Abstract
DNA sequence analysis of five IncW plasmids (R388, pSa, R7K, pIE321, and pIE522) demonstrated that they share a considerable portion of their genomes and allowed us to define the IncW backbone. Among these plasmids, the backbone is stable and seems to have diverged recently, since the overall identity among its members is higher than 95%. The only gene in which significant variation was observed was trwA; the changes in the coding sequence correlated with parallel changes in the corresponding TrwA binding sites at oriT, suggesting a functional connection between both sets of changes. The present IncW plasmid diversity is shaped by the acquisition of antibiotic resistance genes as a consequence of the pressure exerted by antibiotic usage. Sequence comparisons pinpointed the insertion events that differentiated the five plasmids analyzed. Of greatest interest is that a single acquisition of a class I integron platform, into which different gene cassettes were later incorporated, gave rise to plasmids R388, pIE522, and pSa, while plasmids R7K and pIE321 do not contain the integron platform and arose in the antibiotic world because of the insertion of several antibiotic resistance transposons.
Naturally occurring plasmids tend to fall into coherent genetic clusters commonly known as incompatibility groups (21). This is in contrast to the genetic organization of bacteriophages, which show extreme modularity, generating great diversity by the permutation of modules (82). Plasmids of a given incompatibility group normally share the mechanisms of replication (1), copy number control (25), and other maintenance functions (27; for reviews see references 5, 26, and 55). They thus function as a single pool in the bacterial cell and due to random selection for replication or partitioning will segregate cells with one or the other plasmid but not both. They are thus said to be incompatible. From the many incompatibility groups described, a few have been analyzed in detail. Some of the best-known incompatibility groups are those within the IncF complex (IncFI and IncFII) (13, 58), the IncPs (2, 9), the IncQs (62, 63), and the IncH group (33). Although incompatibility is not always associated with sequence identity, some incompatibility groups show a conserved genetic backbone. Their members generally share an essential gene set for plasmid survival, differing in the presence of mobile genetic elements (MGEs) harbored in their genomes. Plasmids can thus be considered vehicles carrying cargos of transposons, integrons, and insertion sequences that move across bacterial populations. IncP, IncN, and IncW groups have a broad host range, so they might serve as MGE shuttles between different species. The role of horizontal gene transfer in bacterial evolution is well documented (23, 56). A planetary-scale experiment in bacterial evolution took place when antibiotics were introduced by humans around 1950 (49). The result was that antibiotic-sensitive bacteria were rapidly replaced by bacteria that had acquired mobile pieces of DNA containing antibiotic resistance genes. Among them, transposons and integrons account for a large portion of the antibiotic resistance genes carried by plasmids and other MGEs (31).
The IncW group takes its name from T. Watanabe, who described the first member of the group, pSa (81). The IncW group includes three “classical” members: pSa (81), R388 (22), and R7K (17). Electron microscope heteroduplex studies (34) and restriction enzyme maps of these plasmids (6, 45, 78, 80) underscored the high DNA sequence homology between them. Physical and genetic maps of these plasmids were drawn based on information from many authors (79). R388 has been studied in the most detail, and its complete sequence has been used to infer many features of the family (28). In this work, we analyzed the DNA sequences of five IncW plasmids in order to ascertain their diversity and evolution. These are pSa, R388, R7K, and two more recent isolates, pIE321 and pIE522 (35). IncW plasmids are less than 40 kb in size (Table 1) and have been isolated from different species of Enterobacteriaceae; pSa was obtained from Shigella (81), R388 from Escherichia coli (41), R7K from Providencia rettgeri (17), pIE321 from Salmonella dublin, and pIE552 from Klebsiella pneumoniae (35). The host range of IncW plasmids, under laboratory conditions, has been shown to comprise most of the species from the phylum Proteobacteria that have been tested so far (for a review, see reference 28).
TABLE 1.
General features of IncW plasmids
| Plasmid | Original host (year of isolation) (reference) | Size (bp) | GC content (%) | No. of genes | No. of indels in the backbone | Antibiotic resistance profilea |
|---|---|---|---|---|---|---|
| R388 | E. coli (1972) (22) | 33,920 | 57.5 | 43 | 1 | Tpr, Sur |
| pIE321 | Salmonella enterica (1996) (35) | 38,151 | 58.4 | 46 | 2 | Tcr, Smr |
| R7K | Providencia rettgeri (1972) (17) | 39,792 | 56.7 | 42 | 3 | Apr, Smr, Spr |
| pIE522 | Klebsiella pneumoniae (1996) (35) | ∼38,000 | 42 | ∼1b | Gmr, Kmr, Sur, Tobr | |
| pSa | Shigella sp. (1968) (82) | ∼39,000 | 47 | ∼1b | Cmr, Gmr, Kmr, Smr, Spr, Sur |
Tpr, resistance to trimethoprim (20 μg/ml); Sur, resistance to sulfonamide (10 μg/ml); Tcr, resistance to tetracycline (10 μg/ml); Smr, resistance to streptomycin (20 μg/ml); Apr, resistance to ampicillin (100 μg/ml); Spr, resistance to spectinomycin (100 μg/ml); Gmr, resistance to gentamicin (10 μg/ml); Kmr, resistance to kanamycin (50 μg/ml); Cmr, resistance to chloramphenicol (25 μg/ml); Tobr, resistance to tobramycin (10 μg/ml).
Since these plasmids have not been completely sequenced, it is possible (but not likely according to the array results and calculated overall size of the plasmids) that they contain additional indels.
In spite of their common genetic backbone, IncW plasmids confer different antibiotic resistance profiles, indicating that these plasmids differ in the antibiotic resistance determinants they carry. Based on the comparison of the DNA sequences of pSa, R388, R7K, pIE321, and pIE522, this work analyzes the recent evolution and diversification of the IncW backbone.
MATERIALS AND METHODS
Standard DNA techniques.
For cloning, plasmid DNA was extracted using a Qiaprep spin miniprep kit (Qiagen). For DNA array, plasmid DNA was extracted using a GenElute plasmid miniprep kit (Sigma), and total genomic E. coli DNA was obtained according to the method described in reference 61. DNA was extracted from agarose gels with a Qiaquick gel extraction kit (Qiagen). Restriction enzyme digestions, agarose gel electrophoresis, DNA cloning, and the transformation of E. coli were all carried out according to the methods described in reference 65.
Conjugation assays.
Matings were carried out using derivatives of strain DH5α [F− endA1 recA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(argF-lacZYA)U169 φ80dlacZΔM15] (38) carrying various plasmids as donors and UB1637 (F− λ− lys his trp rpsL recA56) (24) as a recipient strain. Saturated cultures of donor and recipient strains were mixed in a 1:1 ratio and mated on an LB agar surface for 1 h at 37°C as described previously (48). Transconjugants were selected on trimethoprim (20 μg/ml) for R388, tetracycline (10 μg/ml) for pIE321, chloramphenicol (25 μg/ml) for pSa, ampicillin (100 μg/ml) for R7K, and gentamicin (10 μg/ml) for pIE522. Donors were counterselected on streptomycin (300 μg/ml).
DNA macroarray hybridization. (i) Probe DNA labeling by random priming.
Plasmid DNA (25 ng) was diluted to a 45-μl final volume in Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). DNA was denatured by heating at 95°C for 5 min and placed on ice for an additional 5 min. Denatured DNA was incubated in a reaction solution containing Rediprime II random prime labeling system (Amersham) and 50 μCi of [32P]dCTP (Redivue Amersham) for 10 min at 37°C. The reaction was stopped by adding 0.2 M EDTA (5 μl), heating at 95°C for 5 min, and cooling on ice for an additional 5 min.
(ii) Membrane spotting.
Target DNAs were obtained by PCR amplification of the relevant DNA fragments from either R388 or E. coli DNA. A total of 2.5 μg of each amplified DNA was denatured in alkali and spotted onto a nylon membrane (Zeta-Probe GT; Bio-Rad) by using the high-density replicating tool (HDRT-96) of robot Biomek 2000 (Beckman) and 0.045-in.-wide metal pins. The spotted membrane was neutralized by washing it in Tris-HCl (20 mM, pH 7.0) for 20 min and UV-cross-linking it at 1,200 mJ in a UV Stratalinker 1800 (Stratagene).
(iii) Array hybridization.
The spotted membrane was prehybridized for 2 h with 5 ml buffer (0.25 M sodium phosphate, pH 7.2, 7% sodium dodecyl sulfate [SDS]) at 65°C. Then, 55 μl labeled probe DNA was added and hybridized for 20 h under the same conditions. After hybridization, the membrane was washed twice with 5 ml buffer A (sodium phosphate, 20 mM, pH 7.2) plus 5% SDS for 30 min at 65°C and washed twice again with 5 ml buffer A plus 1% SDS in the same conditions. Treated membranes were scanned using a Molecular Imager FX (Bio-Rad).
DNA sequencing, sequence analysis, and annotation.
The R388 and pIE321 plasmid DNAs, as well as fragments of plasmids pIE522 and pSa, were sequenced by progressive DNA walking, using amplified DNA fragments. The BigDye Terminator v3.0 kit (Applied Biosystems) was used in sequencing reactions. Sequencing products were analyzed in an automatic multicapillary 3700 DNA sequencer with >4-fold DNA coverage. The program suite Vector NTI 5.5 was used for analyzing and annotating the plasmid sequences. ContigExpress was used for assembling the sequences. GC content analysis was carried out with BioPlot. Multiple alignments were done with AlignX. Open reading frames (ORFs) encoding at least 50 amino acids after a translation start codon were identified. DNA and deduced protein sequences were searched by BLAST analysis. ORFs were manually annotated using evidence from BLAST analyses (4) and experimental or other evidence from previous literature. For R7K, the DNA was fragmented by sonication and size fractionated before constructing libraries in pUC19. The sequence was generated from paired-end reads from two pUC19 libraries with insert sizes of 2 to 4 kb and 4 to 6 kb. These sequence reads were performed with ABI BigDye Terminator chemistry on ABI3730 sequencing machines and gave a total coverage for the plasmid of approximately eightfold. All identified repeats were bridged with read pairs or end-sequenced PCR products. Error checking and the finishing of the sequence were performed according to standard criteria using Phrap to assemble the sequence and Gap4 for visualization.
Nucleotide sequence accession numbers.
Complete DNA sequences for R7K and pIE321 are available at EMBL-EBI Bank and GenBank, respectively, under accession numbers AM901564 (R7K) and EF633507 (pIE321). Partial sequences of pIE522 and pSa are available under GenBank accession numbers EU247928 and EU419764, respectively.
RESULTS AND DISCUSSION
A DNA array for the analysis of IncW plasmids.
An array of 79 spots was prepared on nylon membranes by including each of the 43 R388 genes, 14 intergenic regions, 12 E. coli chromosomal genes, and total genomic DNA from E. coli and R388 (see Fig. S1 in the supplemental material). Among other projected applications, this array is useful as a first characterization tool to analyze the diversity of IncW plasmids.
When R388 DNA was hybridized against the array, the intensity of the signals correlated well with the size of the DNA in the spot (data not shown), indicating that association kinetics was the limiting step. No hybridization signal appeared with spots from E. coli ORFs, except an unexplained signal in gene rpoB. When the same array was hybridized against each of the four IncW plasmids shown in Table 1, hybridization to most R388 probes (ORFs and intergenic regions) was observed (see Fig. S1 in the supplemental material). Differences in the hybridization profiles of pIE522 and pSa concerned just the gene cassettes carried by the respective integrons, a consequence of their different antibiotic resistance profiles. Besides, plasmids pIE321 and R7K lacked the six spots (see Fig. S1, 9E to 10D, in the supplemental material) related to the integron, indicating that these two plasmids lack an integron platform. Finally, gene osa was absent in plasmid R7K. The use of the array allowed us to include plasmids pIE321 and pIE522 (from which nearly nothing was known) within the realm of IncW plasmids. Furthermore, based on the array results, we decided to sequence plasmids pIE321 and R7K, which showed the most interesting differences with respect to R388. We did not complete the sequences of the other two because they are practically identical to the R388 backbone.
Analysis of the DNA sequences of five IncW plasmids.
The complete sequences of plasmids pIE321 (accession number EF633507) and R7K (accession number AM901564) were obtained. Plasmid R388 (accession number BR000038) was resequenced, and the 50+ sequence differences were submitted to GenBank for amending. The revised R388 sequence was used in all later analyses. In addition, we used the following pSa sequences available in the databases: L06822 (18, 40), AF143206 (10), X00060 (71), X68227 (11), U04277 and U04278 (68, 77), U30471 (16, 57), and M11444 (72). Additionally, we sequenced 5,600 bp of plasmid pIE522, comprising the backbone region oriT-trwA and the integron insertion (accession number EU247928) as well as the stbA-oriT-trwA region of pSa (accession number EU419764).
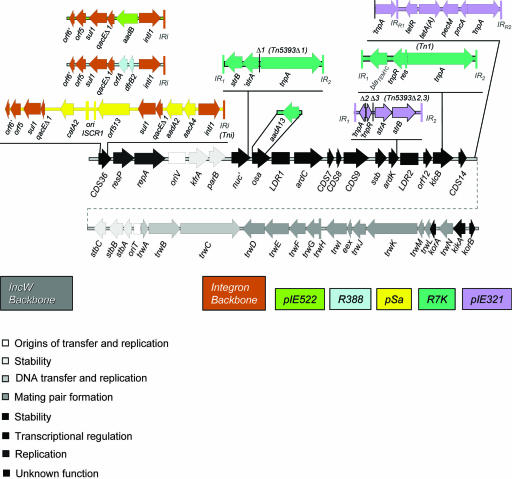
Both the DNA sequences and the previous array data indicate that all five IncW plasmids show extensive genetic conservation. Using the sequence information, the IncW genetic backbone was extrapolated as the genomic sequence in common between its five members and is represented in Fig. 1. The IncW backbone is 29,684 bp long and contains 37 ORFs. Some comparative data among backbones are shown in Table 2. Plasmids R388, pIE321, and R7K diverged significantly (pIE321 and R7K were 97.0 and 97.5% identical to R388, respectively) in comparison to pSa and pIE522 (both had almost 100% identity to R388 in the available backbone sequences). The five IncW plasmids contain one or more indels containing antibiotic resistance genes in an otherwise practically identical backbone (Table 1). They are depicted in Fig. 1. Since all IncW plasmid isolations were carried through biparental matings, based on direct detection of antibiotic resistance markers, IncW cryptic plasmids devoid of transposable elements or integrons have yet to be found.
FIG. 1.
Gene load in the IncW backbone. The insertions of the different MGEs in the IncW backbone that originated the five plasmid isolates are represented. Coding sequences (CDSs) and other sequence features belonging to the IncW backbone are gray, white, or gray-white combinations, depending on their assignment to functional modules. Plasmid-specific genes are represented by the color scheme indicated in the row of rectangles (at right) with inscribed plasmid names. The 5′ and 3′ conserved sequences of the integron platform are brown. When the 5′ or 3′ portion of a given CDS has been deleted, it is indicated by a prime (′) before or after the CDS name, respectively. The deletion sites within transposons are marked by vertical lines.
TABLE 2.
Percent nucleotide identity between five IncW backbonesa
| Plasmid | Overall DNA sequence identity (%)
|
||||
|---|---|---|---|---|---|
| R388 | pIE321 | R7K | pSab | pIE522 | |
| R388 | 100 | 97 (91.2) | 97.5 (87.4) | 99.9 (96.8) | (100) |
| pIE321 | 100 | 97.3 (83.2) | 98 (94.2) | (94) | |
| R7K | 100 | 97.9 (86.8) | (91.7) | ||
| pSab | 100 | (97.8) | |||
| pIE522 | (100) | ||||
The values in parentheses refer to DNA sequence identities (%) in the oriT-trwA region.
For pSa, since the complete DNA sequence is not known, comparisons were made using the region between resP and ardC (8,384 bp).
Sequence divergence in the IncW backbone.
The high degree of sequence conservation and the evidence of recent MGE insertions indicate that IncW is a homogeneous and relatively “young” plasmid cluster. This assertion can be confirmed by using relaxase protein domains as a molecular clock for estimating divergence rates in plasmid backbones, following the seminal work of Francia et al. (30). For instance, IncP plasmids also share an extensive backbone (73). Relaxases of IncP subgroup β are 89% identical. Identity drops to 64% if the relaxases of subgroup α plasmids are considered. Meanwhile, IncW relaxase domains share 100% amino acid identity. The only alterations in the IncW backbone are indels caused by the MGEs described above. No evidence of backbone disruption was observed (see Table S1 in the supplemental material), contrary to what happened in the IncP-β plasmid pB10 (66). It can be assumed that the described IncW plasmids diverged from a common ancestor more recently than IncP plasmids. Perhaps due to their recent origin, IncW plasmids are found less frequently in natural isolates than IncPs (35, 36).
However, <95% identity was observed in four individual genes (see Table S1 in the supplemental material), of which trwA, encoding a relaxosomal protein, shows the highest variation (74 to 87% amino acid identity). These differences explain the lack of PCR and hybridization signals when using trwA probes in several IncW isolates that were positive using oriT and oriV probes (35). trwA sequence divergence indicates that this gene suffered a substitution rate higher than that of other backbone genes. The nonsynonymous-to-synonymous substitution ratio (Ka/Ks) between all pairs of trwA sequences is lower than 1 (R388 versus pIE321, 0.1601; R388 versus R7K, 0.3773; and R7K versus pIE321, 0.2956; calculated with the K-Estimator 6.1 program [http://www.biology.uiowa.edu/comeron/index_files/Page432.htm]) (19, 20). That is, trwA, as well as the remaining IncW genes, is under purifying selection.
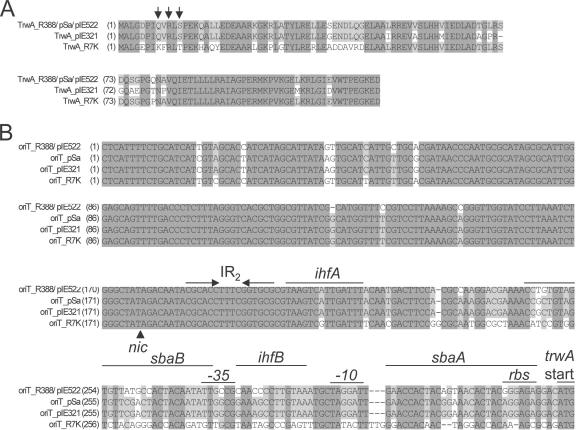
As can be seen in the alignment depicted in Fig. 2A, amino acid differences between TrwAR388 and TrwApIE321 are distributed along the protein lengths. Conversely, TrwAR388 and TrwAR7K exhibit their amino acid differences mainly in the N-terminal part of the protein (Ka = 0.15038 for the 72 N-terminal residues; Ka = 0.00861 for the 48 C-terminal residues). All three TrwA variants retain residue R10, proved to be the most important amino acid for TrwA binding to oriT (51). Interestingly, TrwAR7K shows mutations in two apparently important residues, Q8 and S12, that are involved in DNA recognition (50). Perhaps even more interesting, the oriT region also shows striking differences among these plasmids (Fig. 2B). The sequence recognized by the relaxase domain of TrwC (identical for all these plasmids), comprising the inverted repeat IR2 and the nic site, remains unchanged. Variations fundamentally affect the regions recognized by the proteins integration host factor (IHF) and TrwA (51, 52), as well as the trwA promoter sequence. oriTR7K exhibits the highest variation. IHF binding inhibits nic cleavage catalyzed by TrwC (52), while TrwA binding performs a dual function: stimulation of conjugation by enhancing TrwC nicking and transcriptional repression of the trwABC operon (51). No significant differences were found in conjugation frequencies of these plasmids. It is likely that oriT and TrwA coevolved to conform plasmid-specific interactions. As a result, they could influence nic cleavage and trwABC transcription rates, modeling differences in the conjugation properties in different environments.
FIG. 2.
Alignments of the TrwA and oriT sequences. Alignments of the TrwA (panel A) and oriT (panel B) sequences of R388/pIE522/pSa, pIE321, and R7K are shown. Invariant nucleotides or amino acids in all plasmids are shadowed in dark gray, while those identical in most plasmids are shadowed in light gray. (A) Residues 8, 10, and 12, which are important for oriT recognition by TrwAR388, are indicated by vertical arrows. (B) The oriT DNA strand complementary to that cleaved by the relaxase is shown. The nic site is represented by a black triangle. Structural motif IR2 is represented by convergent horizontal arrows. The sites of interaction of oriTR388 with proteins IHF (ihfA and ihfB) and TrwA (sbaA and sbaB) are marked by horizontal lines. The start codon of gene trwA (trwA start), its putative ribosome binding site (rbs), and the trwA promoter −10 and −35 sequences are indicated.
Natural history of each MGE insertion.
The IncW backbone was loaded with different MGEs in each of the five plasmids (Fig. 1). The integron platform was inserted in R388, pSa, and pIE522. Class II transposons were inserted in R7K (Tn1 and Tn5393Δ1) and pIE321 (Tn1721Δ1 and Tn5393Δ2,3). Interestingly, an integron gene cassette, aadA13, was inserted in a secondary site of plasmid R7K. All insertions occurred outside the conjugation modules. MGEs in IncP plasmids show significant hot spots in oriV-klcA and tra-trb (44, 74). Judging from the representations in Fig. 1 and 3, there are not preferred regions for MGE insertion in the IncW backbone. It may also be an indication of the recent origin of IncW plasmids that all MGE insertions can be explained in one or a few steps. The inserted MGEs themselves are not composite transposons. Figure 1 shows that insertions did not produce extensive reorganizations in the IncW backbone and synteny was maintained.
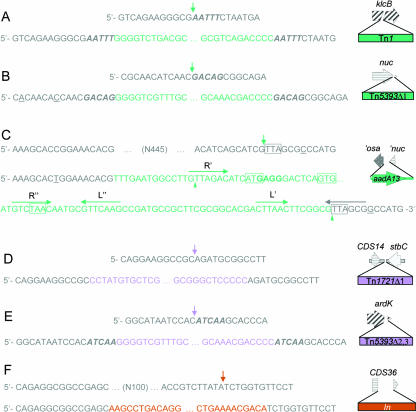
FIG. 3.
Junction sequences of specific insertions. Each insertion is represented at the right by a diagram following the color scheme used in Fig. 1 and is preceded by two DNA sequences. The top one represents the backbone sequence (gray) where the insertion takes place. The exact point of insertion is depicted by a vertical arrow. The bottom sequence contains the corresponding insertion colored according to the same scheme. Target duplications of the backbone sequence are shown in bold italics. Individual base changes with respect to the backbone sequence are underlined. In panel C, two green triangles indicate suggested recombination crossover points flanking the aadA13 integrated cassette. Gene aadA13 start and stop codons as well as the complementary sequence of the nuc stop codon are boxed. A putative ribosome binding site is shown in bold. Core (R′), inverse core (R"), and internal IR (L′ and L") sequences of the potentially recircularized aadA13 cassette are signaled by horizontal green arrows, while the composite core site of the integrated cassette is indicated by a horizontal gray arrow. ′osa, 5′ portion of osa has been deleted; ′nuc, 5′ portion of nuc has been deleted.
Transposon and cassette insertions in plasmid R7K.
In R7K, a 4,949-bp insertion (Fig. 1) disrupts the repressor gene klcB (28). The inserted element is 99.8% identical to Tn1, located in Birmingham IncP-α plasmids RP1/RP4 (accession number L27758) (59), 98.2% to Tn2 (accession numbers X54607 and AY123253) (60), and 98% to Tn3 (accession number V00613). Flanking the inserted element are duplications of the target pentanucleotide AAATT (Fig. 3, panel A), pointing to a clean transposon insertion. Tn1 insertion in plasmid RP4 occurs also within gene klcB, although the insertion site is not identical. Tn1R7K shows perfect 38-bp terminal IRs and retains its three functional genes: tnpA (transposase), tnpR (resolvase), and blaTEM-1c (encoding TEM-1 β-lactamase), as well as the resolution res site. Tn1RP4 and Tn1R7K differ in only 10 nucleotides, resulting in a three-residue change in their TnpAs (A457R, D797G, and D801N) and one residue mutation in TEM proteins (K37Q). Gene blaTEM-1c, as present in R7K, constitutes a new allele, and differs from blaTEM-1a, blaTEM-1b, and blaTEM-2, present in transposons Tn3, Tn2, and Tn1, respectively, by the mutations shown in Table S2 in the supplemental material. Two regions are delimited: one comprising the promoter and the coding region up to position 345, which is identical to that in Tn3, and the other spanning from position 346 to the stop codon, which is identical to that in Tn1. This fact suggests homologous recombination between blaTEM-1a and blaTEM-2, originating blaTEM-1c. The resulting protein, as well as its putative weak promoter, is identical to those encoded by Tn2 and Tn3 (37).
A second transposon, Tn5393Δ1, is inserted in gene nuc, leaving its last 14 codons contiguous to the transposon 3′ end (Fig. 1 and see Fig. S2 in the supplemental material). Tn5393Δ1 is 99% identical to Tn5393 from plasmid pEa34 (accession number M95402) (21) but has an internal deletion (1,982 bp) comprising the resolvase gene, insertion element IS1133, and the first 50 codons of strA. The deletion is clean, without relics of other transposable elements. The transposon is flanked by imperfect IRs (77 nucleotides out of 81) and a direct repeat of GACAG, corresponding to the target duplication at the insertion site (Fig. 3, panel B).
Gene aadA13, coding for aminoglycoside-3′-adenylyltransferase that confers resistance to streptomycin and spectinomycin, is located proximal to Tn5393Δ2 in plasmid R7K (Fig. 1). The presence of 14 codons of the 3′ region of nuc between the insertions of Tn5393Δ1 and aadA13 demonstrates that both elements inserted independently (see Fig. S2 in the supplemental material). Protein AadA13R7K shows 98% identity to reported versions of AadA13 (accession numbers AAV49321, AAY18576, ABG76948, ABG76949, and BAF73713). The AadA family alignment (from AadA1 to AadA15), which contains the N-terminal nucleotidyltransferase domain, shows 33.5% consensus identity. AadA1 is most related to AadA13 (85.3% identity), while AadA14 is the least similar (54.3%).
The aadA13 insertion looks like an integron gene cassette (855 bp long), containing a 3′ attC element as well as a recombination site (G/TTAGAC) upstream of the aadA13 start codon (Fig. 3, panel C). It lacks attI or any other integron component. The attC sequence of the hypothetical circularized cassette (60 bp long) is 100% identical to previously reported aadA13-related attC sequences (accession numbers AY940492, DQ779001, DQ779002, and AB332415) (43). All these homologues are contained in class I integrons. Thus, we assume that the aadA13 cassette integrated in an IntI1 secondary site (G/TTAGCG; the consensus being GWTMW [29]) located in the complementary strand of gene nuc, just downstream of its stop codon (see Fig. 3, panel C). Since the aadB cassette was shown to excise from a secondary site with a completely conserved GTT triplet (67), the composite core site of cassette aadA13 may also be active in IntI1-mediated recombination.
The integration of cassette aadA13 produced a deletion in the osa gene, leaving 114 bp of its 3′ region. Interestingly, this truncated segment shows only 83% identity to the functional osaR388, perhaps underscoring the rapid shift of nonfunctional sequences (see Fig. S2 in the supplemental material). Osa impairs transport of Vir proteins (46) and DNA transfer (14) to plant cells. The repercussion of osa deletion in the ecology of plasmid R7K remains to be investigated.
Gene cassettes do not contain their own promoters and thus are transcribed from the Pant promoter when inserted in the integron, or from a suitable oriented promoter when inserted in a secondary site (64). For example, cassette aadB was integrated in a secondary site of plasmid pRAY and is transcribed from a plasmid promoter (67). In R7K, aadA13 is expressed since the host bacteria are resistant to spectinomycin, a phenotype that is not attributable to the str genes present in transposon Tn5393Δ1. A putative promoter is located upstream of aadA13 in the ′osa region (detected using the bacterial promoter prediction BPROM program at http://www.softberry.com) (see Fig. S2B in the supplemental material). Finally, a 13-bp string of DNA appears between cassette aadA13 and ′osa that is neither part of the gene cassette nor part of the IncW backbone.
Insertions in plasmid pIE321.
Plasmid pIE321 contains insertions of truncated class II transposons Tn1721 and Tn5393c (Fig. 1). The 6,295-bp-long Tn1721Δ1 element is inserted between coding sequence 14 (CDS14) and stbC. Compared to the canonical Tn1721 (11,139 bp long) (accession number X61367) (3), this derivative shows a 4,844-bp deletion that includes the left terminal IR, the gene orfI, the resolution site res, the resolvase gene tnpR, and part of the transposase gene tnpA. No DNA target duplication was observed (Fig. 3, panel D). The transposon keeps the last 256 codons of tnpA, tetR (encoding a tetracycline resistance repressor), tetA(A) (encoding a tetracycline efflux protein that confers resistance to tetracycline), pecM (related to a permease repressor), pncA (a cystein-hydrolase), the second truncated copy of tnpA (encoding the 583 C-terminal residues), and the right IR (IRR2), which appears duplicated in the middle of the element (IRR1), as happens in the original transposon. A simple one-ended transposition event explains the Tn1721Δ1 insertion. Single-ended derivatives of Tn3-like transposons Tn1721, Tn21, and TnA were found to cleanly insert into a recipient plasmid (7, 42, 53). Insertion products contain the entire donor plasmid plus a duplication of the IR (7, 42, 53, 54), or only variable-length portions of the transposable derivative (8, 42, 53). So, transposition termination at a non-IR sequence can yield a simple insertion of a truncated transposon without duplication of the target sequence, like Tn1721Δ1 in plasmid pIE321. This is not an isolated example. A single-end Tn1721 derivative found in the IncP plasmid pB10 (accession number AJ564903) has almost the same truncation (66). Given the fact that single-ended TnpA− Tn21 and TnA derivatives transpose when the cognate transposase is provided in trans (7, 42), the Tn1721Δ1 copy in pIE321 could potentially be mobilized.
The 2,148-bp Tn5393Δ2,3 derivative is inserted between ardK and LDR1 (Fig. 1), duplicating the target sequence ATCAA (Fig. 3, panel E). The element retains the Tn5393 81-bp terminal IRs and genes strA and strB encoding streptomycin resistance (aminoglycoside-3′-phosphotransferase and aminoglycoside-6-phosphotransferase) but has two internal deletions. The first (Δ2; 2,316 bp) includes the N-terminal portions of the transposase (tnpA and tnpR) and resolvase genes, leaving their last 77 and 37 codons, respectively. The second (Δ3; 1,233 bp) comprises IS1133. The three remaining portions, IR-′tnpA, ′tnpR, and strA-strB-IR, are juxtaposed, as happens in the R7K element, and are 100%, 98%, and 100% identical, respectively, to Tn5393 (accession number M95402) (15). Blast analysis of Tn5393Δ2,3 showed 100% nucleotide identity and coverage with an element contained in plasmid pMBSF1, which has exactly the same deletions (accession number AJ518835). This plasmid was obtained from E. coli isolated from pigs and, as with pIE321, also contains a truncated Tn1721 copy but inserted in a different location (12).
Insertion of the integron platform in plasmids R388, pSa, and pIE522.
Complete class I integrons are present in R388, pSa, and pIE522 at exactly the same backbone location (Fig. 3, panel F). They contain the same 5′ and 3′ conserved regions. In all three cases, the integron disrupts orphan gene CDS36, present in integron-free plasmids R7K and pIE321. As a result of the integron insertion, an internal 112-nucleotide deletion was produced in CDS36, leaving 22 codons 5′ and 77 codons 3′ of the inserted integron. Thus, we assume the integron inserted just once in an ancestral IncW backbone, as proposed by Gorai et al. (34) and Stokes et al. (68), as will be discussed later.
Upon integration, gene CDS36 is split in such a way that an in-frame GTG start codon near the integron border gives rise to a chimeric gene in plasmids containing the integron platform. We annotated it as tnpM in R388 (28) by analogy with tnpM of Tn21 (also located after intI1, as shown previously [47]). However, there is practically no similarity with the Tn21 gene, so it should not be called tnpM, to avoid confusion. The DNA sequence that CDS36 and tnpM have in common is responsible for hybridization of the R388 tnpM probe to plasmids R7K and pIE321 in the DNA array (see Fig. S1 in the supplemental material).
The IncW integron platform does not retain the transposition functions responsible for integron movement but conserves the terminal sequence IRi (25 bp long; close to the 3′ end of intI1) of Tni, the vehicle transposon present in Tn21 (47). The integron 3′ conserved region, besides the common qacEΔ1 gene and sul1, the sulfonamide resistance gene, harbors orf5 and a truncated orphan gene, orf6 (coding for its first 57 residues). The integron variable regions contain different gene cassettes. In R388, cassette dhfr codes for dihydrofolate reductase that confers resistance to trimethoprim (70, 83), while cassette orfA (69, 70) has no assigned phenotype. In pIE522, cassette aadB encodes 2-aminoglycoside (2′) adenylyltransferase and confers resistance to gentamicin/kanamycin/tobramycin. In pSa, the inserted integron contains two gene cassettes, aacA4 and aadA2, which confer resistance to gentamicin/kanamycin and streptomycin/spectinomycin, respectively (39, 72).
The existence of different gene cassettes as well as the high conservation of the integrase with that of Tn21 (they differ only in residue 39: His in IntTn21 and Asn in IntIncW plasmids) suggests that the IncW integrase is active in the genesis of new IncW plasmids. According to theory, the first acquired cassette is located in the last position of the integron variable region. Consistent with this, the first inserted cassette in each integron contains the same sequence (GTTAGAT) in its attC core site, derived from the primary attI recombination site in the empty integron.
In the integron version of plasmid pSa, the 3′ conserved genes qacEΔ1 and sul1 have been duplicated (68). In between, there is a chloramphenicol resistance gene, catA2, which does not correspond to a cassette insertion, followed by an ISCR1 element. This insertion sequence contains orf513, a gene found close to many integrons, which encodes a protein related to the IS91 transposase (32, 76). Three copies of the putative 5′ end of the ISCR1 element oriISCR1 appear downstream of orf513. The pSa integron distal 3′ conserved sequence is identical to that in R388. Based on the proposed three-step mechanism (75, 76), a possible scenario for the formation of the pSa integron includes rolling-circle transposition of ISCR1 fused to the 3′ conserved region of a class I integron (containing orf513, sul1, and qacEΔ1) to the vicinity of gene catA2. Subsequent one-ended rolling-circle transposition of this element, now containing catA2, gives rise to circular intermediates that can recombine with the 3′ conserved region of a simple integron platform, like that present in R388.
Concluding remarks.
Several horizontal gene transfer events shaped currently known IncW plasmids. The most parsimonious assumption is to consider that the integron platform was inserted only once in an ancestral IncW plasmid, since its genetic location is exactly the same for the three IncW plasmids harboring it. Different gene cassettes were then incorporated into the integron by integrase-catalyzed reactions, producing the present versions in R388, pIE522, and pSa. A subsequent acquisition of an ISCR1 element in the pSa integron occurred. The diversifications in the integron arrangements must be very recent, considering the 100% nucleotide identity in the integron platform of the three plasmids. Independent events of MGEs loaded in the ancestral plasmid gave rise to the IncW variants pIE321 and R7K. Divergence of those plasmids is older than that of R388/pIE522/pSa, since pIE321 and R7K exhibit small but significant differences in the backbone (see Table S1 in the supplemental material). The IncW group thus provides an example of the recent evolution of a plasmid backbone which acquired different MGEs that were selected by the heavy use of antibiotics.
Supplementary Material
Acknowledgments
Work in the FdlC lab was supported by grant BFU2005-03477/BMC from the Spanish Ministry of Education and Science and contract LSHM-CT-2005_019023 from the E.U. VI Framework Programme. The help of the core sequencing group at the PSU and The Wellcome Trust (grant 063083) are also acknowledged.
We are grateful to K. Smalla for providing plasmids pIE522 and pIE321.
Footnotes
Published ahead of print on 11 February 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Abeles, A. L., K. M. Snyder, and D. K. Chattoraj. 1984. P1 plasmid replication: replicon structure. J. Mol. Biol. 173:307-324. [DOI] [PubMed] [Google Scholar]
- 2.Adamczyk, M., and G. Jagura-Burdzy. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50:425-453. [PubMed] [Google Scholar]
- 3.Allmeier, H., B. Cresnar, M. Greck, and R. Schmitt. 1992. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111:11-20. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin, S., and K. Nordstrom. 1990. Partition-mediated incompatibility of bacterial plasmids. Cell 60:351-354. [DOI] [PubMed] [Google Scholar]
- 6.Avila, P., and F. de la Cruz. 1988. Physical and genetic map of the IncW plasmid R388. Plasmid 20:155-157. [DOI] [PubMed] [Google Scholar]
- 7.Avila, P., F. de la Cruz, E. Ward, and J. Grinsted. 1984. Plasmids containing one inverted repeat of Tn21 can fuse with other plasmids in the presence of Tn21 transposase. Mol. Gen. Genet. 195:288-293. [DOI] [PubMed] [Google Scholar]
- 8.Avila, P., J. Grinsted, and F. de la Cruz. 1988. Analysis of the variable endpoints generated by one-ended transposition of Tn21. J. Bacteriol. 170:1350-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahl, M. I., L. H. Hansen, and S. J. Sorensen. 2007. Impact of conjugal transfer on the stability of IncP-1 plasmid pKJK5 in bacterial populations. FEMS Microbiol. Lett. 266:250-256. [DOI] [PubMed] [Google Scholar]
- 10.Belogurov, A. A., E. P. Delver, O. V. Agafonova, N. G. Belogurova, L. Y. Lee, and C. I. Kado. 2000. Antirestriction protein Ard (type C) encoded by IncW plasmid pSa has a high similarity to the “protein transport” domain of TraC1 primase of promiscuous plasmid RP4. J. Mol. Biol. 296:969-977. [DOI] [PubMed] [Google Scholar]
- 11.Bito, A., and M. Susani. 1994. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob. Agents Chemother. 38:1172-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blickwede, M., and S. Schwarz. 2004. Molecular analysis of florfenicol-resistant Escherichia coli isolates from pigs. J. Antimicrob. Chemother. 53:58-64. [DOI] [PubMed] [Google Scholar]
- 13.Boyd, E. F., C. W. Hill, S. M. Rich, and D. L. Hartl. 1996. Mosaic structure of plasmids from natural populations of Escherichia coli. Genetics 143:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cascales, E., K. Atmakuri, Z. Liu, A. N. Binns, and P. J. Christie. 2005. Agrobacterium tumefaciens oncogenic suppressors inhibit T-DNA and VirE2 protein substrate binding to the VirD4 coupling protein. Mol. Microbiol. 58:565-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiou, C. S., and A. L. Jones. 1993. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J. Bacteriol. 175:732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Close, S. M., and C. I. Kado. 1991. The osa gene of pSa encodes a 21.1-kilodalton protein that suppresses Agrobacterium tumefaciens oncogenicity. J. Bacteriol. 173:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coetzee, J. N., N. Datta, and R. W. Hedges. 1972. R factors from Proteus rettgeri. J. Gen. Microbiol. 72:543-552. [DOI] [PubMed] [Google Scholar]
- 18.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comeron, J. M. 1999. K-estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15:763-764. [DOI] [PubMed] [Google Scholar]
- 20.Comeron, J. M. 1995. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J. Mol. Evol. 41:1152-1159. [DOI] [PubMed] [Google Scholar]
- 21.Couturier, M., F. Bex, P. L. Bergquist, and W. K. Maas. 1988. Identification and classification of bacterial plasmids. Microbiol. Rev. 52:375-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta, N., and R. W. Hedges. 1972. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J. Gen. Microbiol. 72:349-355. [DOI] [PubMed] [Google Scholar]
- 23.de la Cruz, F., and J. Davies. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8:128-133. [DOI] [PubMed] [Google Scholar]
- 24.de la Cruz, F., and J. Grinsted. 1982. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J. Bacteriol. 151:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.del Solar, G., and M. Espinosa. 1992. The copy number of plasmid pLS1 is regulated by two trans-acting plasmid products: the antisense RNA II and the repressor protein, RepA. Mol. Microbiol. 6:83-94. [DOI] [PubMed] [Google Scholar]
- 26.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebersbach, G., D. J. Sherratt, and K. Gerdes. 2005. Partition-associated incompatibility caused by random assortment of pure plasmid clusters. Mol. Microbiol. 56:1430-1440. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Lopez, R., M. P. Garcillan-Barcia, C. Revilla, M. Lazaro, L. Vielva, and F. de la Cruz. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30:942-966. [DOI] [PubMed] [Google Scholar]
- 29.Francia, M. V., F. de la Cruz, and J. M. G. Lobo. 1993. Secondary sites for integration mediated by the Tn21 integrase. Mol. Microbiol. 10:823-828. [DOI] [PubMed] [Google Scholar]
- 30.Francia, M. V., A. Varsaki, M. P. Garcillan-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 31.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 32.Garcillán-Barcia, M. P., I. Bernales, M. V. Mendiola, and F. de la Cruz. 2002. IS91 rolling-circle transposition, p. 891-904. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 33.Gilmour, M. W., N. R. Thomson, M. Sanders, J. Parkhill, and D. E. Taylor. 2004. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52:182-202. [DOI] [PubMed] [Google Scholar]
- 34.Gorai, A. P., F. Heffron, S. Falkow, R. W. Hedges, and N. Datta. 1979. Electron-microscope heteroduplex studies of sequence relationships among plasmids of the W-incompatibility group. Plasmid 2:485-492. [DOI] [PubMed] [Google Scholar]
- 35.Gotz, A., R. Pukall, E. Smit, E. Tietze, R. Prager, H. Tschape, J. D. van Elsas, and K. Smalla. 1996. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol. 62:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gotz, A., and K. Smalla. 1997. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl. Environ. Microbiol. 63:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goussard, S., and P. Courvalin. 1991. Sequence of the genes blaT-1B and blaT-2. Gene 102:71-73. [DOI] [PubMed] [Google Scholar]
- 38.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 40.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriology 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedges, R. W., and N. Datta. 1971. Fi-R factors giving chloramphenicol resistance. Nature 234:220-221. [Google Scholar]
- 42.Heritage, J., and P. M. Bennett. 1985. Plasmid fusions mediated by one end of TnA. J. Gen. Microbiol. 131:1131-1140. [DOI] [PubMed] [Google Scholar]
- 43.Heuer, H., and K. Smalla. 2007. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9:657-666. [DOI] [PubMed] [Google Scholar]
- 44.Heuer, H., R. Szczepanowski, S. Schneiker, A. Puhler, E. M. Top, and A. Schluter. 2004. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1β group without any accessory genes. Microbiology 150:3591-3599. [DOI] [PubMed] [Google Scholar]
- 45.Ireland, C. R. 1983. Detailed restriction enzyme map of crown-gall suppressive IncW plasmid pSa, showing ends of deletion causing chloramphenicol sensitivity. J. Bacteriol. 155:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee, L. Y., S. B. Gelvin, and C. I. Kado. 1999. pSa causes oncogenic suppression of Agrobacterium by inhibiting VirE2 protein export. J. Bacteriol. 181:186-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llosa, M., S. Bolland, and F. de la Cruz. 1991. Structural and functional analysis of the origin of conjugal transfer of the broad-host-range IncW plasmid R388 and comparison with the related IncN plasmid R46. Mol. Gen. Genet. 226:473-483. [DOI] [PubMed] [Google Scholar]
- 49.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moncalian, G., and F. de la Cruz. 2004. DNA binding properties of protein TrwA, a possible structural variant of the Arc repressor superfamily. Biochim. Biophys. Acta 1701:15-23. [DOI] [PubMed] [Google Scholar]
- 51.Moncalian, G., G. Grandoso, M. Llosa, and F. de la Cruz. 1997. OriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J. Mol. Biol. 270:188-200. [DOI] [PubMed] [Google Scholar]
- 52.Moncalian, G., M. Valle, J. M. Valpuesta, and F. de la Cruz. 1999. IHF protein inhibits cleavage but not assembly of plasmid R388 relaxosomes. Mol. Microbiol. 31:1643-1652. [DOI] [PubMed] [Google Scholar]
- 53.Motsch, S., and R. Schmitt. 1984. Replicon fusion mediated by a single-ended derivative of transposon Tn1721. Mol. Gen. Genet. 195:281-287. [DOI] [PubMed] [Google Scholar]
- 54.Motsch, S., R. Schmitt, P. Avila, F. de la Cruz, E. Ward, and J. Grinsted. 1985. Junction sequences generated by one-ended transposition. Nucleic Acids Res. 13:3335-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novick, R. P. 1987. Plasmid incompatibility. Microbiol. Rev. 51:381-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ochman, H. 2001. Lateral and oblique gene transfer. Curr. Opin. Genet. Dev. 11:616-619. [DOI] [PubMed] [Google Scholar]
- 57.Okumura, M. S., and C. I. Kado. 1992. A gene near the plasmid pSa origin of replication encodes a nuclease. Mol. Microbiol. 6:521-527. [DOI] [PubMed] [Google Scholar]
- 58.Osborn, A. M., F. M. D. Tatley, L. M. Steyn, R. W. Pickup, and J. R. Saunders. 2000. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 146:2267-2275. [DOI] [PubMed] [Google Scholar]
- 59.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP-alpha plasmids: compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 60.Partridge, S. R., and R. M. Hall. 2005. Evolution of transposons containing blaTEM genes. Antimicrob. Agents Chemother. 49:1267-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 62.Rawlings, D. E. 2005. The evolution of pTF-FC2 and pTC-F14, two related plasmids of the IncQ-family. Plasmid 53:137-147. [DOI] [PubMed] [Google Scholar]
- 63.Rawlings, D. E., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 65.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 66.Schluter, A., H. Heuer, R. Szczepanowski, L. J. Forney, C. M. Thomas, A. Puhler, and E. M. Top. 2003. The 64 508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology 149:3139-3153. [DOI] [PubMed] [Google Scholar]
- 67.Segal, H., M. V. Francia, J. M. G. Lobo, and G. Elisha. 1999. Reconstruction of an active integron recombination site after integration of a gene cassette at a secondary site. Antimicrob. Agents Chemother. 43:2538-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 69.Sundstrom, L., P. Radstrom, G. Swedberg, and O. Skold. 1988. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol. Gen. Genet. 213:191-201. [DOI] [PubMed] [Google Scholar]
- 70.Swift, G., B. J. McCarthy, and F. Heffron. 1981. DNA sequence of a plasmid-encoded dihydrofolate reductase. Mol. Gen. Genet. 181:441-447. [DOI] [PubMed] [Google Scholar]
- 71.Tait, R. C., T. J. Close, R. L. Rodriguez, and C. I. Kado. 1982. Isolation of the origin of replication of the IncW-group plasmid pSa. Gene 20:39-49. [DOI] [PubMed] [Google Scholar]
- 72.Tait, R. C., H. Rempel, R. L. Rodriguez, and C. I. Kado. 1985. The aminoglycoside-resistance operon of the plasmid pSa: nucleotide sequence of the streptomycin-spectinomycin resistance gene. Gene 36:97-104. [DOI] [PubMed] [Google Scholar]
- 73.Thomas, C. M. 2000. Paradigms of plasmid organization. Mol. Microbiol. 37:485-491. [DOI] [PubMed] [Google Scholar]
- 74.Thorsted, P. A., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. N. Thomas. 1998. Complete sequence of the IncP beta plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 75.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. Common regions e.g., orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 58:1-6. [DOI] [PubMed] [Google Scholar]
- 76.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valentine, C. R., M. J. Heinrich, S. L. Chissoe, and B. A. Roe. 1994. DNA sequence of direct repeats of the sulI gene of plasmid pSa. Plasmid 32:222-227. [DOI] [PubMed] [Google Scholar]
- 78.Valentine, C. R. I. 1985. One-kilobase direct repeats of plasmid pSa. Plasmid 14:167-170. [DOI] [PubMed] [Google Scholar]
- 79.Valentine, C. R. I., and C. I. Kado. 1989. Molecular genetics of IncW plasmids, p. 125-163. In C. M. Thomas (ed.), Promiscuous plasmids of Gram-negative bacteria. Academic Press Inc., San Diego, CA.
- 80.Ward, J. M., and J. Grinsted. 1982. Physical and genetic analysis of the IncW group plasmids R388, Sa, and R7K. Plasmid 7:239-250. [DOI] [PubMed] [Google Scholar]
- 81.Watanabe, T., C. Furuse, and S. Sakaizum. 1968. Transduction of various R factors by phage P1 in Escherichia coli and by phage P22 in Salmonella typhimurium. J. Bacteriol. 96:1791-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weigel, C., and H. Seitz. 2006. Bacteriophage replication modules. FEMS Microbiol. Rev. 30:321-381. [DOI] [PubMed] [Google Scholar]
- 83.Zolg, J. W., and U. J. Hanggi. 1981. Characterization of a R plasmid-associated, trimethoprim-resistant dihydrofolate reductase and determination of the nucleotide sequence of the reductase gene. Nucleic Acids Res. 9:697-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.