regulates T-cell proliferation (22). RGS proteins negatively regulate signaling through GPCRs, including those coupled to the seven-transmembrane domain chemokine receptors (31, 46). The increased RGS2 expression in DV-infected HUVECs may regulate the mechanisms of chemokine response. RSG2 was recently found to be overexpressed in pathogenic hantavirus infections (13).
Our DD and RT-PCR results showed that ESDN mRNA was moderately (twofold) decreased during DV infection. ESDN is a newly characterized gene encoding a neuropilin-like type-1 transmembrane protein that regulates vascular cell growth and mediates chemorepulsion (24). We postulate that this gene may play a role in regulating cell-cell interactions during DV infection.
Another gene that is potentially involved in cell-cell interaction and adhesion during DV infection is Gal-9. Gal-9 belongs to the family of Ca2+-dependent galactoside-binding lectins that modulate a variety of cellular activities such as cell proliferation and/or adhesion, apoptosis, and chemoattraction (4, 25, 34, 38, 39, 53-55). Gal-9 is an eosinophil chemoattractant expressed in HUVEC cells, macrophages, B and T cells, and mast cells (9, 49), all of which may contribute to plasma leakage. IL-1β stimulation enhances the expression of Gal-9 in astrocytes (59). In DV-infected HUVECs, the enhanced expression of Gal-9 at 24 h might be induced by IFNs since the expression of IL-1β is upregulated only by 48 h (Table 4).
In summary, the results of DD, RT-PCR, and microarray analyses demonstrate the activation of broad functional responses including the stress, defense, immune, wounding, inflammatory, and antiviral pathways in DV-infected HUVECs. At least three signaling pathways in the inflammatory response were identified during DV infection of HUVECs: the TNF-α, the IL-1β, and the IFN-α/β pathways. We have proposed novel roles for phospholipid scramblase, Gal-9, ESDN, and RGS2 as antiviral proteins. In terms of the relevance of our findings, it is possible that DV mediates endothelial cell activation via both direct infection and indirectly as a result of cytokine induction. Endothelial cell dysfunction in DV infection, as well as in other hemorrhagic diseases, defines endothelial cells as a target of disease pathogenesis. Our findings here elucidate disease pathogenesis by defining the responses of endothelial cells to DV infection.
Acknowledgments
We acknowledge the Nucleic Acid Facility at UMASS Medical School for the sequencing of DD DNA. We thank Vanessa Melanson and Gregory Raines for critical reading of the manuscript; Angela Ariza and Jurand Janus for excellent technical expertise; Kay Case and the Vascular Research Division, Brigham and Women's Hospital, for HUVEC preparation; Phyllis Spatrick for processing of Affymetrix chips; and the staff of the Banco Municipal de Sangre.
This work was supported by grants R01 AI30624 and U01 AI45440 from the National Institutes of Health.
REFERENCES
- 1.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R., S. Wang, W. Osiowy, and A. Iseekuts. 1997. Activation of endothelial cells via antibody-enhanced dengue virus infection of peripheral blood monocytes. J. Immunol. 71:4226-4434. [DOI] [PMC free article] [PubMed]
- 3.Avirutnan, P., P. Malasit, B. Seliger, S. Bhakdi, and M. Husmann. 1998. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J. Immunol. 161:6338-6346. [PubMed] [Google Scholar]
- 4.Barondes, S. H., V. Castronovo, D. N. Cooper, R. D. Cummings, K. Drickamer, T. Feizi, M. A. Gitt, J. Hirabayashi, C. Hughes, K. Kasai, et al. 1994. Galectins: a family of animal β-galactoside-binding lectins. Cell 76:597-598. [DOI] [PubMed] [Google Scholar]
- 5.Bonner, S. M., and M. A. O'Sullivan. 1998. Endothelial cell monolayers as a model system to investigate dengue shock syndrome. J. Virol. Methods 71:159-167. [DOI] [PubMed] [Google Scholar]
- 6.Bosch, I., H. Melichar, and A. B. Pardee. 2000. Identification of differentially expressed genes from limited amounts of RNA. Nucleic Acids Res. 28:E27. [DOI] [PMC free article] [PubMed]
- 7.Bosch, I., K. Xhaja, L. Estevez, G. Raines, H. Melichar, R. V. Warke, M. V. Fournier, F. A. Ennis, and A. L. Rothman. 2002. Increased production of interleukin-8 in primary human monocytes and in human epithelial and endothelial cell lines after dengue virus challenge. J. Virol. 76:5588-5597. [DOI] [PMC free article] [PubMed]
- 8.Carr, J. M., H. Hocking, K. Bunting, P. J. Wright, A. Davidson, J. Gamble, C. J. Burrell, and P. Li. 2003. Supernatants from dengue virus type-2 infected macrophages induce permeability changes in endothelial cell monolayers. J. Med. Virol. 69:521-528. [DOI] [PubMed] [Google Scholar]
- 9.Chabot, S., Y. Kashio, M. Seki, Y. Shirato, K. Nakamura, N. Nishi, T. Nakamura, R. Matsumoto, and M. Hirashima. 2002. Regulation of galectin-9 expression and release in Jurkat T-cell line cells. Glycobiology 12:111-118. [DOI] [PubMed] [Google Scholar]
- 10.Chu, Z. L., T. A. McKinsey, L. Liu, J. J. Gentry, M. H. Malim, and D. W. Ballard. 1997. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-κB control. Proc. Natl. Acad. Sci. USA 94:10057-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drancourt, M., F. George, P. Brouqui, J. Sampol, and D. Raoult. 1992. Diagnosis of Mediterranean spotted fever by indirect immunofluorescence of Rickettsia conorii in circulating endothelial cells isolated with monoclonal antibody-coated immunomagnetic beads. J. Infect. Dis. 166:660-663. [DOI] [PubMed] [Google Scholar]
- 13.Geimonen, E., S. Neff, T. Raymond, S. S. Kocer, I. N. Gavrilovskaya, and E. R. Mackow. 2002. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc. Natl. Acad. Sci. USA 99:13837-13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halstead, S. B., and E. J. O'Rourke. 1977. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 146:201-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harcourt, J. L., M. K. Hagan, and M. K. Offermann. 2000. Modulation of double-stranded RNA-mediated gene induction by interferon in human umbilical vein endothelial cells. J. Interferon Cytokine Res. 20:1007-1013. [DOI] [PubMed] [Google Scholar]
- 16.Hartmann, R., H. S. Olsen, S. Widder, R. Jorgensen, and J. Justesen. 1998. p59OASL, a 2′-5′ oligoadenylate synthetase-like protein: a novel human gene related to the 2′-5′ oligoadenylate synthetase family. Nucleic Acids Res. 26:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hefti, H. P., M. Frese, H. Landis, C. Di Paolo, A. Aguzzi, O. Haller, and J. Pavlovic. 1999. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La Crosse virus and other lethal viral infections. J. Virol. 73:6984-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horisberger, M. A. 1992. Interferon-induced human protein MxA is a GTPasewhich binds transiently to cellular proteins. J. Virol. 66:4705-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horisberger, M. A. 1995. Interferons, Mx genes, and resistance to influenza virus. Am. J. Respir. Crit. Care Med. 152:S67-S71. [DOI] [PubMed] [Google Scholar]
- 20.Hovnanian, A., D. Rebouillat, E. R. Levy, M. G. Mattei, and A. G. Hovanessian. 1999. The human 2′,5′-oligoadenylate synthetase-like gene (OASL) encoding the interferon-induced 56-kDa protein maps to chromosome 12q24.2 in the proximity of the 2′,5′-OAS locus. Genomics 56:362-363. [DOI] [PubMed] [Google Scholar]
- 21.Huang, L., and A. B. Pardee. 2000. Suberoylanilide hydroxamic acid as a potential therapeutic agent for human breast cancer treatment. Mol. Med. 6:849-866. [PMC free article] [PubMed] [Google Scholar]
- 22.Kehrl, J. H., and S. Sinnarajah. 2002. RGS2: a multifunctional regulator of G-protein signaling. Int. J. Biochem. Cell Biol. 34:432-438. [DOI] [PubMed] [Google Scholar]
- 23.Khanna, M., U. Chaturvedi, M. Sharma, V. Pandey, and A. Mathur. 1990. Increased capillary permeability mediated by a dengue virus-induced lymphokine. Immunology 69:449-453. [PMC free article] [PubMed] [Google Scholar]
- 24.Kobuke, K., Y. Furukawa, M. Sugai, K. Tanigaki, N. Ohashi, A. Matsumori, S. Sasayama, T. Honjo, and K. Tashiro. 2001. ESDN, a novel neuropilin-like membrane protein cloned from vascular cells with the longest secretory signal sequence among eukaryotes, is up-regulated after vascular injury. J. Biol. Chem. 276:34105-34114. [DOI] [PubMed] [Google Scholar]
- 25.Lahm, H., A. Hoeflich, S. Andre, B. Sordat, H. Kaltner, E. Wolf, and H. J. Gabius. 2000. Gene expression of galectin-9/ecalectin, a potent eosinophil chemoattractant, and/or the insertional isoform in human colorectal carcinoma cell lines and detection of frame-shift mutations for protein sequence truncations in the second functional lectin domain. Int. J. Oncol. 17:519-524. [DOI] [PubMed] [Google Scholar]
- 26.Landis, H., A. Simon-Jodicke, A. Kloti, C. Di Paolo, J. J. Schnorr, S. Schneider-Schaulies, H. P. Hefti, and J. Pavlovic. 1998. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J. Virol. 72:1516-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei, H. Y., T. M. Yeh, H. S. Liu, Y. S. Lin, S. H. Chen, and C. C. Liu. 2001. Immunopathogenesis of dengue virus infection. J. Biomed. Sci. 8:377-388. [DOI] [PubMed] [Google Scholar]
- 28.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 29.Lin, C. F., H. Y. Lei, A. L. Shiau, C. C. Liu, H. S. Liu, T. M. Yeh, S. H. Chen, and Y. S. Lin. 2003. Antibodies from dengue patient sera cross-react with endothelial cells and induce damage. J. Med. Virol. 69:82-90. [DOI] [PubMed] [Google Scholar]
- 30.Lin, C. F., H. Y. Lei, A. L. Shiau, H. S. Liu, T. M. Yeh, S. H. Chen, C. C. Liu, S. C. Chiu, and Y. S. Lin. 2002. Endothelial cell apoptosis induced by antibodies against dengue virus nonstructural protein 1 via production of nitric oxide. J. Immunol. 169:657-664. [DOI] [PubMed] [Google Scholar]
- 31.Loetscher, P., and I. Clark-Lewis. 2001. Agonistic and antagonistic activities of chemokines. J. Leukoc. Biol. 69:881-884. [PubMed] [Google Scholar]
- 32.Lukashevich, I. S., R. Maryankova, A. S. Vladyko, N. Nashkevich, S. Koleda, M. Djavani, D. Horejsh, N. N. Voitenok, and M. S. Salvato. 1999. Lassa and Mopeia virus replication in human monocytes/macrophages and in endothelial cells: different effects on IL-8 and TNF-alpha gene expression. J. Med. Virol. 59:552-560. [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, K. J., and A. B. Pardee. 1999. Principles of differential display. Methods Enzymol. 303:234-258. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto, R., H. Matsumoto, M. Seki, M. Hata, Y. Asano, S. Kanegasaki, R. L. Stevens, and M. Hirashima. 1998. Human ecalectin, a variant of human galectin-9, is a novel eosinophil chemoattractant produced by T lymphocytes. J. Biol. Chem. 273:16976-16984. [DOI] [PubMed] [Google Scholar]
- 35.Miettinen, M., T. Sareneva, I. Julkunen, and S. Matikainen. 2001. IFNs activate Toll-like receptor gene expression in viral infections. Genes Immun. 2:349-355. [DOI] [PubMed] [Google Scholar]
- 36.Muller, M., E. L. Winnacker, and G. Brem. 1992. Molecular cloning of porcine Mx cDNAs: new members of a family of interferon-inducible proteins with homology to GTP-binding proteins. J. Interferon Res. 12:119-129. [DOI] [PubMed] [Google Scholar]
- 37.Murgue, B., O. Cassar, and X. Deparis. 2001. Plasma concentrations of sVCAM-1 and severity of dengue infections. J. Med. Virol. 65:97-104. [PubMed] [Google Scholar]
- 38.Oda, Y., Y. Tatsumi, and S. Aonuma. 1991. Mitogenic activity of Tulipa gesneriana lectins on mouse and human lymphocytes. Chem. Pharm. Bull. 39:3350-3352. [DOI] [PubMed] [Google Scholar]
- 39.Paroutaud, P., G. Levi, V. I. Teichberg, and A. D. Strosberg. 1987. Extensive amino acid sequence homologies between animal lectins. Proc. Natl. Acad. Sci. USA 84:6345-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu, J., J. Adam, D. M. Bloxham, K. R. Bruckdorfer, N. G. Miller, N. A. Parkinson, and J. A. Lucy. 2000. Phosphatidylserine-dependent adhesion of T cells to endothelial cells. Biochim. Biophys. Acta 1501:99-115. [DOI] [PubMed] [Google Scholar]
- 41.Rebouillat, D., I. Marie, and A. G. Hovanessian. 1998. Molecular cloning and characterization of two related and interferon-induced 56-kDa and 30-kDa proteins highly similar to 2′-5′ oligoadenylate synthetase. Eur. J. Biochem. 257:319-330. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds, A. M., M. D. Holmes, and R. Scicchitano. 2002. Interleukin-1β and tumour necrosis factor-α increase microvascular leakage in the guinea pig trachea. Respirology 7:23-28. [DOI] [PubMed] [Google Scholar]
- 43.Rothman, A. L., and F. A. Ennis. 1999. Immunopathogenesis of dengue hemorrhagic fever. Virology 257:1-6. [DOI] [PubMed] [Google Scholar]
- 44.Rothman, A. L., and F. A. Ennis. 2000. Toga/flaviviruses: immunopathology. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 45.Ryo, A., Y. Suzuki, M. Arai, N. Kondoh, T. Wakatsuki, A. Hada, M. Shuda, K. Tanaka, C. Sato, M. Yamamoto, and N. Yamamoto. 2000. Identification and characterization of differentially expressed mRNAs in HIV type 1-infected human T cells. AIDS Res. Hum. Retrovir. 16:995-1005. [DOI] [PubMed] [Google Scholar]
- 46.Shepard, L. W., M. Yang, P. Xie, D. D. Browning, T. Voyno-Yasenetskaya, T. Kozasa, and R. D. Ye. 2001. Constitutive activation of NF-κB and secretion of interleukin-8 induced by the G protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus involve G alpha(13) and RhoA. J. Biol. Chem. 276:45979-45987. [DOI] [PubMed] [Google Scholar]
- 47.Silverman, R. H., A. Halloum, A. Zhou, B. Dong, F. Al-Zoghaibi, D. Kushner, Q. Zhou, J. Zhao, T. Wiedmer, and P. J. Sims. 2002. Suppression of ovarian carcinoma cell growth in vivo by the interferon-inducible plasma membrane protein, phospholipid scramblase 1. Cancer Res. 62:397-402. [PubMed] [Google Scholar]
- 48.Sims, P. J., and T. Wiedmer. 2001. Unraveling the mysteries of phospholipid scrambling. Thromb. Haemost. 86:266-275. [PubMed] [Google Scholar]
- 49.Spitzenberger, F., J. Graessler, and H. E. Schroeder. 2001. Molecular and functional characterization of galectin 9 mRNA isoforms in porcine and human cells and tissues. Biochimie 83:851-862. [DOI] [PubMed] [Google Scholar]
- 50.Stehlik, C., R. de Martin, I. Kumabashiri, J. A. Schmid, B. R. Binder, and J. Lipp. 1998. Nuclear factor (NF)-κB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor alpha-induced apoptosis. J. Exp. Med. 188:211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, J., M. Nanjundan, L. J. Pike, T. Wiedmer, and P. J. Sims. 2002. Plasma membrane phospholipid scramblase 1 is enriched in lipid rafts and interacts with the epidermal growth factor receptor. Biochemistry 41:6338-6345. [DOI] [PubMed] [Google Scholar]
- 52.Takatsuka, H., T. Wakae, A. Mori, M. Okada, Y. Fujimori, Y. Takemoto, T. Okamoto, A. Kanamaru, and E. Kakishita. 2003. Endothelial damage caused by cytomegalovirus and human herpesvirus-6. Bone Marrow Transplant. 31:475-479. [DOI] [PubMed] [Google Scholar]
- 52a.Terajima, M., and F. A. Ennis. 2003. Quantitation of antibody-bound and unbound Sin Nombre virus in the plasma of patients with hantavirus pulmonary syndrome. J. Virol. Methods 110:159-162. [DOI] [PubMed]
- 53.Tureci, O., H. Schmitt, N. Fadle, M. Pfreundschuh, and U. Sahin. 1997. Molecular definition of a novel human galectin which is immunogenic in patients with Hodgkin's disease. J. Biol. Chem. 272:6416-6422. [DOI] [PubMed] [Google Scholar]
- 54.Wada, J., and Y. S. Kanwar. 1997. Identification and characterization of galectin-9, a novel β-galactoside-binding mammalian lectin. J. Biol. Chem. 272:6078-6086. [DOI] [PubMed] [Google Scholar]
- 55.Wada, J., K. Ota, A. Kumar, E. I. Wallner, and Y. S. Kanwar. 1997. Developmental regulation, expression, and apoptotic potential of galectin-9, a β-galactoside binding lectin. J. Clin. Investig. 99:2452-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, C. Y., M. W. Mayo, and A. S. Baldwin, Jr. 1996. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science 274:784-787. [DOI] [PubMed] [Google Scholar]
- 57.Wiedmer, T., Q. Zhou, D. Y. Kwoh, and P. J. Sims. 2000. Identification of three new members of the phospholipid scramblase gene family. Biochim. Biophys. Acta 1467:244-253. [DOI] [PubMed] [Google Scholar]
- 58.Wrighton, C. J., R. Hofer-Warbinek, T. Moll, R. Eytner, F. H. Bach, and R. de Martin. 1996. Inhibition of endothelial cell activation by adenovirus-mediated expression of IκBα, an inhibitor of the transcription factor NF-κB. J. Exp. Med. 183:1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida, H., T. Imaizumi, M. Kumagai, K. Kimura, C. Satoh, N. Hanada, K. Fujimoto, N. Nishi, K. Tanji, T. Matsumiya, F. Mori, X. F. Cui, W. Tamo, T. Shibata, S. Takanashi, K. Okumura, T. Nakamura, K. Wakabayashi, M. Hirashima, Y. Sato, and K. Satoh. 2001. Interleukin-1β stimulates galectin-9 expression in human astrocytes. Neuroreport 12:3755-3758. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, Q., J. Zhao, T. Wiedmer, and P. J. Sims. 2002. Normal hemostasis but defective hematopoietic response to growth factors in mice deficient in phospholipid scramblase 1. Blood 99:4030-4038. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zurcher, T., J. Pavlovic, and P. Staeheli. 1992. Mechanism of human MxA protein action: variants with changed antiviral properties. EMBO J. 11:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]