Abstract
The diketo acid L-708,906 has been reported to be a selective inhibitor of the strand transfer step of the human immunodeficiency virus type 1 (HIV-1) integration process (D. Hazuda, P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller, Science 287:646-650, 2000). We have now studied the development of antiviral resistance to L-708,906 by growing HIV-1 strains in the presence of increasing concentrations of the compound. The mutations T66I, L74M, and S230R emerged successively in the integrase gene. The virus with three mutations (T66I L74M S230R) was 10-fold less susceptible to L-708,906, while displaying the sensitivity of the wild-type virus to inhibitors of the RT or PRO or viral entry process. Chimeric HIV-1 strains containing the mutant integrase genes displayed the same resistance profile as the in vitro-selected strains, corroborating the impact of the reported mutations on the resistance phenotype. Phenotypic cross-resistance to S-1360, a diketo analogue in clinical trials, was observed for all strains. Interestingly, the diketo acid-resistant strain remained fully sensitive to V-165, a novel integrase inhibitor (C. Pannecouque, W. Pluymers, B. Van Maele, V. Tetz, P. Cherepanov, E. De Clercq, M. Witvrouw, and Z. Debyser, Curr. Biol. 12:1169-1177, 2002). Antiviral resistance was also studied at the level of recombinant integrase. Single mutations did not appear to impair specific enzymatic activity. However, 3′ processing and strand transfer activities of the recombinant integrases with two (T66I L74M) and three (T66I L74M S230R) mutations were notably lower than those of the wild-type integrase. Although the virus with three mutations was resistant to inhibition by diketo acids, the sensitivity of the corresponding enzyme to L-708,906 or S-1360 was reduced only two- to threefold. As to the replication kinetics of the selected strains, the replication fitness for all strains was lower than that of the wild-type HIV-1 strain.
The replication of human immunodeficiency virus type 1 (HIV-1) in infected patients can be reduced considerably by treatment with potent combinations of drugs with multiple viral targets (31). Drugs that have been officially approved for anti-HIV treatment belong either to the class of nucleoside and nonnucleoside reverse transcriptase (RT) inhibitors or to protease (PRO) inhibitors (11). Due to low-level residual replication, long-term therapeutic success of highly active antiretroviral therapy may be jeopardized by the emergence of virus strains resistant to the currently used antiviral drugs. Therefore, it is essential to develop drugs targeting alternative steps of the viral replication cycle. An attractive target, in addition to RT and PRO, is the viral enzyme integrase (IN). The integration of retrotranscribed viral DNA into the host cell chromosome is an essential step in the replication cycle of retroviruses (20, 33). After integration, the proviral DNA is replicated and genetically transmitted as part of the cellular genome. Therefore, integration defines a point of no return in the life cycle of HIV. Since no human counterpart of the enzyme is known, there is substantial interest in developing effective and selective inhibitors of HIV IN (9, 37).
The 288 amino acids of IN (32 kDa) are encoded by the 3′ end of the pol gene (4). The enzyme is produced by PRO-mediated cleavage of the Gag-Pol precursor during virion maturation. IN recognizes specific sequences in the long terminal repeats (LTRs) of the viral retrotranscribed DNA. The terminal 15 bp of the LTRs are necessary and sufficient for site-specific endonucleolytic activity and integration. The highly conserved dinucleotide CA immediately upstream of the cleavage site is critical for enzymatic activity. In the first step of the integration reaction, referred to as 3′-end processing, the terminal GT dinucleotides are removed from each 3′ end to produce new 3′ hydroxyl ends (CA-3′-OH). This reaction occurs in the cytoplasm within a large viral nucleoprotein complex, the preintegration complex (PIC) (21). After the PIC is transported through the nuclear pore, the processed viral double-stranded DNA is joined to the host DNA. The joining reaction, termed strand transfer, includes a coupled 5-bp staggered cleavage of the target DNA and ligation of the processed CA-3′-OH viral DNA ends to the 5′ O-phosphate ends of the target DNA. The 3′ ends of the target DNA remain unjoined after strand transfer. The integration reaction is completed by the removal of the two unpaired nucleotides at the 5′ end of the viral DNA and the repair of the single-stranded gaps created between the viral and target DNA. This repair is probably accomplished by host cell DNA repair enzymes, although retroviral enzymes have been implicated as well (8, 36). Staggered strand transfer and gap repair result in the duplication of host cell sequences immediately flanking the inserted proviral DNA.
IN inhibition is typically assessed in oligonucleotide-based assays using LTR mimics to evaluate both processing and joining reactions in vitro (5, 34). A vast series of compounds have been reported to inhibit IN activity in these oligonucleotide assays (29). However, most of these compounds do not exhibit antiviral activity or are too toxic in cell culture. For most of the IN inhibitors that did show antiviral activity in cell culture, it was not unambiguously demonstrated that the integration step was targeted. Moreover, for two classes of IN inhibitors with antiviral activity, the G quartets (24) and l-chicoric acid derivatives (32), it has been demonstrated by means of selecting and sequencing drug-resistant HIV-1 strains that viral entry and not integration is the main antiviral target in cell culture (13, 27). In contrast, Ojwang et al. (24) reported that G quartets generated an increase in two-LTR circles and a decrease in integrated DNA at concentrations that inhibit viral replication. Regarding chicoric acid, King and Robinson (17) reported that antiviral resistance to l-chicoric acid resulted from a single mutation, G140S, in IN. In a recent study, King et al. claimed that this mutation confers cross-resistance to diketo acids (DKA) and that the G140S mutation attenuates IN activity (18).
For at least two classes of compounds, inhibition of HIV-1 integration in cell culture has been clearly demonstrated (14, 25). DKA were the first class of IN inhibitors discovered. They were reported to demonstrate antiviral activity in cell culture as a consequence of their effect on integration, i.e., via specific inhibition of the DNA strand transfer step (14). The DKA L-708,906 showed activity against various HIV-1, HIV-2, and simian immunodeficiency virus (SIV) strains at micromolar concentrations (14, 28). No cross-resistance was observed with HIV strains resistant to inhibitors of viral entry, reverse transcription, or proteolytic processing. A series of heterocyclic compounds in the class of DKA that are metabolically more stable are represented by L-870,810. L-870,810 showed good pharmacokinetic characteristics in animal studies. Subsequent to safety assessment studies, the compound has moved into phase I clinical trials for evaluation in healthy volunteers (S. Young and the HIV Integrase Discovery Team, Merck & Co., Inc., Abstr. XIV Int. AIDS Conf., abstr. LbPeA9007, 2002). S-1360, a structural analogue of the DKAs, was reported to inhibit IN in nanomolar concentrations and viral replication in micromolar concentrations. S-1360 has already been the subject of a phase II study in healthy volunteers. The results of this study support clinical studies with S-1360 in HIV seropositive subjects (2). Recently, a series of 5H-pyrano[2,3-d:-6,5-d′] dipyrimidines (PDPs) has been identified as the second class of inhibitors that interfere with HIV-1, HIV-2, and SIV integration in cell culture (25). The most potent congener of this class, V-165, inhibited HIV-1 replication in cell culture and IN activity in an oligonucleotide-based enzymatic assay at micromolar concentrations. V-165 displayed antiviral activity against virus strains that were resistant to inhibitors of viral entry or reverse transcription. When V-165 was combined in cell culture with approved anti-HIV drugs, a subsynergistic effect with the nucleoside RT inhibitor zidovudine (AZT) and the PRO inhibitor nelfinavir and a synergistic effect with the nonnucleoside RT inhibitor nevirapine were observed. Mechanism of action studies revealed that V-165 interferes with DNA-IN complex formation (25).
Little is known about the development of resistance to HIV IN inhibitors. HIV strains that were selected in the presence of the DKA L-708,906 or L-731,988 carried the mutations T66I, S153Y, and M154I that are located near the metal coordinating residues D64 and E152 in the active site of the IN gene (14). We have now investigated in detail the development of resistance of HIV to the DKA L-708,906 through the selection of HIV-1 strains in the presence of increasing concentrations of L-708,906. A genotypic analysis revealed novel mutations. The selected strains and recombinant strains containing the mutant IN gene were analyzed phenotypically. The replication kinetics of the resistant strains has been investigated, and the mutations were examined individually and in combination at the enzymatic level. Finally, cross-resistance to other IN inhibitors, S-1360 and V-165, was evaluated.
MATERIALS AND METHODS
Compounds.
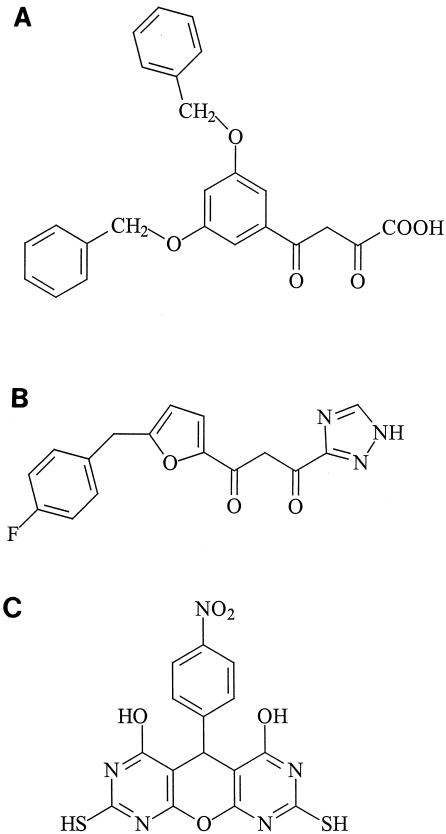
The DKA L-708,906 was synthesized at the National Cancer Institute and kindly provided by T. R. Burke (National Institutes of Health, Bethesda, Md.). AZT was synthesized by the method of Horwitz et al. (15). Nevirapine (BI-RG587) was obtained from Boehringer Ingelheim (Ridgefield, Conn.). Ritonavir (ABT538) was obtained from J. M. Leonard, Abbott Laboratories (Abbott Park, Ill.). AMD3100 was provided by G. Bridger and G. Henson, AnorMED (Langley, British Columbia, Canada) and was synthesized as described previously (3). S-1360 was synthesized at Gilead Sciences (Foster City, Calif.). The PDP derivative V-165 was obtained from Ampharm Inc. (Ramsey, N.J.) (Fig. 1). All compounds were dissolved in dimethyl sulfoxide.
FIG. 1.
Chemical structures of the HIV-1 IN inhibitors evaluated. (A) L-708,906, (B) S-1360, and (C) V-165.
Cells.
MT-4 cells (22) and MOLT-4 cell clone 8 (16) were grown in a humidified atmosphere with 5% CO2 at 37°C and maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 0.1% sodium bicarbonate, and 20 μg of gentamicin per ml.
Virus strains.
The origin of HIV-1(IIIB) has been described elsewhere (30). The plasmid pNL4.3 (1) is a molecular clone obtained from the National Institutes of Health.
Selection of antiviral resistance.
The selection of resistance to HIV-1(IIIB) to L-708,906 was initiated at a low multiplicity of infection (0.01) in MT-4 cells and a drug concentration equal to the 50% inhibitory concentration (IC50), as determined in the MT-4/MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. Every 3 to 4 days, the MT-4 cell culture was monitored for the appearance of HIV-induced cytopathic effect (CPE). When a CPE was observed, the cell-free culture supernatant was used to infect fresh, uninfected MT-4 cells in the presence of an equal or higher concentration of the compound. When no virus breakthrough was observed, the infected cell culture was subcultivated in the presence of the same concentration of the compound. The compound concentration was gradually increased.
PCR amplification and sequencing of the IN-coding region. (i) PCR amplification of IN-coding sequences.
Proviral DNA extraction of MT-4 cells infected with different passages of HIV-1(IIIB) that were selected in the presence of L-708,906 was performed using the QIAamp blood kit (Qiagen, Hilden, Germany). A 2,576-nucleotide fragment (corresponding to positions 2998 to 5574) was amplified by PCR using the Expand High Fidelity PCR system (Roche, Mannheim, Germany), which is composed of an enzyme mix containing thermostable Taq DNA polymerase and Pwo DNA polymerase with 3′→5′ exonuclease proofreading capacity. The PCR was performed using primers MW1 (5′-CCA CAR GGA TGG AAA GGA TCA CC-3′; corresponding to positions 2998 to 3018) and MW2 (5′-CTG GGG CTT GTT CCA TCT RTC YTC T-3′; corresponding to positions 5557 to 5574). Primer positions correspond to the positions in HIV-1(HXB2) (GenBank accession no. K03455). The cycling conditions were as follows: (i) a denaturation step of 2 min at 95°C; (ii) 40 cycles of amplification, with 1 cycle consisting of 15 s at 95°C, 30 s at 60°C, and 3 min at 68°C; and (iii) a final extension step of 10 min at 72°C.
(ii) Sequencing of the IN-coding region.
PCR products were purified using the PCR purification kit (Qiagen). To perform the sequencing reaction, the ABI PRISM dye terminator cycle sequencing core kit (Perkin-Elmer, Brussels, Belgium) was used. The primers used to sequence the MW1-MW2 amplified region were as follows: MW3 (5′-TAT GTA GGR TCT GAY TTA GAA ATA GGG-′; corresponding to positions 3111 to 3137), AV59 (5′-GGG GCA AGG CCA ATG GAC-′; corresponding to positions 3545 to 3562), AV60 (5′-TTT CAG ATT TTT AAA TGG-′; corresponding to positions 3582 to 3599), IN-PCRA (5′-GGA GGA AAT GAA CAA GTA GAT-′; corresponding to positions 4176 to 4196), HP4392C (5′-TCT ACT TGT CCA TGC ATG GCT TC-′; corresponding to positions 4371 to 4393), IN-SEQ1 (5′-CTT AAG ATG TTC AGC CTG ATC T-′; corresponding to positions 4727 to 4748), IN-SEQ3 (5′-GGA TAT ATA GAA GCA GAA GTA A-′; corresponding to positions 4472 to 4494), IN-SEQ4 (5′-GAA CAT CTT AAG ACA GCA GTA-′; corresponding to positions 4737 to 4757), IN-PCRB (5′-CCC TGA AAC ATA CAT ATG GT-′; corresponding to positions 5120 to 5140) and MW4 (5′-TAA CAC TAG GCA RAG GTG GCT T-′; corresponding to positions 5518 to 5539). Primer positions correspond to HIV-1(HXB2) (GenBank accession no. K03455). The samples were loaded on the ABI PRISM 310 genetic analyzer (Perkin-Elmer). The sequences were analyzed using the software program Geneworks 2.5.1 (Intelligenetics Inc., Oxford, United Kingdom). Mutations present in more than 25% of the global virus population can be detected by means of population sequencing.
Construction of an HIV-1 clone with a deletion of the IN gene.
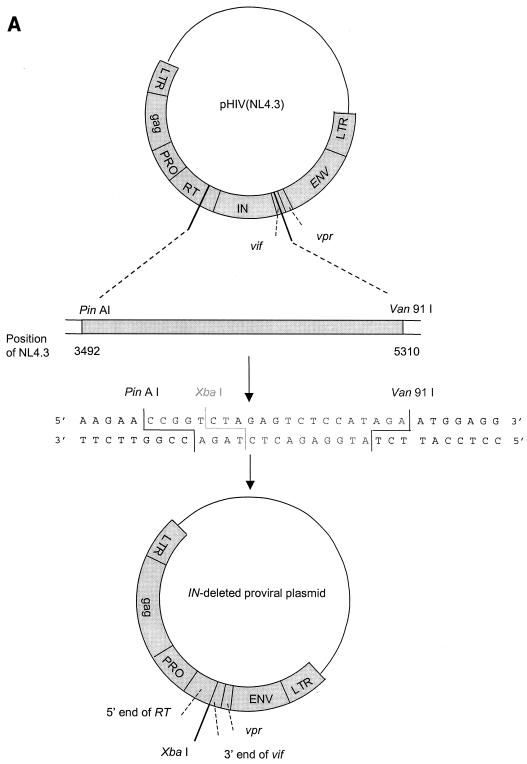
To generate the clone in which the IN gene had been deleted, the proviral molecular clone pNL4.3 (1) was used as starting material (Fig. 2A). This clone consists of the plasmid pUC18 in which the complete HIV-1 genome flanked by chromosomal DNA is inserted. pNL4.3 was digested with the restriction enzymes AgeI and Van91I. The vector was purified by gel extraction (using β-agarase I) and subsequent phenol-chloroform extraction. A linker sequence containing the XbaI restriction site was ligated into the vector to recircularize the plasmid (Fig. 2A). Escherichia coli DH5α was transformed with this clone in which the IN gene had been deleted.
FIG.2.
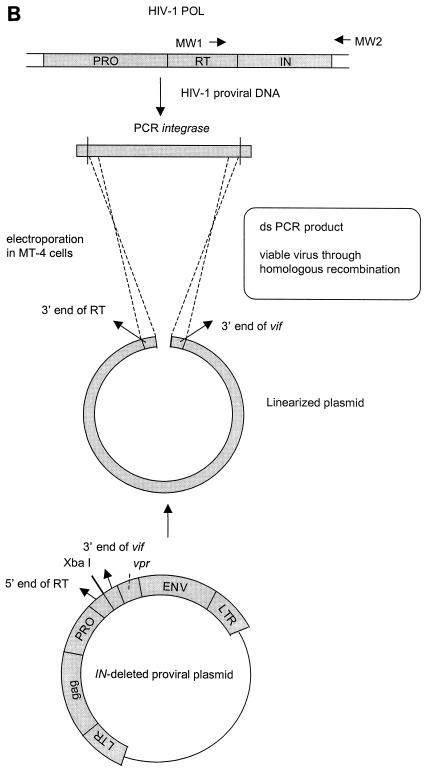
IN chimeric virus technology. (A) Construction of the pNL4.3 clone with the IN gene deleted. To generate the clone with the IN gene deleted, the proviral molecular clone pNL4.3 was digested with AgeI and Van91I. A linker sequence containing the XbaI restriction site was ligated into the vector. (B) Construction of HIV strains with recombined IN genes. For IN recombination experiments, MT-4 cells were cotransfected with the XbaI-linearized clone in which the IN gene had been deleted and MW1-MW2 PCR product. ds, double-stranded.
IN recombination assay.
MT-4 cells were subcultured at a density of 500,000 cells/ml one day before transfection. Cells were pelleted and resuspended in phosphate-buffered saline at a concentration of 3.13 × 106 cells/ml. For each transfection, 2.5 × 106 cells (0.8 ml) was used. Transfection was performed by electroporation using EASYJECT (Eurogentec, Seraing, Belgium) and electroporation cuvettes (Eurogentec). For IN recombination experiments, MT-4 cells were cotransfected with 10 μg of XbaI-linearized clone with the IN gene deleted and 2 μg of purified and concentrated MW1-MW2 PCR product (PCR purification kit; Qiagen) (Fig. 2B). Electroporation was done at 300 μF and 300 V. After 30-min incubation at room temperature, the transfected cells suspended in 5 ml of culture medium were incubated at 37°C in a humidified atmosphere with 5% CO2. When full CPE was observed in the culture (about 6 days posttransfection), the recombinant virus was harvested by centrifugation and stored in 1-ml aliquots at −80°C for subsequent infectivity and drug susceptibility determinations (IC50) in the MT-4/MTT assay.
Drug susceptibility assay.
The inhibitory effects of antiviral drugs on the HIV-induced CPE in human lymphocyte MT-4 cell culture were determined by the MTT assay (26). This assay is based on the reduction of the yellow-colored MTT by mitochondrial dehydrogenase of metabolically active cells to a blue formazan derivative, which can be measured spectrophotometrically. The 50% cell culture infective doses (CCID50) of the HIV strains were determined by titration of the virus stock using MT-4 cells. For the drug susceptibility assays, MT-4 cells were infected with 100 to 300 CCID50 of the HIV strains in the presence of fivefold serial dilutions of the antiviral drugs. The concentration of the compound achieving 50% protection against the CPE of HIV, which is defined as the IC50, was determined. In MOLT-4 cells cultured in the presence of various compound concentrations, anti-HIV activity was determined by monitoring the appearance of HIV-1-induced CPE 5 days postinfection. MOLT-4 cells (2 × 105/ml) were infected with different HIV-1 strains at 200 CCID50.
Construction of IN mutants and enzyme purification.
The expression plasmid pRP1012 (R. H. A. Plasterk, Dutch Cancer Institute, Amsterdam, The Netherlands), encoding HIV-1 IN (strain HTLV III), was used to generate the following IN mutants: IN with the single mutations T66I, L74M, and S230R, the double mutation T66I L74M, and the triple mutation T66I L74M S230R. Site-directed mutagenesis was performed using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Briefly, supercoiled double-stranded pRP1012 and two synthetic primers containing the desired mutation were used. To generate single mutants, the primers, each complementary to one of the two strands of the vector, T66I upper (5′-GAA TAT GGC AAC TAG ATT GTA TAC ATT TAG AAG GAA AAG-3′) and T66I lower (5′ CTT TTC CTT CTA AAT GTA TAC AAT CTA GTT GCC ATA TTC 5′) and the combinations L74 M upper and L74 M lower (5′-GGA AAA GTT ATC ATG GTA GCA GTA CAT GTA GCC AGT GG-3′ and 5′-CCA CTG GCT ACA TGT ACT GCT ACC ATG ATA ACT TTT CC-3′, respectively) and S230R upper and S230R lower (5′-CGG GTT TAT TAC AGG GAC AGG AGA AAT CCA C-3′ and 5′-GTG GAT TTC TCC TGT CCC TGT AAT AAA CCC G-3′, respectively) were used. These primer sets were extended during temperature cycling by using Pfu DNA polymerase. Subsequently, the product was digested with DpnI, which selects for the synthesized DNA containing the mutations. Expression plasmids containing single mutations were mutagenized to obtain the double mutant and subsequently the triple mutant. The presence of the expected mutations was confirmed by sequencing of the complete IN gene. Recombinant His-tagged HIV-1 IN enzymes were produced by transforming E. coli PC2 [BL21(DE3) (pLysS) ΔendA::Tcr], which was resistant to phage T1, with the wild-type and different mutant expression plasmids. Enzymes were purified on a nickel-nitrilotriacetic acid column (Qiagen), followed by a Hi Trap heparin column (Pharmacia) (7, 10).
Substrate and target DNA used in the enzymatic IN assay.
The following high-performance liquid chromatography-purified deoxyoligonucleotides, corresponding to the U5 end of the HIV-1 LTR, were purchased from Amersham Biosciences (Piscataway, N.J.): INT1 (5′-TGT GGA AAA TCT CTA GCA GT) and INT2 (5′-ACT GCT AGA GAT TTT CCA CA). The INT1 oligonucleotide was purified through a 20% denaturing polyacrylamide-urea gel and was 5′ end labeled using polynucleotide T4 kinase and [γ-32P]ATP (Amersham Biosciences). The DNA substrate for the IN reactions was made by annealing INT1 and INT2 oligonucleotides. An equimolar mixture of the two oligonucleotides in the presence of 100 mM NaCl was briefly heated to 95°C and allowed to cool slowly to room temperature. Likewise, annealing of oligonucleotides SK70 and T35 resulted in a 35-bp double-stranded DNA molecule that was used as a target DNA molecule (T35/SK70).
3′ processing, overall integration, and strand transfer assays.
The enzymatic integration reaction was performed as described previously with minor modifications (7, 10).
The final reaction mixture for the 3′ processing assay contained 20 mM HEPES (pH 7.5), 5 mM dithiothreitol, 10 mM MgCl2, 75 mM NaCl, 15% (vol/vol) polyethylene glycol 8000, 15% dimethyl sulfoxide, 20 nM concentration of the oligonucleotide substrate, and 1 μM concentration of the His-tagged IN (final volume of 10 μl). Reactions were started by the addition of the enzyme. To determine the susceptibility to different inhibitors, compounds were briefly incubated with the reaction components before the addition of IN. Reactions were allowed to proceed at 37°C for 7 min and were stopped by the addition of formamide loading buffer (95% formamide, 30 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue, 0.1% sodium dodecyl sulfate). In the overall integration assay, this reaction mixture was allowed to proceed for 60 min before the addition of formamide dye. Strand transfer was assayed in the following way: 20 nM DNA substrate was preincubated with 1 μM IN at 37°C for 7 min to allow the cleavage reaction to occur. The composition of the reaction mixture was identical to that in the processing assay. After 7 min, 1 μl of excess target DNA (final concentration, 250 nM) with or without inhibitor was added, and the samples were incubated at 37°C for 1 h. This excess target DNA competitively blocks further binding of IN to the oligonucleotide. Subsequent products were separated in a 15% denaturing polyacrylamide-urea gel. The results were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Specific activities of different enzyme preparations (1 μM) were determined in this assay. The extent of 3′ processing or DNA strand transfer was based on the amounts of −2 bands (−2 refers to size of uncleaved DNA minus 2 (nucleotides) or strand transfer products relative to the intensity of the total radioactivity present in the lane as determined using OptiQuant acquisition and analysis software (Perkin-Elmer Corp., Fremont, Calif.).
Determination of replication fitness of HIV-1 strains selected in the presence of L-708,906.
Inoculants of various HIV-1 strains containing equal amounts of HIV-1 p24 antigen (250, 50, 10, and 2 pg/ml) were added to MT-4 cells. Starting at 3 days postinfection, cells were examined daily for the appearance of HIV-1-induced cytopathogenicity. In addition, aliquots of cell-free supernatants were taken for the determination of viral p24 levels.
Construction of the 3D model of mutant HIV-1 IN.
On the basis of the published partial X-ray IN structures 1EX4 (35) and 1K6Y (6), a three-dimensional (3D) IN model has been assembled by standard homology modeling techniques. All computations were done using the Brugel package. Differences between the target and template sequences were incorporated using the dead-end elimination method. A long 9-residue loop (from residues 47 to 55) missing from the template structures was inserted using the Brugel spare-part approach. Subsequently all mutants were modeled using the dead-end elimination method (12).
RESULTS
Selection of HIV-1 strains resistant to DKA.
DKA-resistant HIV-1 strains were selected by serial passage of HIV-1(IIIB) in the presence of increasing concentrations of L-708,906. After 15 passages, the selected strain was able to grow at a compound concentration of 27.5 μM, a concentration that is 4.8-fold higher than the concentration required to inhibit the replication of wild-type HIV-1(IIIB) by 50% (IC50) (5.7 μM). A higher concentration proved to be cytotoxic. To increase the selective pressure even further without increasing the compound concentration, the volume of virus-containing supernatant used to reinfect fresh MT-4 cells was gradually decreased from 500 to 25 μl in a total culture volume of 2 ml during the 55 passages of the selection process.
Genotypic analysis of the IN genes of the selected HIV-1 strains.
The IN-coding regions of the HIV-1(IIIB) strains selected in the presence of L-708,906 (IIIB/L-708,906) were sequenced. Several mutations were detected in comparison with the DNA sequence of the wild-type HIV-1(IIIB) strain (Table 1). Part of the HIV-1(IIIB) virus population that was passaged 25 times [IIIB/L-708,906(#25)] in the presence of DKA carried the T66I mutation. After 35 passages in the presence of L-708,906, the T66I mutation was present in the entire virus population, whereas the L74M mutation was present as a mixture with the wild-type L74. Selection for 40 passages resulted in a strain containing a mixture of S230 and S230R in addition to T66I and L74M. After 60 passages, all three mutations were present in the entire virus population. No additional mutations were selected during selection up to 70 passages. Once the mutations were present, 25 more passages of IIIB/L-708,906(#60) in the absence of drug did not result in a reversal to wild-type IN sequences (Table 1). The IN gene sequences observed after IN recombination were similar to those of the respective wild-type or in vitro-selected IIIB/L-708,906 strains (data not shown).
TABLE 1.
Genotypic analysis of the IN genes from HIV-1(IIIB) strains selected in the presence of L-708,906 relative to the wild-type HIV-1(IIIB) strain
| HIV-1 straina | Amino acid at position
|
||
|---|---|---|---|
| 66 | 74 | 230 | |
| HIV(IIIB) | T | L | S |
| IIIB/L-708,906(#25) | T/Ic | L | S |
| IIIB/L-708,906(#35) | I | L/M | S |
| IIIB/L-708,906(#40) | I | M | S/R |
| IIIB/L-708,906(#60) | I | M | R |
| IIIB/L-708,906(#70) | I | M | R |
| IIIB/L-708,906(#60+#25 w/o L-708,906)b | I | M | R |
| Mutation type | Transition (ACA→ATA) | Transversion (CTG→ATG) | Transversion (AGC→AGA) |
HIV-1(IIIB) strains selected in MT-4 cells in the presence of L-708,906 during a certain number of passages (25, 35, 40, 60, or 70 passages).
HIV-1(IIIB) strain selected in MT-4 cells in the presence of L-708,906 during 60 passages, followed by 25 additional passages of this virus in the absence of L-708,906.
Part of the virus population (at least 30%) contains a certain mutation, while the wild-type amino acid is still present at this position in the rest of the population as determined by population sequencing.
Evaluation of phenotypic (cross)-resistance of the different selected HIV-1 strains.
To confirm that the HIV strains selected to grow in the presence of high DKA concentrations were indeed resistant to the drug, we determined the antiviral activity of L-708,906 against strains HIV-1(IIIB), IIIB/L-708,906(#25), IIIB/L-708,906(#35), IIIB/L-708,906(#40), and IIIB/L-708,906(#60) in MT-4 and MOLT-4 cells. In parallel, the sensitivities to two other HIV IN inhibitors, namely, the diketo compound S-1360 and the PDP derivative V-165, were determined in both cell lines (Table 2). The inhibitory effects of the nucleoside RT inhibitor AZT, the nonnucleoside RT inhibitor nevirapine, the PRO inhibitor ritonavir, and the entry inhibitor AMD3100 were evaluated in MT-4 cells, whereas nevirapine was evaluated in MOLT-4 cells as well.
TABLE 2.
Susceptibilities of selected and recombinant HIV-1 strains to various antiviral compounds evaluated in MT-4 and MOLT-4 cells
| Antiviral compound | Cell | IC50 (μM) (fold resistance) in HIV-1 strainsa
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
Passaged 35 times
|
Passaged 40 times
|
Passaged 60 times
|
||||||
| HIV(IIIB) | RIN/IIIB | IIIB/L-708,906(35#) | RIN/L-708,906(35#) | IIIB/L-708,906(40#) | RIN/L-708,906(40#) | IIIB/L-708,906(60#) | RIN/L-708,906(60#) | ||
| IN inhibitors | |||||||||
| L-708,906 | MT-4 | 5.7 ± 4.7 | 5.4 ± 3.9 | 16.6 ± 5.4 (2.9) | 23.8 ± 21.0 (4.4) | 25.0 ± 21.8 (4.4) | 24.1 ± 15.3 (4.5) | >55.9 ± 20.0 (>9.8) | >62.6 ± 0.5 (>11.6) |
| MOLT-4 | 1.1 ± 0.9 | ND | 5.0 ± 2.1 (4.5) | ND | 11.9 ± 0.9 (10.8) | ND | 20.0 ± 7.0 (18.2) | ND | |
| S-1360 | MT-4 | 1.6 ± 0.9 | 3.1 ± 1.6 | 10.5 ± 2.5 (6.6) | 13.3 ± 1.2 (4.3) | 10.8 ± 2.2 (6.8) | 13.3 ± 1.6 (4.3) | 13.6 ± 1.6 (8.5) | 21.7 ± 13.0 (7.0) |
| MOLT-4 | 0.3 ± 0.03 | ND | 3.4 ± 0.7 (11.3) | ND | 8.5 ± 8.4 (28.3) | ND | 7.8 ± 7.0 (26.0) | ND | |
| V-165 | MT-4 | 12.7 ± 6.2 | 10.2 ± 3.7 | 11.5 ± 4.8 (0.9) | 9.3 ± 1.6 (0.9) | 9.4 ± 3.5 (0.7) | 9.2 ± 0.7 (0.9) | 8.2 ± 2.5 (0.6) | 9.7 ± 0.7 (1.0) |
| MOLT-4 | 7.5 ± 2.5 | ND | 16.8 ± 3.5 (2.2) | ND | 16.3 ± 12.4 (1.3) | ND | 15.1 ± 12.4 (2.0) | ND | |
| Entry inhibitor | |||||||||
| AMD3100 | MT-4 | 0.019 ± 0.013 | 0.002 ± 0.002 | 0.012 ± 0.001 (0.6) | 0.001 ± 0.001 (0.5) | 0.008 ± 0.004 (0.4) | 0.001 ± 0.001 (0.5) | 0.012 ± 0.005 (0.6) | 0.002 ± 0.001 (1.0) |
| RT inhibitors | |||||||||
| AZT | MT-4 | 0.001 ± 0.0003 | 0.001 ± 0.0004 | 0.001 ± 0.0005 (1.0) | 0.001 ± 0.0003 (1.0) | 0.001 ± 0.0004 (1.0) | 0.002 ± 0.0005 (2.0) | 0.001 ± 0.0002 (1.0) | 0.001 ± 0.0004 (1.0) |
| Nevirapine | MT-4 | 0.056 ± 0.020 | 0.128 ± 0.063 | 0.049 ± 0.012 (0.9) | ND | 0.034 ± 0.022 (0.6) | ND | 0.034 ± 0.011 (0.6) | ND |
| MOLT-4 | 0.024 | ND | 0.024 (1.0) | ND | 0.052 (2.2) | ND | 0.052 (2.2) | ND | |
| PRO inhibitor | |||||||||
| Ritonavir | MT-4 | 0.041 ± 0.029 | 0.088 ± 0.010 | 0.056 ± 0.011 (1.4) | 0.085 ± 0.023 (1.0) | 0.041 ± 0.024 (1.0) | 0.065 ± 0.031 (0.7) | 0.031 ± 0.019 (0.8) | 0.061 ± 0.025 (0.7) |
IC50, 50% inhibitory concentration or concentration required to inhibit the CPE of different HIV strains by 50% in MT-4 cells. Fold increase in the IC50 of the compound against the in vitro-selected HIV-1(IIIB) or recombined selected strain compared to the IC50 of the compound against the parental HIV-1(IIIB) strain or the recombined HIV-1(IIIB) strain, respectively, is shown in parentheses. HIV-1 strain designations were explained in Results. Boldface values indicate reduced susceptibility compared to the susceptibility of the wild-type strain. ND, not determined.
In MT-4 cells, IIIB/L-708,906(#25) displayed the same susceptibility as the HIV-1(IIIB) strain to L-706,906 (data not shown), while partial resistance was observed with the strains passaged 35 or 40 times (2.9- or 4.4-fold increase in IC50, respectively). IIIB/L-708,906(#60), however, was completely resistant to the inhibitory effect of L-708,906 (>9.8-fold drop in sensitivity). The sensitivity to S-1360 decreased by 6.6-, 6.8-, or 8.5-fold after 35, 40, or 60 passages, respectively. When evaluated in MOLT-4 cells, resistance to L-708,906 was observed with the strains passaged 35, 40, or 60 times (4.5-, 10.8-, or 18.2-fold increase in IC50, respectively). The sensitivity to S-1360 had decreased by 11.3-, 28.3-, or 26.0-fold after 35, 40, or 60 passages, respectively, when evaluated in MOLT-4 cells. All IIIB/L-708,906 strains remained fully sensitive to V-165 as determined in both MT-4 and MOLT-4 cells. The other inhibitors evaluated retained their full activity against the different selected strains in both cell lines (Table 2).
Construction and evaluation of HIV-1 strains with recombined IN genes.
To evaluate the importance of the described mutations in the IN gene for the observed phenotype, we constructed recombinant HIV-1 strains carrying the wild-type or resistant IN gene from HIV-1(IIIB) in an NL4.3 backbone. Therefore, a novel IN recombination assay was established. Briefly, the IN recombination assay was performed by cotransfecting concentrated PCR product derived from the different HIV-1(IIIB) strains using primers MW1 and MW2 with the XbaI-linearized NL4.3 clone in which the IN gene had been deleted (Fig. 2). IN recombination was performed with the following strains: HIV-1(IIIB) and IIIB/L-708,906(#35), IIIB/L-708,906(#40), and IIIB/L-708,906(#60)]. The recombinant strains are referred to as RIN/IIIB, RIN/L-708,906(#35), RIN/L-708,906(#40), and RIN/L-708,906(#60), respectively. The IN sequences after recombination were verified to be identical to those of the respective wild-type or in vitro-selected parental IIIB/L-708,906 strains (data not shown). Antiviral susceptibilities of the strains with recombined IN genes was determined by the standard MT-4/MTT assay (Table 2). The loss in sensitivity of the different strains with recombined IN genes, RIN/L-708,906(#35), RIN/L-708,906(#40,) and RIN/L-708,906(#60) compared to that of the wild-type recombined strain RIN/IIIB mirrored the decreased sensitivity of the corresponding parental selected strains compared to HIV-1(IIIB) for all antiviral compounds evaluated (AZT, nevirapine, ritonavir, AMD3100, L-708,906, S-1360, and V-165). These data indicate that the observed mutations in the IN gene are responsible for the observed phenotypic resistance to the DKA L-708,906 and the cross-resistance to the diketo compound S-1360.
Analysis of drug-induced mutations in recombinant IN.
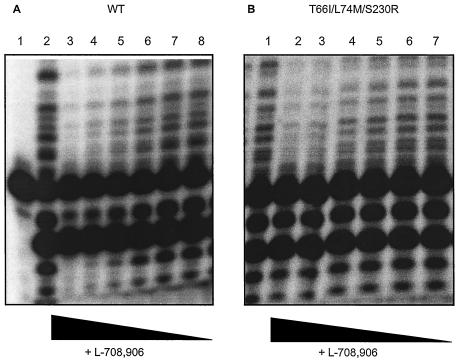
To determine the impact of the different mutations on enzymatic activity and drug sensitivity, five recombinant IN enzymes were produced by site-directed mutagenesis. The enzymes with single mutations T66I, L74M, and S230R and the double (T66I L74M) and triple (T66IL74MS230R) mutations were purified by liquid chromatography in parallel with wild-type HIV-1 IN, and the enzymatic activities were determined in the oligonucleotide-based 3′ processing and strand transfer assay. No reduced specific activity was observed for the enzymes with a single mutation (T66I, L74M, or S230R) in both assays. However, in the strand transfer assay, the enzymes with double and triple mutations both displayed a 2.5-fold reduction in 3′ processing and strand transfer activity compared to that of the wild-type IN enzyme. As evaluated in the 3′ processing assay, these mutants showed a fivefold decrease in 3′ processing (Table 3). L-708,906 inhibited mutant IN enzymes with the L74M mutation and T66I L74M S230R mutations to levels that were 1.8- and 2.2-fold lower than that of wild-type IN (Fig. 3 and Table 4). The triple mutant was inhibited to a lesser extent than wild-type IN by S-1360 (3.6-fold), while no increase in the IC50 was observed for the other mutants. The mutant IN enzymes with the T66I mutation (3.9-fold) and, to a lesser extent, the S230R mutation (2.3-fold) were less sensitive to the inhibitory effect of V-165, whereas the mutant enzymes with double and triple mutations remained fully sensitive to this compound (Table 4).
TABLE 3.
Specific enzymatic activities of the IN mutants as evaluated by the 3′ processing and strand transfer assays
| IN enzyme | 3′ Processing (%)a
|
Strand transfer (%) by strand transfer assaya | |
|---|---|---|---|
| 3′ Processing assay | Strand transfer assay | ||
| Wild type | 6.8 ± 0.3 | 4.6 ± 0.2 | 16.6 ± 0.5 |
| Mutant | |||
| T66I | 7.4 ± 0.4 (1.1) | 4.7 ± 0.1 (1.0) | 14.5 ± 0.1 (0.9) |
| L74M | 7.7 ± 0.4 (1.1) | 3.8 ± 0.1 (0.8) | 14.7 ± 0.9 (0.9) |
| S230R | 6.0 ± 0.6 (0.9) | 3.9 ± 0.6 (0.9) | 11.5 ± 0.7 (0.7) |
| T66I L74M | 1.2 ± 0.1 (0.2) | 2.0 ± 0.2 (0.4) | 6.1 ± 0.2 (0.4) |
| T66I L74M S230R | 1.1 ± 0.6 (0.2) | 1.8 ± 0.1 (0.4) | 7.0 ± 0.5 (0.4) |
Specific activities of different enzyme preparations (1 μM) determined by both the 3′ processing and strand transfer assays. The extent of 3′ processing or DNA strand transfer was based on the amounts of −2 bands or strand transfer products relative to the intensity of the total radioactivity present in the lane. Fold increase as a percentage of the specific activity of the mutant enzyme compared to the specific activity of the wild-type enzyme is shown in parentheses. Boldface values indicate reduced specific activity compared to that of the wild type.
FIG. 3.
Inhibition of recombinant IN by L-708,906. The inhibitory effect of L-708,906 against IN with three mutations (T66I L74M S230R) (B) was evaluated and compared to that of the wild-type (WT) IN enzyme (A). In panel A, lane 1, DNA substrate control; lanes 2 to 8, DNA substrate in the presence of 0, 2.48, 0.82, 0.27, 0.09, 0.03, and 0.01 μM L-708,906. In panel B, lanes 1 to 7, DNA substrate in the presence of 0, 2.48, 0.82, 0.27, 0.09, 0.03, and 0.01 μM L-708,906.
TABLE 4.
Susceptibilities of wild-type and mutant IN to IN inhibitors
| IN enzyme | IC50 (μM) (fold resistance)a
|
||
|---|---|---|---|
| L-708,906 | S-1360 | V-165 | |
| Wild type | 0.069 ± 0.015 | 1.05 ± 0.19 | 4.78 ± 0.16 |
| Mutant | |||
| T66I | 0.061 ± 0.008 (0.9) | 0.68 ± 0.33 (0.6) | 18.71 ± 2.30 (3.9) |
| L74M | 0.121 ± 0.051 (1.8) | 0.66 ± 0.22 (0.6) | 3.95 ± 0.82 (0.8) |
| S230R | 0.084 ± 0.040 (1.2) | 1.36 ± 0.09 (1.3) | 11.22 ± 3.85 (2.3) |
| T66I L74M | 0.104 ± 0.012 (1.5) | 1.08 ± 0.38 (1.0) | 4.57 ± 0.05 (1.0) |
| T66I L74M S230R | 0.154 ± 0.010 (2.2) | 3.78 ± 0.34 (3.6) | 6.48 ± 1.34 (1.4) |
IC50 or concentration of the compound required to inhibit the overall integrase activity of each enzyme by 50%. Enzymes were used at concentrations resulting in comparable enzymatic activities. Fold increase in the IC50 of the compound against the mutant enzyme compared to the IC50 of the compound against wild-type IN is shown in parentheses. Boldface values indicate reduced susceptibility compared to that of the wild-type IN.
Replication kinetics of HIV-1 strains selected in the presence of L-708,906.
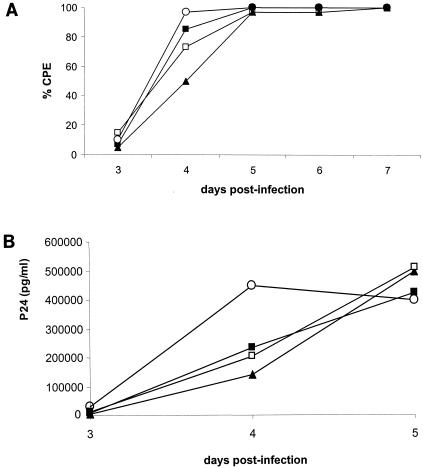
To evaluate whether drug-induced mutations in IN affect viral fitness, the HIV-1 strains selected in the presence of L-708,906 were examined for their ability to replicate in MT-4 cells in comparison with the parental HIV-1(IIIB) strain (Fig. 4). Replication kinetics of IIIB/L-708,906(#35), IIIB/L-708,906(#40), and IIIB/L-708,906(#60), as evaluated by microscopic evaluation of CPE and p24 measurements, were delayed relative to the replication of the parental HIV-1(IIIB) strain. HIV-1(IIIB) appeared fitter than IIIB/L-708,906(#35), IIIB/L-708,906(#60), and IIIB/L-708,906(#40). This relative order was observed in two independent experiments.
FIG. 4.
Replication kinetics of HIV-1 strains selected in the presence of L-708,906. MT-4 cells were inoculated with equivalent amounts (in terms of p24 antigen) of wild-type or mutant viruses. For the data shown, the virus inoculate was equivalent to 10 pg of HIV p24 antigen per ml. Cells were evaluated microscopically for HIV-induced cytopathogenicity (CPE) (A), and viral p24 levels were determined in cell-free supernatant (B). Symbols: ○, wild-type HIV-1(IIIB); ▪, IIIB/L-708,906(35#); ▴, IIIB/L-708,906(40#); □, IIIB/L-708,906(60#).
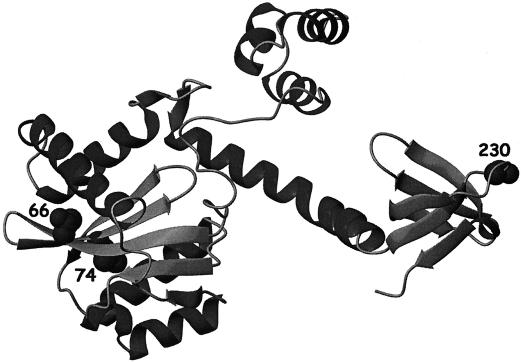
3D model of mutant HIV-1 IN.
A 3D IN model containing the described mutations has been built (Fig. 5). Positions 66 and 74 are situated in close proximity to each other and to the active site, while position 230 is located in the DNA-binding domain of IN.
FIG. 5.
3D model of HIV-1 IN. Model of the structure of the IN enzyme with three mutations showing the positions of the mutations associated with resistance to L-708,906. This model has been constructed on the basis of published partial X-ray IN structures 1EX4 (35) and 1K6Y (6).
DISCUSSION
The DKA were the first compounds reported to interfere with HIV replication through a specific inhibition of the integration step (14). Since the diketo derivative S-1360 is now being evaluated in clinical trials in HIV seropositive individuals (2) and the DKA L-870,810 has moved into clinical trials in healthy volunteers (Young et al., Abstr. XIV Int. AIDS Conf.), studying the development of resistance of HIV to IN inhibitors is warranted. In line with the emergence of resistant strains in patients treated with the current anti-HIV therapies based on RT and PRO inhibitors, the development of resistance to IN inhibitors is expected. However, the region of the HIV genome encoding IN shows more conservation than the regions encoding RT and PRO (19).
Hazuda et al. (14) previously selected HIV strains in the presence of the DKA L-708,906 or L-731,988. These selected stains carried combinations of the T66I, S153Y, and M154I mutations. Substitutions at position 66 of IN have been reported to be associated with in vitro resistance to S-1360 (T. Yoshinaga, A. Sato, T. Fujishita, and T. Fujiwara, Abstr. 9th Conf. Retrovir. Opportun. Infect., oral abstr. 8, 2002). To further study the development of resistance of HIV against DKA, we have selected HIV-1(IIIB) strains in the presence of increasing concentrations of the DKA L-708,906 (IIIB/L-708,906) and analyzed the selected strains genotypically and phenotypically at different stages throughout the selection process. An accumulation of the T66I, L74M, and S230R mutations was observed during this selection for resistance. After 35, 40, or 60 passages in the presence of L-708,906, the T66I mutation or the T66I L74M or T66I L74M S230R mutations, respectively, were present in the total virus population (Table 1). Positions 66 and 74 are in close proximity to the active site, and position 230 is located in the DNA-binding domain of IN (Fig. 5). This is the first time that the L74M and S230R mutations have been described in relation to resistance against IN inhibitors. The S153Y mutation was observed after 35 passages in the presence of L-708,906 in a minority of sequences as a result of cloning in order to sequence this strain. Population sequencing did not reveal the presence of this mutation in any of the analyzed strains.
To confirm that the selection in the presence of high concentrations of DKA indeed rendered the strains drug resistant, we determined the antiviral activities of the IN inhibitors L-708,906, S-1360, and V-165 against these strains using the MT-4 cell line. In parallel, the sensitivities to the nucleoside RT inhibitor AZT, the nonnucleoside RT inhibitor nevirapine, the PRO inhibitor ritonavir, and the entry inhibitor AMD3100 were determined (Table 2). Selection up to 40 passages rendered the double mutant IIIB/L-708,906(#40) only four times less susceptible to L-708,906 compared to the wild-type strain. However, the triple mutant IIIB/L-708,906(#60) showed a decrease in susceptibility to L-708,906 of more than 10-fold. Of interest, cross-resistance (decrease of more than sixfold) to the IN inhibitor S-1360 was already observed for IIIB/L-708,906(#35), suggesting that the single mutation T66I is sufficient to cause cross-resistance to S-1360. All viral mutants remained fully susceptible to inhibition by V-165 (Table 2).
Since it has been reported that HIV may bypass integration in some cell lines (23), it is conceivable that the mutant viruses increasingly bypass the strand transfer step inhibited by DKA in MT-4 cells. We evaluated this hypothesis by analyzing the susceptibilities of the resistant strains to the different IN inhibitors compared to that of wild-type strain in MOLT-4 cells. The MOLT-4 cell line is not productively infected with human T-cell leukemia virus type 1 and is believed not to support viral replication of mutant strains with defective IN function (23). In MOLT-4 cells, the susceptibility of the wild-type strain to DKA was fivefold higher than in MT-4 cells. The fold resistance of the IIIB/L-708,906 strains to L-708,906 and S-1360 was two- to fourfold more pronounced when evaluated in MOLT-4 cells. In line with the observation in MT-4 cells, all selected strains remained susceptible to inhibition by V-165 (Table 2). Since the resistance found in MT-4 cells was confirmed using MOLT-4 cells, we conclude that both cell lines are suitable models for resistance screening and that the MT-4 cell line is a appropriate alternative to the more cumbersome MOLT-4 cell evaluation system.
To assess the significance of the observed mutations for the resistance profile of the selected strains, a recombination assay, termed chimeric virus technology, was established for the HIV-1 IN gene. This technology allows the recombination of IN sequences derived from different strains into a proviral wild-type HIV-1(NL4.3) clone in which the corresponding IN gene had been deleted (Fig. 2). This technique was used to bring the different mutated IN genes of the IIIB/L-708,906 strains into the wild-type background. The resulting recombined strains displayed the same loss in sensitivity to the IN inhibitors as their original in vitro-selected strains, indicating that the mutations in the recombined region are responsible for the (cross-)resistance phenotype of the strains selected in the presence of L-708,906. All IN inhibitor-resistant strains, either selected in vitro or with recombined IN genes, remained sensitive to the inhibitory effects of inhibitors interfering with RT and PRO and entry processes of the replication cycle (Table 2). Besides mutations in the IN genes, the A502V substitution was found in the RT gene of IIIB/L-708,906 strains after 20 passages. This mutation remained present after continuous passaging (up to 70 passages). In addition, the D113N mutation was observed in the vif gene of IIIB/L-708,906 strains after 40 passages. During construction of the clone with the IN gene deleted, the 3′ end of the RT gene and the 5′ end of vif were also deleted. Since these regions will be reconstituted after recombination, we cannot rule out the possibility that both additional mutations contributed to the observed (cross-)resistance of IIIB/L-708,906 and RIN/L-708,906. Therefore, we have subsequently developed a second-generation IN recombination assay designed to recombine the IN gene without its flanking regions. The recombined IN strain containing only IN mutations T66I, L74M, and S230R was fivefold less sensitive to L-708,906 than the wild-type strain with a recombined IN gene (data not shown). The potential contribution of the A502V mutation in the RT gene and/or the D113N mutation in vif to the resistant phenotype needs further investigation.
To analyze the effects of the drug-induced mutations in IN on viral fitness, the selected strains were examined for their ability to replicate in MT-4 cells compared to the wild-type strain. All mutant strains showed a substantially reduced replication fitness compared to wild-type HIV-1(IIIB). The order of replication fitness of viral mutants (in decreasing order) was T66I > T66I L74M S230R > T66I L74M, pointing to a positive role for S230R in the replication ability of the triple mutant (Fig. 4).
In addition, IN enzymes containing the mutations separately and in combination were generated in order to evaluate the effects of the different mutations on enzymatic activity and drug sensitivity in the oligonucleotide-based assay. Individual mutations did not appear to impair 3′ processing or strand transfer activity. In contrast, the IN enzymes with double and triple mutations displayed a 2.5-fold reduction in both 3′ processing and strand transfer activities compared to the activities of the wild-type IN enzyme in the assay that measures strand transfer specifically. When evaluated in the 3′ processing assay, both mutants displayed a fivefold reduction in this specific activity (Table 3). Next, the inhibition of the different IN mutants by L-708,906, S-1360, and V-165 was determined. In the overall integration reaction, L-708,906 and S-1360 inhibited the IN enzyme with three mutations to a level 2.2- to 3.6-fold lower than that of the wild-type IN, whereas no significant loss in susceptibility to these compounds was observed for the IN enzymes with one and two mutations (Fig. 3). However, in apparent contrast with the cell culture data, V-165 inhibited the IN enzymes with the single mutations T66I and S230R to a level 3.9- and 2.3-fold lower than that of the wild-type IN, although the IN with three mutations had full activity (Table 4).
A comparable attenuation of viral growth and IN enzymatic activity was recently reported for IN mutations associated with antiviral resistance against DKA (14) and chicoric acid (18).
Both cell culture and enzymatic data indicate that three mutations in the IN gene are required for reduced sensitivity to L-708,906. The T66I mutation was sufficient to cause resistance to S-1360 in cell culture, although reduced susceptibility in the enzymatic assay required three mutations. Interestingly, the extent of resistance caused by the mutations to DKA in cell culture (reduction in inhibitory activity of at least fivefold) could not be entirely reproduced at the enzymatic level (twofold reduction in inhibitory activity). A possible explanation for this observation could be related to the artificial nature of the oligonucleotide-based assay. The isolated enzymatic assay does not account for concerted integration of both LTR ends or other functionalities of HIV IN, such as nuclear import or interaction with cellular cofactors. An analogous divergence in the extent of resistance obtained by cell culture versus enzymatic assay has been obtained by King et al. (18). They observed only a modest decrease in sensitivity of the recombinant IN with the G140S mutation to chicoric acid and DKA compared to the loss in susceptibility of the mutant virus (17). However, we have to point out that the resistance to DKA observed by Hazuda et al. (14) was more pronounced in their enzymatic assay than in their cellular assay. A single mutation (T66I, S153Y, or M154I) in the IN gene induced a five- to sixfold reduction of activity of the compound in the IN enzymatic assay and a two- to threefold reduction in the HIV infectivity assay. IN with the T66I S153Y and T66I M154I double mutations displayed a more than 20-fold decrease in susceptibility to DKA in the IN assay and a 6-fold decrease in the cellular assay. The discrepancy between these results may be explained by the fact that Hazuda et al. assessed the inhibitory effect of DKA in a oligonucleotide-based assay that measures DNA strand transfer specifically.
The observation that resistance against DKA in cell culture does emerge after a time period of approximately 30 weeks is of clinical significance. From a clinical point of view, HIV strains resistant to L-708,906 are likely to be cross-resistant to the diketo derivative S-1360. Of interest in this context is the noteworthy reduction in viral fitness associated with the emergence of the observed mutations. Although the T66I mutation reduced sensitivity to V-165 at the enzymatic level, the L74M and S230R mutations apparently reversed the resistance phenotype to V-165. Thus, the DKA-resistant strain remained sensitive to inhibition by V-165. The distinct antiviral resistance profiles of the different IN inhibitors support the use of IN inhibitors in addition to current antiretroviral compounds in the next generation of anti-HIV therapy.
Acknowledgments
This work was supported in part by the Belgian Geconcerteerde Onderzoeksacties (GOA 00/12, Vlaamse Gemeenschap) and by the Fonds voor Wetenschappelijk Onderzoek, FWO-Vlaanderen (grant 0104.98). Z. Debyser has a postdoctoral fellowship from the Flemish Fund for Scientific Research (FWO). B. Van Maele and J. Vercammen were funded by a grant from the Flemish Institute supporting Science-Technological Research in Industry (IWT). M. Witvrouw had a Marie Curie Host fellowship (Marie Curie Training Site) of the European Commission (HPMT-CT-2001-00355). C. Gurnari worked in our research group as a Marie Curie fellow.
We thank Liesbet De Dier for excellent technical assistance.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billich, A. 2003. S-1360 Shionogi-GlaxoSmithKline. Curr. Opin. Investig. Drugs 4:206-209. [PubMed] [Google Scholar]
- 3.Bridger, G. J., R. T. Skerlj, S. Padmanabhan, S. A. Martellucci, G. W. Henson, S. Struyf, M. Witvrouw, D. Schols, and E. De Clercq. 1995. Synthesis and structure-activity relationship of phenylenebis(methylene)-linked bis-tetraazamacrocycles that inhibit HIV replication. Effect of heteroaromatic linkers on the activity of bicyclams. J. Med. Chem. 38:366-378. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. O. 1997. Integration of retroviral DNA. Curr. Top. Microbiol. Immunol. 157:19-47. [DOI] [PubMed] [Google Scholar]
- 5.Bushman, F. D., and R. Craigie. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. USA 88:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J. C., J. Krucinski, L. J. Miercke, J. S. Finer-Moore, A. H. Tang, A. D. Leavitt, and R. M. Stroud. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA 97:8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P., J. A. Esté, R. F. Rando, J. O. Ojwang, G. Reekmans, R. Steinfeld, G. David, E. De Clercq, and Z. Debyser. 1997. Mode of interaction of G-quartets with the integrase of human immunodeficiency virus type 1 (HIV-1). Mol. Pharmacol. 52:771-780. [DOI] [PubMed] [Google Scholar]
- 8.Chow, S. A., K. A. Vincent, V. Ellison, and P. O. Brown. 1992. Reversal of integration and DNA splicing mediated by integrase of HIV. Science 255:723-726. [DOI] [PubMed] [Google Scholar]
- 9.Debyser, Z., P. Cherepanov, B. Van Maele, E. De Clercq, and M. Witvrouw. 2002. In search of authentic inhibitors of HIV-1 integration. Antivir. Chem. Chemother. 13:1-15. [DOI] [PubMed] [Google Scholar]
- 10.Debyser, Z., P. Cherepanov, W. Pluymers, and E. De Clercq. 2001. Assays for the evaluation of HIV-1 integrase inhibitors. Methods Mol. Biol. 160:139-155. [DOI] [PubMed] [Google Scholar]
- 11.De Clercq, E. 1995. Toward improved anti-HIV chemotherapy: therapeutic strategies for intervention with HIV infections. J. Med. Chem. 38:2491-2517. [DOI] [PubMed] [Google Scholar]
- 12.De Maeyer, M., J. Desmet, and I. Lasters. 2000. The dead-end elimination theorem: mathematical aspects, implementation, optimizations, evaluation, and performance. Methods Mol. Biol. 143:265-304. [DOI] [PubMed] [Google Scholar]
- 13.Esté, J. A., D. Schols, K. De Vreese, P. Cherepanov, M. Witvrouw, C. Pannecouque, Z. Debyser, R. F. Rando, B. Clotet, J. Desmyter, and E. De Clercq. 1998. Human immunodeficiency virus glycoprotein gp120 as the primary target for the antiviral action of AR177 (zintevir). Mol. Pharmacol. 53:340-345. [DOI] [PubMed] [Google Scholar]
- 14.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 15.Horwitz, J. P., J. Chua, and M. Noel. 1964. Nucleosides. V. The monomesylates of 1-(2′-deoxy-β-d-lyxofuranosyl)thymidine. J. Org. Chem. 29:2076-2078. [Google Scholar]
- 16.Kikukawa, R., Y. Koyanagi, S. Harada, N. Kobayashi, M. Hatanaka, and N. Yamamoto. 1986. Differential susceptibility to the acquired immunodeficiency syndrome retrovirus in cloned cells of human leukemic T-cell line MOLT-4. J. Virol. 57:1159-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King, P. J., and W. E. Robinson, Jr. 1998. Resistance to the anti-human immunodeficiency virus type 1 compound l-chicoric acid results from a single mutation at amino acid 140 of integrase. J. Virol. 72:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King, P. J., D. J. Lee, R. A. Reinke, J. G. Victoria, K. Beale, and W. E. Robinson, Jr. 2003. Human immunodeficiency virus type 1 integrase containing a glycine to serine mutation at position 140 is attenuated for catalysis and resistant to integrase inhibitors. Virology 306:147-161. [DOI] [PubMed] [Google Scholar]
- 19.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinsky. Human retroviruses and AIDS 2000: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 20.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi, I., H. Taguchi, I. Kobonishi, S. Yoshimoto, Y. Ohtsuki, and Y. Shiraishi. 1982. Type C virus-producing cell lines derived from adult T cell leukemia. Gann. Monogr. 28:219-228. [Google Scholar]
- 23.Nakajima, N., R. Lu, and A. Engelman. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J. Virol. 75:7944-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ojwang, J. O., R. W. Buckheit, Y. Pommier, A. Mazumder, K. De Vreese, J. A. Esté, D. Reymen, L. A. Pallansch, C. Lackman-Smith, T. L. Wallace, E. De Clercq, M. S. McGrath, and R. F. Rando. 1995. T30177, an oligonucleotide stabilized by an intramolecular guanosine octet, is a potent inhibitor of laboratory strains and clinical isolates of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 39:2426-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pannecouque, C., W. Pluymers, B. Van Maele, V. Tetz, P. Cherepanov, E. De Clercq, M. Witvrouw, and Z. Debyser. 2002. New class of HIV integrase inhibitors that block viral replication in cell culture. Curr. Biol. 12:1169-1177. [DOI] [PubMed] [Google Scholar]
- 26.Pauwels, R., J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter, and E. De Clercq. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20:309-321. [DOI] [PubMed] [Google Scholar]
- 27.Pluymers, W., N. Neamati, C. Pannecouque, V. Fikkert, C. Marchand, T. R. Burke, Y. Pommier, D. Scols, E. De Clercq, Z. Debyser, and M. Witvrouw. 2000. Viral entry as the primary target for the anti-HIV activity of chicoric acid and its tetra-acetyl esters. Mol. Pharmacol. 58:641-648. [DOI] [PubMed] [Google Scholar]
- 28.Pluymers, W., G. Pais, C. Pannecouque, V. Fikkert, Y. Pommier, T. R. Burke, Jr., E. De Clercq, M. Witvrouw, N. Neamati, and Z. Debyser. 2002. Inhibition of human immunodeficiency virus type 1 (HIV-1) integrase activity and HIV-1 replication by a series of diketo derivatives. Antimicrob. Agents Chemother. 46:3292-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pommier, Y., and N. Neamati. 1999. Inhibitors of human immunodeficiency virus integrase. Adv. Virus Res. 52:427-458. [DOI] [PubMed] [Google Scholar]
- 30.Popovic, M., M. G. Sarngadharan, E. Read, and R. C. Gallo. 1984. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 224:497-500. [DOI] [PubMed] [Google Scholar]
- 31.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 32.Robinson, W. E., Jr., M. Cordeiro, S. Abdel-Malek, Q. Jia, S. A. Chow, M. G. Reinecke, and W. M. Mitchell. 1996. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus integrase: inhibition of the core catalytic domain of human immunodeficiency virus integrase. Mol. Pharmacol. 50:846-855. [PubMed] [Google Scholar]
- 33.Sakai, H., M. Kawamura, J. Sakugari, S. Sakugari, R. Shibata, A. Ishimoto, N. Ono, S. Ueda, and A. Adachi. 1993. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 67:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman, P. A., and J. A. Fyfe. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA 87:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, J. Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young, S. D. 2001. Inhibition of HIV-1 integrase by small molecules: the potential for a new class of AIDS chemotherapeutics. Curr. Opin. Drug Discov. Dev. 4:402-410. [PubMed] [Google Scholar]