Abstract
Summary: Nearly 2,000 ribotyping-based studies exist, ranging from epidemiology to phylogeny and taxonomy. None precisely reveals the molecular genetic basis, with many incorrectly attributing detected polymorphisms to rRNA gene sequences. Based on in silico genomics, we demonstrate that ribotype polymorphisms result from sequence variability in neutral housekeeping genes flanking rRNA operons, with rRNA gene sequences serving solely as conserved, flank-linked tags. We also reveal that from such an informatics perspective, it is readily feasible a priori to design an interpretable ribotyping scheme for a genomically sequenced microbial species, and we discuss limitations to the basic restriction fragment length polymorphism-based method as well as alternate PCR ribotyping-based schemes.
INTRODUCTION
Nearly 2,000 publications involving ribotyping have followed the initial description of the basic method in 1986 (25). The more than 200 microbial genera so analyzed range from fungi (19, 52, 53, 84, 92) to gram-positive cocci (13, 18, 66, 82) and bacilli (36, 38, 55, 57, 63, 71) and more than 50 gram-negative bacterial genera, with particular focus on human commensal and/or disease-causing species.
Rationales for the application of ribotype-based differentiation of independent isolates within a species have included taxonomic classification (25, 46), epidemiological tracking (32, 48, 78, 81), geographical distribution (14, 32, 81), and population biology and phylogeny (1, 32, 62, 81). Such wide interest has led to the development of variant schemes from that initially described. While the molecular genetic basis of conventional ribotyping is the focus of this review, variant approaches will also be addressed.
The name “ribotyping” has inadvertently proven to be a misnomer, leading to a misconception that observed polymorphisms arise directly from rRNA gene sequences. We reveal, based on in silico genomic analyses, that resolved DNA polymorphisms rather reflect restriction fragment length polymorphisms (RFLPs) of the neutrally evolving housekeeping genes typically found to flank chromosomal rRNA gene sequences. We also demonstrate that with this fundamental insight into the molecular genetic basis of ribotype polymorphisms, it is now feasible a priori, in the age of genomics, to rationally design a ribotyping scheme in silico, consequently allowing for interpretation of RFLPs based on evolution of polymorphic sites found within housekeeping genes flanking the ribosomal operons.
UNDERSTANDING THE MOLECULAR GENETIC BASIS OF RIBOTYPING
Typically, each ribosomal operon consists of the three genes encoding the structural rRNA molecules, 16S, 23S, and 5S, cotranscribed as a polycistronic operon. Among bacterial species, the average lengths of the structural rRNA genes are 1,522 bp, 2,971 bp, and 120 bp, for 16S, 23S, and 5S, respectively (R. R. Gutell, presented at The Origin and Evolution of Prokaryotic and Eukaryotic Cells, Shimoda, Japan, 1992). The copy numbers, overall ribosomal operon sizes, nucleotide sequences, and secondary structures of the three rRNA genes are highly conserved within a bacterial species (49) due to their fundamental role in polypeptide synthesis (89). Because the 16S rRNA gene is the most conserved of the three rRNA genes, 16S rRNA gene sequencing has been established as the “gold standard” for identification and taxonomic classification of bacterial species (41, 61, 90). Knowledge of intraspecies conservation of the 16S rRNA gene sequence (21) and basic 16S-23S-5S ribosomal operon structure (17) led Grimont and Grimont (25) to the first insights into its usefulness in developing ribotyping for bacterial classification.
Based on these fundamental insights, we sought to further elucidate the molecular genetic basis of conventional ribotyping. As depicted in Fig. 1, conventional ribotyping is based on restriction endonuclease cleavage of total genomic DNA followed by electrophoretic separation, Southern blot transfer (75), and hybridization of transferred DNA fragments with a radiolabeled ribosomal operon probe. Following autoradiography, only those bands containing a portion of the ribosomal operon are visualized. The number of fragments generated by ribotyping is a reflection of the multiplicity of rRNA operons present in a bacterial species. Copy numbers of rRNA operons have been found to range from 1 (e.g., for Chlamydia trachomatis) to 15 (e.g., for Photobacterium profundum) (40; http://www.ncbi.nlm.nih.gov:80/genomes/static/eub_g.html) (Fig. 2).
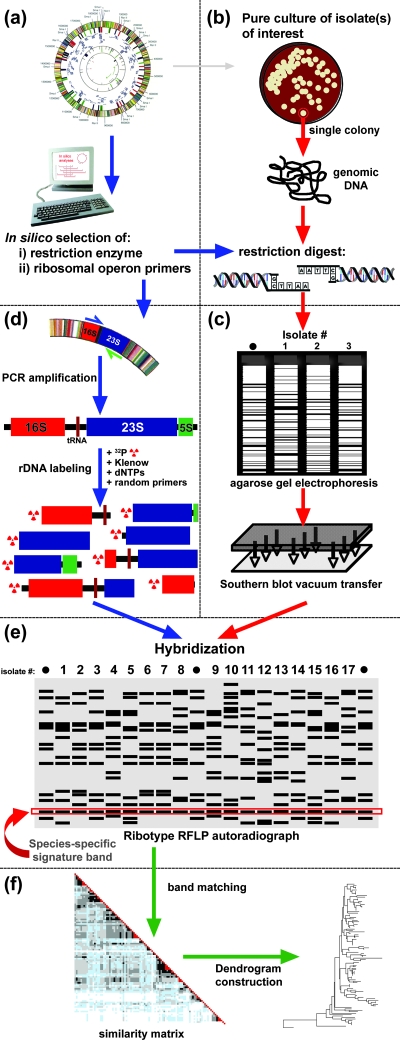
FIG. 1.
Ribotyping methodology. (a) The genomic sequence(s) of species of interest is used to identify, in silico, an appropriate restriction enzyme for ribotyping, ideally one cutting once within the 16S rRNA gene and once within the 23S rRNA gene. The ribosomal operon sequence may also be used for design of primers to amplify a species-specific ribosomal probe for later steps of the protocol. (b) A single colony of the strain of interest is grown in liquid medium, genomically extracted, and digested with the selected restriction enzyme. (c) Restricted genomic DNA, including a genomically sequenced reference isolate as a size standard (•), is electrophoresed through a 0.8% agarose gel and then transferred by Southern blot (75) vacuum transfer to a nylon membrane. (d) Primers designed during initial in silico analyses are used to amplify the entire 16S-23S-5S ribosomal operon. The amplified product is run through a preparative low-melting-point agarose gel for size confirmation and cut directly from the agarose gel. The agarose-embedded product is boiled to solubilize the DNA fragments. Twenty to 50 nanograms of ribosomal template is then used to generate a radiolabeled ([α-32P]dCTP) probe with DNA polymerase I large (Klenow) fragment and random primers. (e) The transferred membrane containing genomic DNA digests is hybridized with the radiolabeled ribosomal operon probe and exposed to autoradiographic film. Ribotype RFLP bands are analyzed manually or with the aid of appropriate fingerprint analysis software. (f) Fingerprint analysis software is applied for band identification, normalization, and matching of band positions across strains both within the same autorad and between multiple autorads. The final output includes a similarity matrix defining, in this case by color shading, the percentage of bands shared among each strain in the collection and a dendrogram or some other pictorial representation of the relatedness of isolates within a collection.
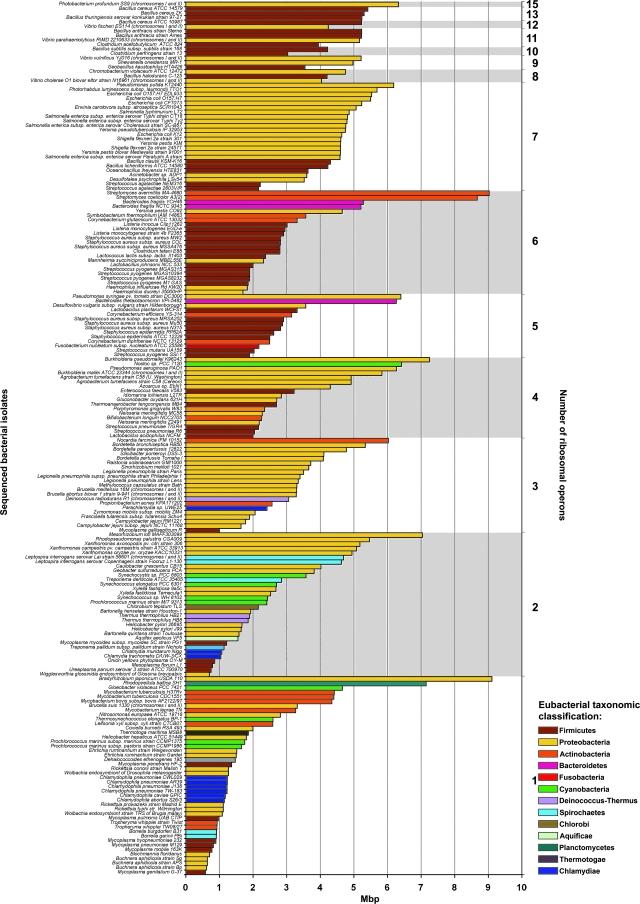
FIG. 2.
Genome sizes and ribosomal operon copy numbers of 190 genomically sequenced bacterial strains. Genomically sequenced bacterial isolates are categorized by number of ribosomal operons (right y axis). Within each ribosomal number category, bacterial isolates are organized by decreasing genome size, in Mbp (x axis), demonstrating a lack of concordance between number of ribosomal operons and genome size. Eubacterial genomes are color coded according to their taxonomic classification (phyla) as assigned in the NCBI genome database and noted in the color key.
We reasoned that use of a single restriction endonuclease with a conserved cleavage site in the 16S and 23S rRNA genes would enable detection of DNA sequence polymorphisms in immediately adjacent upstream and downstream genes flanking each ribosomal operon following hybridization of the electrophoresed chromosomal digest with a labeled rRNA gene operon probe. It then follows that the total number of RFLP bands so detected would be equal to twice the number of ribosomal operons, with an additional fragment(s) reflecting the 16S-23S internal spacer (Fig. 3).
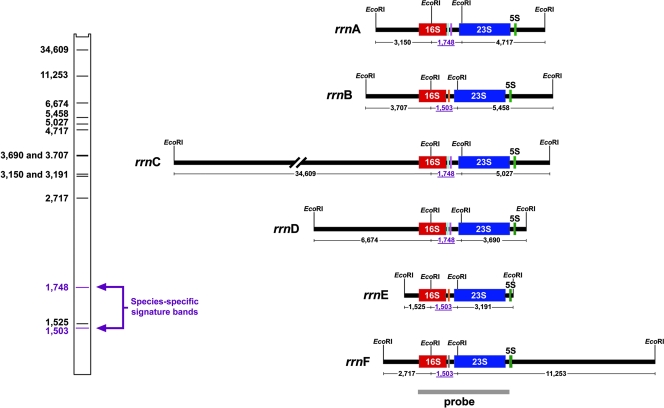
FIG. 3.
Conservation of rRNA operon sequences and variability of chromosomal flank sequences are responsible for ribotyping polymorphisms, as revealed by in silico analysis (Gene Construction Kit; Textco BioSoftware, Inc.) of the six ribosomal operons of the genomically sequenced H. influenzae strain Rd. Evolutionarily diverse isolates within a bacterial species display ribotype fragments of differing sizes comprising either the 5′ end of the 16S rRNA gene and neighboring flank DNA or the 3′ end of 23S-5S rRNA genes and neighboring flank sequence. The six ribosomal operons of strain Rd, labeled A through F, are depicted, along with EcoRI restriction sites within the rrn genes and the first flanking EcoRI site adjacent to each ribosomal operon. Based on the sizes of EcoRI fragments generated, the bar code at the left depicts the predicted EcoRI ribotype profile for strain Rd. Twelve of the 14 bands in the profile result from polymorphisms in the chromosomal flanks of the six rrn operons; the remaining two bands are the species-specific signature bands, comprising the ISR between the 16S and 23S rRNA genes and size dependent on the presence of one or two tRNA sequences. Species-specific signature band sizes are depicted beneath each operon genetic map in mauve, underlined numerals. Ribosomal operons A, C, and D contain two tRNAs (tRNAIle shown in cyan and tRNAAla shown in mauve). Ribosomal operons B, E, and F contain one tRNA (tRNAGlu shown in orange). The gray bar beneath the linear maps of the ribosomal operons depicts the location of the probe used to identify rRNA gene-containing fragments.
Well before the first genomic sequence became available in 1995, the linear Escherichia coli linkage map (5) revealed that chromosomal genes immediately flanking (∼50 kb) the seven ribosomal operons of this species consist primarily of evolutionarily neutral (housekeeping) genes (see “In Silico and Experimental Resolution of Variable Ribosomal Operon Flanks” below), the same category of genes used for phylogenic analysis by multilocus enzyme electrophoresis (MLEE) (68) and more recently by multilocus sequence typing (MLST) (50). This perspective therefore suggested to us that RFLPs generated by ribotyping could be used to characterize the evolutionary genetic relatedness of independent isolates (8, 9, 16, 31, 32, 81) as well as for simple fingerprinting to differentiate independently isolated nonclonal bacterial strains within a species (4, 24, 31, 32, 78).
IN SILICO-BASED EXPERIMENTAL DESIGN AND INTERPRETATION USING HAEMOPHILUS INFLUENZAE AS THE PROTOTYPE
Knowledge of 16S rRNA sequence conservation (21) and the presence of neutral genes flanking the seven ribosomal operons of Escherichia coli (5) allowed us, in 1990, to develop a hypothetical model of the molecular genetic basis of ribotype RFLPs. However, our current, detailed understanding began some 5 years later with the in silico analysis of the first available bacterial genomic sequence in 1995, that of H. influenzae strain Rd (20), containing six ribosomal operons. In silico analysis of H. influenzae Rd ribosomal flanking sequences confirmed the presence of neutral genes as being responsible for the ribotyping polymorphisms, while also providing the tool necessary for rational design of a ribotyping protocol, as described below.
In Silico Resolution of Conserved Internal rRNA Gene Cleavage Sites
Our initial step in designing a ribotype protocol for H. influenzae involved an in silico survey of the genomic sequence of strain Rd (20) to search for conserved restriction endonuclease cleavage sites within the six ribosomal operons. The ideal restriction enzyme would cut once within the 16S rRNA gene and once within the 23S rRNA gene, so as to create an internal species-specific fragment (Fig. 1e and 3; also see “In Silico and Experimental Resolution of Variable Ribosomal Operon Flanks” below). Other parameters considered for selection of an appropriate restriction enzyme included length, GC content, and specificity of the recognition site, all of which contribute to the sensitivity of ribotyping.
Candidate restriction cleavage sites fulfilling the above criteria were then scanned for in all publicly available 16S and 23S H. influenzae rRNA gene sequences in addition to those of closely related species to confirm conservation of the restriction site. This in silico-based rational selection allows for a priori choice of restriction sites likely to be conserved among all members of the species, a prerequisite for designing a reliable and readily interpretable species-wide ribotyping scheme (Fig. 3). Following restriction enzyme selection, genomic DNAs of isolates to be ribotyped are digested, separated by electrophoresis, transferred onto a nylon membrane by Southern blotting, and hybridized to a labeled probe consisting of ribosomal operon sequence, enabling resolution of ribotype RFLP patterns (Fig. 1). Of note here, an appropriate choice of the ribosomal operon probe sequence is essential for efficient, specific hybridization. Early ribotype studies (80) used purified 16S and 23S rRNAs from E. coli as probe substrates. Subsequently, pKK3535, a recombinant plasmid consisting of the cloning vector pBR322 and the ribosomal operon B rRNA gene from E. coli K-12 (10), was adopted for this purpose (4). However, it has been our experience that species phylogenically distant from E. coli (e.g., Neisseria meningitidis, Streptococcus pneumoniae, Pseudomonas aeruginosa, and Burkholderia cepacia) will hybridize inefficiently to this ribosomal E. coli probe due to low sequence homology. We eliminate this hybridization problem by using as our labeled probe PCR-amplified rRNA genes of the species being ribotyped (Fig. 1 and 3).
In Silico and Experimental Resolution of Variable Ribosomal Operon Flanks
The high degree of conservation of ribosomal operons suggests that in fact ribotype RFLP variability is a reflection of polymorphisms not in the ribosomal operons themselves but rather in flanking chromosomal genes (Fig. 3). In silico analysis of 50,000 bp immediately upstream and downstream of the six ribosomal operons of H. influenzae Rd (20) revealed that DNA flanks are primarily (see Table S1 in the supplemental material) composed of neutral housekeeping genes encoding proteins not subject to diversifying selection. Genes in this category, defined according to Kimura's neutral theory of molecular evolution (39), evolve through point mutations. New alleles become prevalent in the population through random genetic drift, not through diversifying Darwinian selection. As such, neutral genes typically evolve at a predictable pace (58). Given the astronomical numbers of bacteria that exist for any given species, the number of variant alleles for a housekeeping gene is vast (54) and provides the basis for polymorphisms detected by ribotyping (Fig. 4). This is the same category of genes providing the molecular genetic basis for MLEE (69)- and MLST (50)-based analyses of microbial population structures.
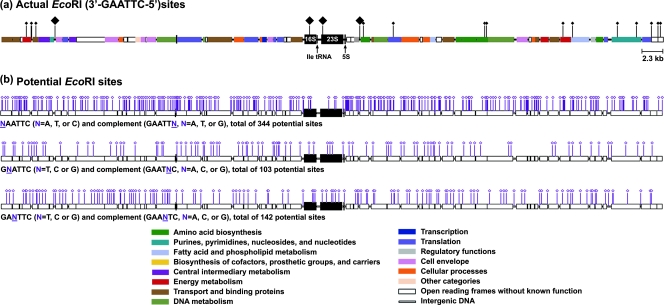
FIG. 4.
In silico analysis (Gene Construction Kit) of potential EcoRI sites in 50 kb upstream and downstream of the H. influenzae strain Rd ribosomal operon C (rrnC). Flanks of rrnC, as well as flanks of the remaining five ribosomal operons (data not shown) in H. influenzae strain Rd, are composed primarily (90.3%) of neutral housekeeping genes. Because of high intraspecies conservation of rRNA gene sequences, ribotype RFLPs found among independent H. influenzae isolates result from changes in restriction sites in the chromosomal flanks in which there exist a vast array of “potential” EcoRI sites. (a) In silico analysis of rrnC of H. influenzae strain Rd and contiguous ∼50 kb upstream and downstream sequences. Genes encoding rRNA (16S, 23S, and 5S) are depicted as large, black boxes; genes flanking the ribosomal operon are color coded according to their products’ cellular functions. Functional classification indicates that the majority of flank sites are evolutionarily neutral. Black diamonds indicate existing EcoRI restriction sites, with the four larger diamonds indicating EcoRI sites responsible for the actual RFLP profile of H. influenzae Rd. (b) In silico analysis of the ∼600 “potential” EcoRI sites that could give rise to a novel cut site by single point mutation, with each of the three maps depicting point mutations at different positions of the palindromic recognition sequence. The locations of the small diamonds reveal that these potential EcoRI sites are distributed along the two 50-kb flank sequences. EcoRI site-generating point mutations of this type in the ribosomal operon chromosomal flanks are the primary molecular genetic basis for the diversity of ribotype profiles attained. Similar in silico analysis of the remaining ribosomal operons (data not shown) allowed us to calculate the maximum possible number of ribotype RFLP profiles as 30012 = 5.3 × 1029, where 300 is the number of potential EcoRI sites for each flank and 12 is the total number of upstream and downstream flanks in any given H. influenzae isolate. As explained above in the text (see “Molecular Basis of Species Diversity Resolved by Conventional Ribotyping: Single-Point-Mutation-Based Model”), 1029 is hypothetical (i.e., the potential number of polymorphisms available by this analysis), while in reality, because of numerous biological constraints, the actual number is much smaller than the possible number.
Other RFLP-based methods used to resolve the relatedness of independent bacterial isolates, e.g., pulsed-field gel electrophoresis (PFGE) (3), are often broadly focused on the bacterial genome as whole rather than a particular category of genes. Because PFGE RFLP cut sites occur across the genome, they may reflect diversifying selection typical of genes encoding virulence factors and surface-exposed proteins, potentially confounding RFLP interpretation as to the level of evolutionary relatedness of any two isolates. In contrast, the tight genetic linkage of evolutionarily neutral gene sequences to the highly conserved ribosomal operons makes ribotyping analysis readily interpretable. In essence, the rRNA gene sequences are exploited as “linked tags” to the adjacent neutral gene sequences. Because rRNA gene operon sequences per se are so highly conserved within a species, polymorphisms are revealed in the flank sequences while no such changes are found in the anchoring rRNA gene. The availability of completed genomic sequences for 155 bacterial species (http://www.ncbi.nlm.nih.gov:80/genomes/static/eub_g.html) allowed for in silico analysis confirming that ribosomal flanks (∼50,000 bp) consist on average of 93% (standard error, 0.0033) neutral housekeeping genes (ranging from 79.2% in Coxiella burnetii to 98% in Corynebacterium glutamicum [see Table S1 in the supplemental material]).
While MLST (50) and ribotyping both appear to be based on neutral gene polymorphisms, establishment of a new MLST scheme for a particular species calls for a significant empirical effort in screening candidate neutral genes to determine whether their evolution indeed is neutral for a particular species. Evolution of seemingly neutral genes may be subject to hitchhiking effects of nonneutral neighboring genes under diversifying selective pressure involving recombination-based mutation, e.g., those coding for outer membrane proteins (83). In contrast, because rRNA gene chromosomal flanks are composed for the most part of densely clustered neutral genes (Fig. 4), such confounding effects on neutral gene evolution are minimized. We refer to such protected rRNA gene flanking regions of low recombination as recombinationally quiescent.
For any bacterial isolate with a known genomic sequence, the precise ribotype RFLP profile for a restriction enzyme can be determined a priori. For the case of H. influenzae strain Rd, we determined in silico that EcoRI would have two unique cutting sites, one within the 16S rRNA gene and one within the 23S rRNA gene. Such a digest results in two RFLP band categories: 12 RFLP bands corresponding to two flank fragments for each of the six ribosomal operons, plus two bands comprising the internal spacer regions (ISR) located between the 16S and 23S genes of the six rRNA gene operons (Fig. 3). Interpretation of ribotyping depends in part on size variation of these ISR bands containing tRNA-encoding DNA sequence(s). For the most part, ISR size is related to the number of tRNAs found within this region, typically one or two, as in the case of H. influenzae, thereby explaining presence of two variant size ISR bands. Vibrio vulnificus (12), with eight ribosomal operons, displays the largest known range of ISR size (422 to 743 bp) and tRNA number (one to four). Since the sizes of these internal fragments are conserved within most species (2, 23), we have named them species-specific signature bands (Fig. 3).
By including strain Rd as a size standard on every H. influenzae ribotype gel, all polymorphic changes found within the ribosomal operon flanks of independent isolates can be compared to the prototypic Rd RFLP profile to extrapolate RFLP band sizes.
Molecular Basis of Species Diversity Resolved by Conventional Ribotyping: Single-Point- Mutation-Based Model
The diversity of polymorphic ribotype fragments relies on the assumption that as strains evolve they acquire random mutations throughout their genome and, as such, is dependent on the rate of point mutations occurring in the genes flanking the ribosomal operons (Fig. 4). A single base pair change in a typical 6-bp restriction endonuclease recognition site will result in the loss of the cutting site and consequently in a change in the RFLP fingerprint profile. Given that ribosomal operons are flanked by ∼50,000 bp of DNA carrying neutrally evolving genes, ribotype RFLP variation is a reflection of neutral gene evolution.
In order to estimate the potential diversity of polymorphic ribotype fragment sizes, we developed a model based on single nucleotide point mutations using the Haemophilus influenzae Rd genome sequence. The flanking 50-kb upstream and downstream regions of the six ribosomal operons were scanned for 6-bp imperfect (potential) EcoRI recognition sites, i.e., sites one point mutation away from becoming an actual EcoRI site: 5′-GCATTC-3′, 5′-GGATTC-3′, 5′-GTATTC-3′, 5′-GACTTC-3′, 5′-GAGTTC-3′, 5′-GATTTC-3′, 5′-CAATTC-3′, 5′-AAATTC-3′, and 5′-TAATTC-3′ (where an underlined nucleotide indicates the position of a point mutation from the actual EcoRI recognition site, 5′-GAATTC-3′) (Fig. 4b). Assuming that independent isolates of the species acquire novel sites by different single point mutations, we were able to estimate the number of possible combinations of polymorphic fragment sizes. In this one-step point mutation model, the number of possible ribotypes was estimated to be ∼1029 within 12 flanks (∼600 kb total) for the case of H. influenzae (see the legend to Fig. 4).
Experimental studies allowed testing of our model as to the degree to which all possible new EcoRI sites are actually found in the natural population structure of H. influenzae. By plotting the accumulated number of novel ribotypes resolved versus the accumulated number of strains analyzed, we show that, in fact, in a ∼600-isolate collection, the number of possible ribotypes tends to be limited to a number well below the estimated 1029 (Fig. 5). This likely reflects biological constraints to mutations, as some point mutations giving rise to a new EcoRI site will be lethal. For example, while both 16S and 23S rRNA genes contain potential EcoRI sites (Fig. 4b), in the ∼600 isolates analyzed to date the actual EcoRI sites remain conserved, based on the consistent size of signature bands. This is expected due to secondary structure constraints allowing very little variation within these ribosomal sequences, such that mutations giving rise to new EcoRI sites have been eliminated by purifying selection (47). Analogously, in the flanking region, some EcoRI sites will be fixed because they are located in essential regions of flanking genes. An additional limitation on the estimated number of possible ribotype patterns reflects the reality that in order to resolve ribotype RFLP fragments resulting from potential restriction sites distant from the ribosomal operon (∼30 to 50 kb), all actual EcoRI sites located closer to the ribosomal operon would have to undergo point mutations making them no longer recognizable by EcoRI.
FIG. 5.
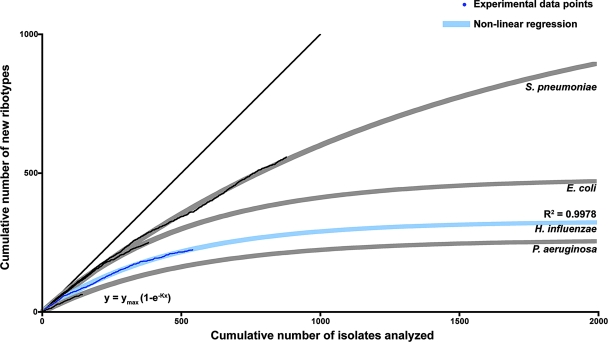
Estimation of the number of H. influenzae isolates necessary to analyze by EcoRI ribotyping in order to reach species diversity. A randomized list of ribotyped isolates was generated. For each isolate (x axis), the y value increases by 1 if the isolate represents a new ribotype profile not previously identified in the randomized set. The resulting plot is depicted in dark blue, with fitted nonlinear regression in light blue. The model used for the nonlinear regression is the one-phase exponential association function y = ymax(1 − e−Kx). As the data set increases and becomes more representative of species diversity, fewer new clusters appear, and the tangent to the regression curve tends toward a slope of 0, at which point one can estimate both the number of possible ribotype profiles attainable and the approximate number of isolates required in order to achieve this maximal representation of the diversity of the species. This analysis was performed for three other bacterial species, shown here for comparison purposes in light gray (fitted nonlinear regression) and black (experimental data). For the four bacterial species, the nonlinear regression analysis of ribotype polymorphisms is in accord with the previously established degree of clonality/panmixia: significantly more polymorphic profiles are predicted for the panmictic S. pneumoniae than for the relatively clonal H. influenzae.
The fundamental in silico approach described above for the design and interpretation of a conventional ribotyping scheme for H. influenzae has since been successfully implemented for an additional eight bacterial species, both gram negative and gram positive: E. coli, P. aeruginosa, B. cepacia, S. pneumoniae, N. meningitidis, Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica.
CAVEATS
Enzyme Selection
While ideal restriction enzyme selection has been discussed above, we recognize that identification of such an enzyme is not always possible. For the case of S. pneumoniae, the most promising enzyme with a unique restriction site within both the 16S and 23S rRNA genes is SmaI. However, the SmaI recognition sequence (5′-CCCGGG-3′) differs greatly in GC content from that of S. pneumoniae (39%). For this reason, SmaI is the restriction endonuclease most commonly used for PFGE analyses of S. pneumoniae, a technique requiring an infrequently cutting restriction enzyme, thus generating very large fragments (>400 kb) (3) not suitable for ribotyping. This necessitated a modified scheme for S. pneumoniae utilizing two frequently cutting enzymes each having a unique recognition site within the ribosomal operon, HindIII (5′-AAGCTT-3′) and PvuII (5′-GAGCTG-3′). While neither enzyme alone gives sufficient discrimination, together they provide a level of discrimination equivalent to that resolved for H. influenzae (38% GC content) using EcoRI (5′-GAATTC-3′ recognition site).
IVSs within 16S and 23S rRNA genes
rRNA genes are typically transcribed from the ribosomal operon as 30S rRNA precursor molecules which are then cleaved by RNase III into 16S, 23S, and 5S rRNA molecules (17, 59, 77). Winkler was the first to observe fragmentation of 23S rRNA molecules of Salmonella enterica serovar Typhimurium into several smaller molecules during maturation (88). Later studies revealed that rRNA molecules of other bacterial species are likewise fragmented due to the presence of intervening sequences (IVSs) within either the 16S rRNA or the 23S rRNA (see studies cited in Table 3 of reference 6). Cotranscribed as part of the rRNA precursor, IVSs form extended stem-loop secondary structures that are cleaved during the rRNA maturation process. The presence of fragmented rRNAs among closely related isolates is sporadic, found in some strains of a given bacterial species, and not always present in all copies of the ribosomal operon within the same bacterium.
The presence of IVSs within the 16S or 23S rRNA gene could potentially confuse interpretation of ribotyping RFLP patterns by (i) containing endonuclease recognition sequences of the selected restriction enzyme and/or (ii) altering the size of the signature band. The appearance of an additional restriction site within the IVS would result either in truncation of the species-specific signature band or in alteration of the flanking DNA band, depending on the position of the new cut site. IVSs reported to date range in size from 72 to 759 bp, with most being approximately 100 bp (6). When we developed a ribotyping scheme for H. influenzae, the presence of IVSs in this species had not yet been identified. In 1999, Song et al. (74) reported the sporadic presence of two types of IVSs in the 23S rRNA gene sequences of some isolates of H. influenzae, IVS1 (112 bp) and IVS2 (121 to 123 bp), with none in the six ribosomal operons of the genomically sequenced strain Rd. A review of our RFLP patterns and in silico analysis of the relative positions of IVS1 and IVS2 within the 23S rRNA gene revealed that the signature band size was not affected. As well, the presence of IVS1 or IVS2 did not noticeably affect the size of flank RFLP fragments due to their relatively small size (112 to 123 bp).
Limited Number of Ribosomal Operons
The copy number of ribosomal operons in bacterial species has been found to range from 1 to 15 (Fig. 2) and to be typically constant within species. As ribotype RFLPs are based on identification of fragments immediately upstream and downstream of each ribosomal operon, the multiplicity of these operons will determine the number of ribotype RFLP bands generated and thus the sensitivity of the method. The minimum number of ribosomal operons within a genome for which ribotyping is being carried out in our laboratory is four (P. aeruginosa and S. pneumoniae). While more than one restriction enzyme could be used to generate multiple ribotype RFLP profiles in bacterial species with a lower ribosomal operon copy number in order to increase discrimination, the ribosome-flanking region investigated would still remain limited.
LIMITED APPLICABILITY OF ALTERNATE RIBOTYPE SCHEMES
Attempts at developing more rapid, less labor-intensive typing schemes, also referred to as ribotyping, have appeared in the literature. As discussed below, while these alternatives are seemingly more practical for the clinical microbiology laboratory than conventional ribotyping and have all been found to be effective techniques for identifying bacteria to the species level, the highly conserved nature of the rRNA genes makes these alternatives insufficiently discriminatory for rigorous intraspecies epidemiological differentiation. While these alternatives may in fact answer the question “identical, or not?,” anyone applying these alternatives needs to be satisfied with an answer that cannot be precisely interpreted as to the evolutionary genetic relatedness of any two isolates.
PCR Ribotyping
PCR ribotyping was developed by Kostman et al. (42, 43) and Gurtler et al. (27, 28) in the 1990s as a response in part to the need in the clinical microbiology laboratory setting for expeditious epidemiological discrimination among pathogenic microorganisms without the use of probes, thus making the analysis more widely applicable.
Using primers complementary to the 3′ end of the 16S rRNA gene and the 5′ end of the 23S rRNA gene, PCR ribotyping reveals length heterogeneity of the PCR-amplified ISR (27, 42). Although developed for epidemiological analysis and for discrimination of pathogens, PCR ribotyping proved not to be universally applicable (11).
In silico analysis of ISR length variability in 27 genomically sequenced bacterial species (data not shown) revealed that while in some species ISR length is variable within and between isolates, in others ISR lengths are limited to one or two sizes, usually dependent on the number of tRNAs present (15, 70). For example, while the multiple copies of the S. enterica serovar Typhimurium, serovar Infantis, and serovar Derby ISR are polymorphic, they are conserved in other Salmonella species, as well in Listeria, Streptococcus, and certain species of Staphylococcus (23, 28, 45, 51). This consideration suggests that PCR ribotyping would be a generally more effective technique for identifying bacteria to the species level and less reliable for intraspecies epidemiological classification at the strain level.
One species with which this methodology has found some success is Clostridium difficile, having a total of 11 ribosomal operons, with differing tRNAs and ISR lengths found among ribosomal operons of the same organism (67) (GenBank accession number AM180355). As of the date of submission, ∼43% of all published PCR ribotyping studies have been performed with C. difficile (7, 37, 64, 79). An analysis of 45 isolates of C. difficile from which the IVS had been sequenced did not reveal striking sequence length diversity (data not shown); however, it appears that the selection of PCR ribotyping as a typing technique relies more on the comparative ease of the technique and repeatability rather than on the discriminatory ability (7), due to the occurrence of DNA degradation in C. difficile interfering with PFGE analysis. More recent studies have begun to recognize the comparative lack of discrimination of this technique and are favoring more discriminatory techniques, such as PFGE and multiple-locus variable-number tandem repeat analysis, for differentiating isolates having identical PCR ribotype profiles (37, 86). C. difficile serves as an example of the fact that differences in specific bacterial properties and the levels of discrimination needed by a particular bacterial species must be taken into account when selecting the most applicable typing technique.
PCR Ribotyping Followed by Restriction Endonuclease Subtyping
In addition to length variation, many species demonstrate high degrees of sequence variability among multiple copies of the ISR (28). Such variability is due, in part, to the fact that intergenic regions often encode one or two tRNAs (2, 15). As well, certain organisms (e.g., E. coli) possess multiple alleles of the rRNA gene cluster, with considerable interallelic variation occurring in the lengths and sequences of the ISRs (22) both at the level of operons in the same genome and between operons within a species.
In an attempt to further subtype isolates for which PCR ribotyping was nondiscriminatory, Ryley et al. (65) and Shreve et al. (72) performed PCR amplification of ISRs, as described above, followed by restriction endonuclease digestion. While additional discriminatory power was found in some cases, the method proved to be inherently limited, typically generating only two to four bands. Further, for some species tested, e.g., H. influenzae (35) and group A streptococci (76), strains distinguished by conventional ribotyping were resolved as identical by this method.
ARDRA
An alternate variation of PCR ribotyping, amplified rRNA gene restriction analysis (ARDRA) is based on PCR amplification of the 16S rRNA gene followed by restriction digestion. Jayarao et al. (34) developed this technique to determine subspecies of Streptococcus uberis, as a means to avoid methods involving DNA hybridization or sequencing. As 16S rRNA gene sequence variation for interspecies differentiation of isolates (i.e., species determination) is well established (21, 87, 91), it is not surprising that ARDRA, even more so than PCR ribotyping, has proven useful for species differentiation rather than intraspecies epidemiological discrimination or phylogenic organization of independent isolates.
Some laboratories have likewise used the 23S rRNA gene to differentiate isolates (35, 85), with the rationale that the 23S rRNA gene, being 60% larger and with a more frequent rate of sequence change than the 16S rRNA gene (60), would be more polymorphic. Despite the increased length and variability, 23S ARDRA proved to be significantly less discriminatory than conventional ribotyping where so compared, e.g., with nontypeable H. influenzae (35).
Long PCR Ribotyping
As a means to enhance the differentiating capacity of PCR ribotyping followed by restriction digestion (see above), Smith-Vaughan et al. (73) developed long PCR ribotyping. This technique is based on PCR amplification of the entire 16S-23S-5S ribosomal operon (∼5.5 kb) followed by restriction endonuclease digestion (73). This method should be more discriminatory than 16S sequencing alone in that it covers the more highly variable ISR yet possesses the epidemiologic advantages of the species-specific conservation of 16S and, to a lesser degree, 23S rRNA genes (26, 30, 60). While it has been applied only to H. influenzae, given the considerations discussed above regarding lack of heterogeneity of the ISR in many species and the known conservation of 16S and 23S rRNA genes, it can be assumed that this technique will require the same caveats as PCR ribotyping.
AUTOMATED RIBOTYPING
As described by Dupont Qualicon, the RiboPrinter automated ribotyping system is a technological breakthrough with respect to convenience, reproducibility, and speed. Our limited experience, however, revealed a downside associated with the speed obtained using the relatively shorter-length agarose gel format standard with the device. In this case, band separation proved not as discriminatory as that resolved using the more standard, larger (16-cm length) gels depicted in the protocol shown in Fig. 1 and 3. In addition, the cost of ribotyping is significantly greater using the automated system. Nonetheless, if lesser discriminatory power provides the degree of resolution of ribotype RFLPs needed to address questions of clinical epidemiology (e.g., “identical, or not?”), then this would be an ideal system in the clinical microbiology laboratory because of its speed and reliability. The speed in this case appears to be ∼2- to 3-fold greater than that for manual ribotyping, with far less labor-intensive procedures.
BEYOND “IDENTICAL OR NOT?”
Having published the first report on the application of pulsed-field electrophoresis to molecular epidemiology (3), we subsequently used this technique in studies involving both ribotype and PFGE analysis for the same sets of isolates of a number of different bacterial species (3, 4, 24, 32, 78, 81). Both approaches provided ample discriminatory power and were for the most part in accord. However, as mentioned above, unlike conventional ribotyping, the molecular genetic basis for a detected PFGE profile is inherently imprecise for a number of reasons: (i) restriction endonuclease cut sites giving rise to polymorphisms revealed by PFGE are unpredictably scattered throughout the chromosome, and (ii) unlike in ribotyping, detected polymorphisms may involve any functional category of nonneutral genes, including those under the pressure of diversifying antigenic selection.
As such, while PFGE results can address the question “identical or not?,” indexing the degree of identity between any two isolates with variant PFGE profiles remains at best uncertain. As an understanding of the evolutionary genetic relationships between bacterial isolates provides valuable insight into the emergence and spread of pathogenic organisms, we have come to rely on ribotyping or MLST to provide this picture, which cannot be accurately obtained from PFGE analysis or alternate ribotyping schemes.
CONCLUDING REMARKS
Our studies reveal that the ribosomal operon flanks in bacteria are composed principally of housekeeping genes and that genetic variation in these genes is primarily responsible for ribotype polymorphisms. The evolutionary neutrality of housekeeping genes constitutes a major factor in the interpretation of ribotype RFLPs. Elucidation of the basis of ribotyping at this molecular genetic level has since allowed us to confidently use this technique to study bacterial population genetics and species diversity in H. influenzae (9, 16), S. pneumoniae (47a), B. cepacia (32, 78, 81), P. aeruginosa (R. Z. Jiang, L. Sun, A. Agodi, S. Steinbach, and R. Goldstein, presented at the 6th Annual Meeting of the North American Cystic Fibrosis Foundation, Washington, DC, October 1992), and E. coli (4). For two of these species, H. influenzae and S. pneumoniae, where comparative ribotype and MLST-based dendrograms were constructed with the same large sets of >500 isolates each, these proved to be highly congruent (33).
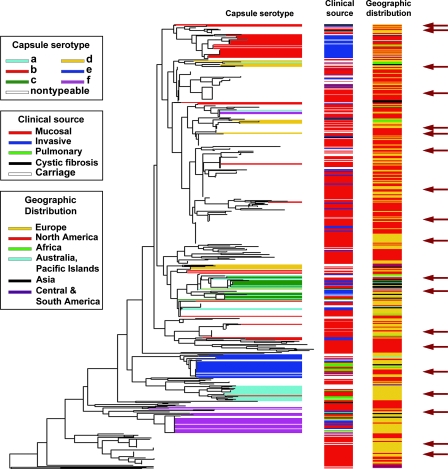
Figure 6 depicts such a species-level ribotyping dendrogram consisting of ∼600 independently isolated strains of typeable and nontypeable H. influenzae (9, 16). Possession of the collection of genetically organized isolates so correlated with this dendrogram has enabled us to select a manageable subset of strains representative of species diversity for identification of shared virulence factors using an animal model (9), for lipopolysaccharide structural analysis (16), and for vaccine target conservation survey and testing (8).
FIG. 6.
Ribotype RFLP phylogenic tree (BioNumerics; Applied Maths) based on ribotyping of ∼600 H. influenzae isolates of diverse origins. Capsular serotypes, clinical sources, and geographic distribution are depicted as horizontal barcodes. The choice of isolates to create a manageable, phylogenetically representative subset for further studies is indicated by dark arrows. Independent confirmation of this dendrogram was obtained based on capsular operon gene polymorphism analysis (44) and congruence of a ribotype-based phylogenetic tree of a subset of the type a to f isolates with results from MLEE analysis (56). Further validation was obtained by comparative gene sequencing of recA and 16S rRNA genes among a representative subset of 50 of the isolates and MLST analysis of a representative set involving 51 of the nontypeable isolates.
Supplementary Material
Acknowledgments
We thank Porter Anderson, Michel Arthur, Ru-Zhang Jiang, Magali Leroy, Antoinella Agodi, Marc Lipsitch, William Hanage, Brian Spratt, Sanjay Ram, Richard Moxon, Peter Rice, and Stephen Pelton for thoughtful discussions and critical comments. We gratefully acknowledge Thomas M. Johnson for encouragement, enthusiasm, and support.
These studies were supported in part by NIH NIDCD awards DC04583, DC05564, and DC005855 to R.G.; NIH NIAID award AI048935; award GOLDST99G0 from the Cystic Fibrosis Foundation; and a grant from The Shereta R. Seelig Charitable Foundation Trust (Boston, MA).
Footnotes
Supplemental material for this article may be found at http://cmr.asm.org/.
REFERENCES
- 1.Aarestrup, F. M. 2001. Comparative ribotyping of Staphylococcus intermedius isolated from members of the Canoidea gives possible evidence for host-specificity and co-evolution of bacteria and hosts. Int. J. Syst. Evol. Microbiol. 51:1343-1347. [DOI] [PubMed] [Google Scholar]
- 2.Anton, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1998. Sequence diversity in the 16S-23S intergenic spacer region (ISR) of the rRNA operons in representatives of the Escherichia coli ECOR collection. J. Mol. Evol. 47:62-72. [DOI] [PubMed] [Google Scholar]
- 3.Arbeit, R. D., M. Arthur, R. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230-235. [DOI] [PubMed] [Google Scholar]
- 4.Arthur, M., R. D. Arbeit, C. Kim, P. Beltran, H. Crowe, S. Steinbach, C. Campanelli, R. A. Wilson, R. K. Selander, and R. Goldstein. 1990. Restriction fragment length polymorphisms among uropathogenic Escherichia coli isolates: pap-related sequences compared with rrn operons. Infect. Immun. 58:471-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachmann, B. J. 1990. Linkage map of Escherichia coli K-12, edition 8. Microbiol. Rev. 54:130-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, B. J., P. Hugenholtz, S. C. Dawson, and J. F. Banfield. 2003. Extremely acidophilic protists from acid mine drainage host Rickettsiales-lineage endosymbionts that have intervening sequences in their 16S rRNA genes. Appl. Environ. Microbiol. 69:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidet, P., V. Lalande, B. Salauze, B. Burghoffer, V. Avesani, M. Delmee, A. Rossier, F. Barbut, and J. C. Petit. 2000. Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 38:2484-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolduc, G. R., V. Bouchet, R. Z. Jiang, J. Geisselsoder, Q. C. Truong-Bolduc, P. A. Rice, S. I. Pelton, and R. Goldstein. 2000. Variability of outer membrane protein P1 and its evaluation as a vaccine candidate against experimental otitis media due to nontypeable Haemophilus influenzae: an unambiguous, multifaceted approach. Infect. Immun. 68:4505-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouchet, V., D. W. Hood, J. Li, J. R. Brisson, G. A. Randle, A. Martin, Z. Li, R. Goldstein, E. K. Schweda, S. I. Pelton, J. C. Richards, and E. R. Moxon. 2003. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. USA 100:8898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosius, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6:112-118. [DOI] [PubMed] [Google Scholar]
- 11.Cartwright, C. P. 1995. Polymerase chain reaction ribotyping: a “universal” approach? J. Infect. Dis. 172:1638-1639. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesneau, O., A. Morvan, S. Aubert, and N. el Solh. 2000. The value of rRNA gene restriction site polymorphism analysis for delineating taxa in the genus Staphylococcus. Int. J. Syst. Evol. Microbiol. 50 Pt. 2:689-697. [DOI] [PubMed] [Google Scholar]
- 14.Chisholm, S. A., P. B. Crichton, H. I. Knight, and D. C. Old. 1999. Molecular typing of Salmonella serotype Thompson strains isolated from human and animal sources. Epidemiol. Infect. 122:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen, H., K. Jorgensen, and J. E. Olsen. 1999. Differentiation of Campylobacter coli and C. jejuni by length and DNA sequence of the 16S-23S rRNA internal spacer region. Microbiology 145:99-105. [DOI] [PubMed] [Google Scholar]
- 16.Cody, A. J., D. Field, E. J. Feil, S. Stringer, M. E. Deadman, A. G. Tsolaki, B. Gratz, V. Bouchet, R. Goldstein, D. W. Hood, and E. R. Moxon. 2003. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 3:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doolittle, W. F., and N. R. Pace. 1971. Transcriptional organization of the ribosomal RNA cistrons in Escherichia coli. Proc. Natl. Acad. Sci. USA 68:1786-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eldar, A., S. Lawhon, P. F. Frelier, L. Assenta, B. R. Simpson, P. W. Varner, and H. Bercovier. 1997. Restriction fragment length polymorphisms of 16S rDNA and of whole rRNA genes (ribotyping) of Streptococcus iniae strains from the United States and Israel. FEMS Microbiol. Lett. 151:155-162. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Espinar, M. T., V. Lopez, D. Ramon, E. Bartra, and A. Querol. 2001. Study of the authenticity of commercial wine yeast strains by molecular techniques. Int. J. Food Microbiol. 70:1-10. [DOI] [PubMed] [Google Scholar]
- 20.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 21.Fox, G. E., E. Stackebrandt, R. B. Hespell, J. Gibson, J. Maniloff, T. A. Dyer, R. S. Wolfe, W. E. Balch, R. S. Tanner, L. J. Magrum, L. B. Zablen, R. Blakemore, R. Gupta, L. Bonen, B. J. Lewis, D. A. Stahl, K. R. Luehrsen, K. N. Chen, and C. R. Woese. 1980. The phylogeny of prokaryotes. Science 209:457-463. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Martinez, J., A. Martinez-Murcia, A. I. Anton, and F. Rodriguez-Valera. 1996. Comparison of the small 16S to 23S intergenic spacer region (ISR) of the rRNA operons of some Escherichia coli strains of the ECOR collection and E. coli K-12. J. Bacteriol. 178:6374-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giannino, V., M. Santagati, G. Guardo, C. Cascone, G. Rappazzo, and S. Stefani. 2003. Conservation of the mosaic structure of the four internal transcribed spacers and localisation of the rrn operons on the Streptococcus pneumoniae genome. FEMS Microbiol. Lett. 223:245-252. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein, R., L. Sun, R. Z. Jiang, U. Sajjan, J. F. Forstner, and C. Campanelli. 1995. Structurally variant classes of pilus appendage fibers coexpressed from Burkholderia (Pseudomonas) cepacia. J. Bacteriol. 177:1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimont, F., and P. A. Grimont. 1986. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann. Inst. Pasteur Microbiol. 137B:165-175. [DOI] [PubMed] [Google Scholar]
- 26.Gurtler, V. 1993. Typing of Clostridium difficile strains by PCR-amplification of variable length 16S-23S rDNA spacer regions. J. Gen. Microbiol. 139:3089-3097. [DOI] [PubMed] [Google Scholar]
- 27.Gurtler, V., and H. D. Barrie. 1995. Typing of Staphylococcus aureus strains by PCR-amplification of variable-length 16S-23S rDNA spacer regions: characterization of spacer sequences. Microbiology 141:1255-1265. [DOI] [PubMed] [Google Scholar]
- 28.Gurtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Hillis, D. M., and M. T. Dixon. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66:411-453. [DOI] [PubMed] [Google Scholar]
- 31.Holmes, A., and R. Goldstein. 1995. Beyond ‘identical or not?’ The potential of molecular epidemiology. Pediatr. Pulmonol. S. 12:166-169. [Google Scholar]
- 32.Holmes, A., R. Nolan, R. Taylor, R. Finley, M. Riley, R. Z. Jiang, S. Steinbach, and R. Goldstein. 1999. An epidemic of Burkholderia cepacia transmitted between patients with and without cystic fibrosis. J. Infect. Dis. 179:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huot, H., V. Bouchet, M. Leroy, S. Pelton, K. L. O'Brien, M. Santosham, K. O'Neill, M. Lipsitch, and R. Goldstein. 2006. Congruence of pneumococcal population structure resolved by ribotyping and MLST in a defined community, abstr. PO5.20, p. 205. 5th Int. Symp. Pneumococci Pneumococcal Dis. ISPPD5 Ltd., Sydney, Australia.
- 34.Jayarao, B. M., J. J. Dore, Jr., G. A. Baumbach, K. R. Matthews, and S. P. Oliver. 1991. Differentiation of Streptococcus uberis from Streptococcus parauberis by polymerase chain reaction and restriction fragment length polymorphism analysis of 16S ribosomal DNA. J. Clin. Microbiol. 29:2774-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordens, J. Z., and N. I. Leaves. 1997. Source of variation detected in ribotyping patterns of Haemophilus influenzae: comparison of traditional ribotyping, PCR-ribotyping and rDNA restriction analysis. J. Med. Microbiol. 46:763-772. [DOI] [PubMed] [Google Scholar]
- 36.Joung, K. B., and J. C. Cote. 2002. Evaluation of ribosomal RNA gene restriction patterns for the classification of Bacillus species and related genera. J. Appl. Microbiol. 92:97-108. [DOI] [PubMed] [Google Scholar]
- 37.Kikkawa, H., S. Hitomi, and M. Watanabe. 2007. Prevalence of toxin A-nonproducing/toxin-B-producing Clostridium difficile in the Tsukuba-Tsuchiura district, Japan. J. Infect. Chemother. 13:35-38. [DOI] [PubMed] [Google Scholar]
- 38.Kilic, U., B. Schalch, and A. Stolle. 2002. Ribotyping of Clostridium perfringens from industrially produced ground meat. Lett. Appl. Microbiol. 34:238-243. [DOI] [PubMed] [Google Scholar]
- 39.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, United Kingdom.
- 40.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolbert, C. P., and D. H. Persing. 1999. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr. Opin. Microbiol. 2:299-305. [DOI] [PubMed] [Google Scholar]
- 42.Kostman, J. R., M. B. Alden, M. Mair, T. D. Edlind, J. J. LiPuma, and T. L. Stull. 1995. A universal approach to bacterial molecular epidemiology by polymerase chain reaction ribotyping. J. Infect. Dis. 171:204-208. [DOI] [PubMed] [Google Scholar]
- 43.Kostman, J. R., T. D. Edlind, J. J. LiPuma, and T. L. Stull. 1992. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J. Clin. Microbiol. 30:2084-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kroll, J. S., S. Ely, and E. R. Moxon. 1991. Capsular typing of Haemophilus influenzae with a DNA probe. Mol. Cell. Probes 5:375-379. [DOI] [PubMed] [Google Scholar]
- 45.Lagatolla, C., L. Dolzani, E. Tonin, A. Lavenia, M. Di Michele, T. Tommasini, and C. Monti-Bragadin. 1996. PCR ribotyping for characterizing Salmonella isolates of different serotypes. J. Clin. Microbiol. 34:2440-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laurent, F., A. Carlotti, P. Boiron, J. Villard, and J. Freney. 1996. Ribotyping: a tool for taxonomy and identification of the Nocardia asteroides complex species. J. Clin. Microbiol. 34:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, W.-H., and D. Graur. 2000. Gene duplication, exon shuffling, and concerted evolution, p. 249-322. In A. D. Sinauer (ed.), Fundamentals of molecular evolution, 2nd ed. Sinauer Associates, Sunderland, MA.
- 47a.Lipsitch, M., K. O'Neill, D. Cordy, B. Bugalter, K. Trzcinski, C. M. Thompson, R. Goldstein, S. Pelton, H. Huot, V. Bouchet, R. Reid, M. Santosham, and K. L. O'Brien. 2007. Strain characteristics of Streptococcus pneumoniae carriage and invasive disease isolates during a cluster-randomized clinical trial of the 7-valent pneumococcal conjugate vaccines. J. Infect. Dis. 196:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LiPuma, J. J., J. E. Mortensen, S. E. Dasen, T. D. Edlind, D. V. Schidlow, J. L. Burns, and T. L. Stull. 1988. Ribotype analysis of Pseudomonas cepacia from cystic fibrosis treatment centers. J. Pediatr. 113:859-862. [DOI] [PubMed] [Google Scholar]
- 49.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marsou, R., M. Bes, M. Boudouma, Y. Brun, H. Meugnier, J. Freney, F. Vandenesch, and J. Etienne. 1999. Distribution of Staphylococcus sciuri subspecies among human clinical specimens, and profile of antibiotic resistance. Res. Microbiol. 150:531-541. [DOI] [PubMed] [Google Scholar]
- 52.McCullough, M. J., K. V. Clemons, J. H. McCusker, and D. A. Stevens. 1998. Intergenic transcribed spacer PCR ribotyping for differentiation of Saccharomyces species and interspecific hybrids. J. Clin. Microbiol. 36:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Messner, R., and H. Prillinger. 1995. Saccharomyces species assignment by long range ribotyping. Antonie van Leeuwenhoek 67:363-370. [DOI] [PubMed] [Google Scholar]
- 54.Milkman, R. 1973. Electrophoretic variation in Escherichia coli from natural sources. Science 182:1024-1026. [DOI] [PubMed] [Google Scholar]
- 55.Miteva, V., I. Boudakov, G. Ivanova-Stoyancheva, B. Marinova, V. Mitev, and J. Mengaud. 2001. Differentiation of Lactobacillus delbrueckii subspecies by ribotyping and amplified ribosomal DNA restriction analysis (ARDRA). J. Appl. Microbiol. 90:909-918. [DOI] [PubMed] [Google Scholar]
- 56.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narayanan, S., T. G. Nagaraja, J. Staats, M. M. Chengappa, and R. D. Oberst. 1998. Biochemical and biological characterizations and ribotyping of Actinomyces pyogenes and Actinomyces pyogenes-like organisms from liver abscesses in cattle. Vet. Microbiol. 61:289-303. [DOI] [PubMed] [Google Scholar]
- 58.Nei, M. 1987. Molecular evolutionary genetics, p. 287-326. Columbia University Press, New York, NY.
- 59.Nikolaev, N., L. Silengo, and D. Schlessinger. 1973. A role for ribonuclease 3 in processing of ribosomal ribonucleic acid and messenger ribonucleic acid precursors in Escherichia coli. J. Biol. Chem. 248:7967-7969. [PubMed] [Google Scholar]
- 60.Olsen, G. J., and C. R. Woese. 1993. Ribosomal RNA: a key to phylogeny. FASEB J. 7:113-123. [DOI] [PubMed] [Google Scholar]
- 61.Pace, N. R., G. J. Olsen, and C. R. Woese. 1986. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell 45:325-326. [DOI] [PubMed] [Google Scholar]
- 62.Pitt, T. L., S. Trakulsomboon, and D. A. Dance. 2000. Molecular phylogeny of Burkholderia pseudomallei. Acta Trop. 74:181-185. [DOI] [PubMed] [Google Scholar]
- 63.Popovic, T., I. K. Mazurova, A. Efstratiou, J. Vuopio-Varkila, M. W. Reeves, A. De Zoysa, T. Glushkevich, and P. Grimont. 2000. Molecular epidemiology of diphtheria. J. Infect. Dis. 181(Suppl. 1):S168-S177. [DOI] [PubMed] [Google Scholar]
- 64.Rupnik, M., J. S. Brazier, B. I. Duerden, M. Grabnar, and S. L. Stubbs. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439-447. [DOI] [PubMed] [Google Scholar]
- 65.Ryley, H. C., L. Millar-Jones, A. Paull, and J. Weeks. 1995. Characterisation of Burkholderia cepacia from cystic fibrosis patients living in Wales by PCR ribotyping. J. Med. Microbiol. 43:436-441. [DOI] [PubMed] [Google Scholar]
- 66.Salmenlinna, S., and J. Vuopio-Varkila. 2001. Recognition of two groups of methicillin-resistant Staphylococcus aureus strains based on epidemiology, antimicrobial susceptibility, hypervariable-region type, and ribotype in Finland. J. Clin. Microbiol. 39:2243-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 68.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Selander, R. K., J. M. Musser, D. A. Caugant, M. N. Gilmour, and T. S. Whittam. 1987. Population genetics of pathogenic bacteria. Microb. Pathog. 3:1-7. [DOI] [PubMed] [Google Scholar]
- 70.Severino, P., A. L. Darini, and V. D. Magalhaes. 1999. The discriminatory power of ribo-PCR compared to conventional ribotyping for epidemiological purposes. APMIS 107:1079-1084. [DOI] [PubMed] [Google Scholar]
- 71.Shangkuan, Y. H., J. F. Yang, H. C. Lin, and M. F. Shaio. 2000. Comparison of PCR-RFLP, ribotyping and ERIC-PCR for typing Bacillus anthracis and Bacillus cereus strains. J. Appl. Microbiol. 89:452-462. [DOI] [PubMed] [Google Scholar]
- 72.Shreve, M. R., S. J. Johnson, C. E. Milla, C. L. Wielinski, and W. E. Regelmann. 1997. PCR ribotyping and endonuclease subtyping in the epidemiology of Burkholderia cepacia infection. Am. J. Respir. Crit. Care Med. 155:984-989. [DOI] [PubMed] [Google Scholar]
- 73.Smith-Vaughan, H. C., K. S. Sriprakash, J. D. Mathews, and D. J. Kemp. 1995. Long PCR-ribotyping of nontypeable Haemophilus influenzae. J. Clin. Microbiol. 33:1192-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song, X. M., A. Forsgren, and H. Janson. 1999. Fragmentation heterogeneity of 23S ribosomal RNA in Haemophilus species. Gene 230:287-293. [DOI] [PubMed] [Google Scholar]
- 75.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 76.Sriprakash, K. S., and D. L. Gardiner. 1997. Lack of polymorphism within the rRNA operons of group A streptococci. Mol. Gen. Genet. 255:125-130. [DOI] [PubMed] [Google Scholar]
- 77.Srivastava, A. K., and D. Schlessinger. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44:105-129. [DOI] [PubMed] [Google Scholar]
- 78.Steinbach, S., L. Sun, R. Z. Jiang, P. Flume, P. Gilligan, T. M. Egan, and R. Goldstein. 1994. Transmissibility of Pseudomonas cepacia infection in clinic patients and lung-transplant recipients with cystic fibrosis. N. Engl. J. Med. 331:981-987. [DOI] [PubMed] [Google Scholar]
- 79.Stubbs, S. L., J. S. Brazier, G. L. O'Neill, and B. I. Duerden. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stull, T. L., J. J. LiPuma, and T. D. Edlind. 1988. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J. Infect. Dis. 157:280-286. [DOI] [PubMed] [Google Scholar]
- 81.Sun, L., R. Z. Jiang, S. Steinbach, A. Holmes, C. Campanelli, J. Forstner, U. Sajjan, Y. Tan, M. Riley, and R. Goldstein. 1995. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat. Med. 1:661-666. [DOI] [PubMed] [Google Scholar]
- 82.Syrogiannopoulos, G. A., C. Doit, I. N. Grivea, P. Geslin, and E. Bingen. 2001. Clonal relationships among penicillin-susceptible, multiresistant serotype 6B Streptococcus pneumoniae isolates recovered in Greece and France. Eur. J. Clin. Microbiol. Infect. Dis. 20:61-64. [DOI] [PubMed] [Google Scholar]
- 83.Thampapillai, G., R. Lan, and P. R. Reeves. 1994. Molecular evolution in the gnd locus of Salmonella enterica. Mol. Biol. Evol. 11:813-828. [DOI] [PubMed] [Google Scholar]
- 84.Uijthof, J. M., G. S. de Hoog, A. W. de Cock, K. Takeo, and K. Nishimura. 1994. Pathogenicity of strains of the black yeast Exophiala (Wangiella) dermatitidis: an evaluation based on polymerase chain reaction. Mycoses 37:235-242. [DOI] [PubMed] [Google Scholar]
- 85.Van Camp, G., S. Chapelle, and R. De Wachter. 1993. Amplification and sequencing of variable regions in bacterial 23S ribosomal RNA genes with conserved primer sequences. Curr. Microbiol. 27:147-151. [DOI] [PubMed] [Google Scholar]
- 86.van den Berg, R. J., I. Schaap, K. E. Templeton, C. H. Klaassen, and E. J. Kuijper. 2007. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J. Clin. Microbiol. 45:1024-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winkler, M. E. 1979. Ribosomal ribonucleic acid isolated from Salmonella typhimurium: absence of the intact 23S species. J. Bacteriol. 139:842-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woese, C. R. 1996. The world of ribosomal RNA, p. 23-48. In R. A. Zimmermann and A. E. Dahlberg (ed.), Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. CRC Press, Boca Raton, FL.
- 90.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yurlova, N. A., J. M. Uijthof, and G. S. de Hoog. 1996. Distinction of species in Aureobasidium and related genera by PCR-ribotyping. Antonie van Leeuwenhoek. 69:323-329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.