Abstract
This study demonstrated the capacity of bacteriocin-producing lactic acid bacteria (LAB) to reduce intestinal colonization by vancomycin-resistant enterococci (VRE) in a mouse model. Lactococcus lactis MM19 and Pediococcus acidilactici MM33 are bacteriocin producers isolated from human feces. The bacteriocin secreted by P. acidilactici is identical to pediocin PA-1/AcH, while PCR analysis demonstrated that L. lactis harbors the nisin Z gene. LAB were acid and bile tolerant when assayed under simulated gastrointestinal conditions. A well diffusion assay using supernatants from LAB demonstrated strong activity against a clinical isolate of VRE. A first in vivo study was done using C57BL/6 mice that received daily intragastric doses of L. lactis MM19, P. acidilactici MM33, P. acidilactici MM33A (a pediocin mutant that had lost its ability to produce pediocin), or phosphate-buffered saline (PBS) for 18 days. This study showed that L. lactis and P. acidilactici MM33A increased the concentrations of total LAB and anaerobes while P. acidilactici MM33 decreased the Enterobacteriaceae populations. A second in vivo study was done using VRE-colonized mice that received the same inocula as those in the previous study for 16 days. In L. lactis-fed mice, fecal VRE levels 1.73 and 2.50 log10 CFU/g lower than those in the PBS group were observed at 1 and 3 days postinfection. In the P. acidilactici MM33-fed mice, no reduction was observed at 1 day postinfection but a reduction of 1.85 log10 CFU/g was measured at 3 days postinfection. Levels of VRE in both groups of mice treated with bacteriocin-producing LAB were undetectable at 6 days postinfection. No significant difference in mice fed the pediocin-negative strain compared to the control group was observed. This is the first demonstration that human L. lactis and P. acidilactici nisin- and pediocin-producing strains can reduce VRE intestinal colonization.
Although vancomycin has been used in human medicine since 1958 (12), the first strain of vancomycin-resistant enterococci (VRE) was isolated in Europe in 1986 (33) and a VRE strain was subsequently isolated in the United States in 1987 (28). Since then, the proportion of VRE has risen throughout medical centers and has made VRE a serious clinical problem in many countries (6). The dissemination of VRE can lead to clinical isolates resistant to all antibiotics, because enterococci have become important nosocomial pathogens and a reservoir for resistance genes. The dissemination of glycopeptide resistance to other pathogenic bacteria, such as Staphylococcus and Streptococcus species, has occurred previously because there is no barrier to heterospecific expression or gene transfer among gram-positive cocci (6, 27). Thus, alternatives to traditional antibiotics are needed. To date, few effective therapies are available to prevent and control intestinal colonization by VRE.
Probiotic bacteria are live microorganisms that, when administered in adequate amounts, confer a health benefit on the host (8). The antagonistic activities of the probiotics against food-borne and clinical pathogens resistant to antibiotics have been observed previously during in vitro (9, 19, 20) and in vivo (7) studies using animal infection models. Bacteriocins are bactericidal peptides secreted by many species of bacteria. For example, nisin kills bacteria by pore formation and uses lipid II as a docking molecule, which undermines the ability of the bacteria to synthesize the cell wall (10). Nisin may increase the bacterial sensitivity to antibiotics by improving the entrance of the antibiotic into the cell. Few studies using probiotics have reported a capacity for preventing or reducing VRE colonization. A randomized and placebo-controlled human study has demonstrated that a yogurt containing Lactobacillus rhamnosus GG treats gastrointestinal carriage of VRE (13). However, no study has implicated bacteriocin production by probiotics in the prevention of infections with antibiotic-resistant bacteria. Such strategies offer the prospect of developing antimicrobials to which certain pathogens cannot adapt.
Lactococcus lactis MM19 and Pediococcus acidilactici MM33 were originally isolated from human stool, and this was the first time that bacteriocin-producing strains of these species had been isolated from the human gut (17). The bacteriocin secreted by P. acidilactici MM33 was purified and identified as pediocin PA-1/AcH (18). The bacteriocin synthesized by L. lactis MM19 is active against a wide spectrum of lactic acid bacteria (LAB) but also against vancomycin-sensitive Enterococcus faecium and methicillin-sensitive Staphylococcus aureus. It can be inactivated by various proteolytic enzymes. Moreover, the antimicrobial activity of the supernatant is resistant to heating at 70°C for up to 15 min and stable at pHs ranging from 2 to 8 (17).
Thus, it was hypothesized that the oral administration of bacteriocin-producing strains of LAB would decrease the intestinal VRE population in a rodent model.
The objectives of this study were as follows: (i) to evaluate the potential of these strains to inhibit the growth of VRE in vitro and in vivo using a mouse model of VRE intestinal colonization; (ii) to identify the bacteriocin produced by L. lactis MM19; (iii) to evaluate the resistance of both bacteriocin-producing L. lactis MM19 and P. acidilactici MM33 to gastrointestinal conditions; and (iv) to demonstrate the safety and the capacities of these bacteriocin-producing strains in modulating intestinal microbiota in healthy mice.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. lactis subsp. lactis MM19 and P. acidilactici MM33 isolates previously obtained from human stool (17) were used. P. acidilactici MM33A, a mutant derived from P. acidilactici MM33 that has lost its ability to produce pediocin through a plasmid curing procedure described in detail elsewhere (18), was also used in this study. Lactobacillus rhamnosus GG ATCC 53103, a well-known probiotic acid- and bile-tolerant bacterium, was used as a positive control for the in vitro gastrointestinal resistance experiments. A clinical isolate of vancomycin-resistant E. faecium was provided by the Centre Hospitalier de l'Université de Montréal microbiology laboratory (Montréal, QC, Canada). Previous PCR studies using primers (26) specific to the vancomycin resistance genes have revealed that this isolate is a VanA-type VRE strain. The LAB and E. faecium strains were maintained at −80°C in lactobacillus MRS medium (Difco Laboratories, Detroit, MI) and brain heart infusion (BHI) medium (Difco), respectively, each containing 10% glycerol (wt/vol). Before being used, all strains evaluated were propagated for 2 days in MRS or BHI broth overnight at 37°C without agitation.
In vitro VRE inhibition.
An agar well diffusion assay was performed as described previously (16) to verify the antimicrobial capacities of the neutralized cell-free supernatants (CFS) against a VRE clinical isolate. CFS was obtained by centrifuging a culture at 6,000 × g for 30 min at 4°C and neutralizing the material to pH 6.5 by the addition of NaOH (5 M). The resulting CFS was then filtered through a 0.2-μm-pore-size filter (Sarstedt, Montréal, QC, Canada). A volume of 30 ml of sterile BHI agar (45°C), containing 0.75% agar, was inoculated with 107 CFU of VRE/ml, poured into 100-by-15-mm standard petri dishes, and allowed to solidify for 30 min at room temperature. Wells of 6 mm in diameter were cut out, and 80 μl of the antimicrobial agent was placed into each well. All plates were then placed at 4°C for 30 min, incubated at 37°C for 24 h, and examined for inhibition zones. Inhibition was scored as positive if the width of the clearing zone around the well was ≥0.5 mm. Antibiotics were also evaluated with this method to ascertain the resistance of the VRE strain. Vancomycin and clindamycin (Sigma-Aldrich, Oakville, ON, Canada) were used in concentrations ranging from 0 to 800 μg/ml. When LAB supernatants were assayed, proteases from Streptomyces griseus type XIV (3 mg/ml; Sigma) were added to the soft agar in order to verify whether the inhibition was caused by bacteriocin (16).
PCR and DNA sequencing of the nisin gene.
The PCR amplification was performed as described by Millette et al. (18). The primers were designed from nisin and pediocin PA-1/AcH structural genes. The nisin gene primers were complementary to regions 17 bp upstream (5′ CCGGAATTCATAAGGAGGCACTCAAAATG 3′) and 2 bp downstream (3′ CGGGGTACCTACTATCCTTTGATTTGGTT 5′) of the coding region for nisin (24), and the pediocin gene primers were as described by Millette et al. (18). The EcoRI and KpnI restriction sites were added at the 5′ ends of primer 1 and primer 2, respectively, for cloning purposes.
The amplified PCR products were visualized and purified from a 2% agarose gel by using a QIAquick gel extraction kit (Qiagen, Mississauga, ON, Canada), and the nucleotide sequences were determined by Génome Québec (Montréal, QC, Canada).
Acid tolerance of LAB.
Simulated gastric fluid (SGF) was formulated according to the guidelines of the U.S. Pharmacopeia. Briefly, SGF was composed of 3.2 g of pepsin (Sigma)/liter and 2.0 g of NaCl/liter, and the pH was adjusted to 1.5, 2.0, 2.5, or 3.0 by the addition of HCl (5 M). Volumes of 1 ml of overnight MRS broth cultures of LAB were added to 19 ml of SGF at 37°C under mild agitation (200 rpm) in a G24 environmental incubator shaker (New Brunswick Scientific Co. Inc., NJ). After 30 min of incubation of the gastric solution, 1 ml was collected, mixed in sterile phosphate-buffered saline (PBS; pH 7.4), and immediately diluted in sterile peptone water (0.1%, wt/vol) and plated (by the pour-plate method) onto lactobacillus MRS agar. Plates were incubated under aerobic conditions at 37°C for 48 h. The average number of CFU from triplicate analyses was determined by using a dark-field Quebec colony counter. A similar process was carried out for bacteria without SGF treatment in order to determine the initial concentrations of LAB. Lactobacillus rhamnosus GG was used as a positive control because it is a probiotic bacterium well known for its resistance to gastrointestinal conditions (31).
Bile salt tolerance of LAB.
The bile salt tolerance of LAB was ascertained in MRS agar containing a commercial preparation of bile salts normally used to inhibit the growth of gram-positive bacteria in broth. A bile salt mixture (Sigma B-3426) was added in concentrations varying between 0 and 10% with increments of 1%. Another bile salt preparation (LP0055; Oxoid, Nepean, ON, Canada) was also evaluated in concentrations from 0 to 24% with increments of 4%. The MRS agar containing the bile salts was then autoclaved for 15 min at 121°C, cooled, and finally plated. Aliquots of overnight MRS broth cultures (100 μl of bacteria in the stationary phase obtained after 24 h of growth) were inoculated onto the surface of the bile salt-containing MRS agar, and the plates were incubated at 37°C for 72 h under anaerobic conditions. The presence of a bacterial lawn indicated good growth and thus good resistance of the bacteria to bile salts, while the presence of small and isolated colonies indicated poor resistance to bile salts. The absence of colonies indicated that LAB did not tolerate the bile salts at the concentrations tested. The MIC was defined as the lowest concentration of bile salts needed to totally inhibit the growth of colonies as judged by visual examination.
Animals.
Six- to eight-week-old female C57BL/6 mice (Charles River Laboratories, St.-Constant, QC, Canada) were used for the evaluation of the fecal microbiota in the microbiota modulation experiment, while 25- to 30-g female CF-1 mice (Charles River) were used for the VRE intestinal colonization experiment. Mice were housed in groups of three to five per plastic cage and kept under specific-pathogen-free conditions with free access to a commercial diet (lab diet 5001; Ren's Feed & Supplies, Oakville, ON, Canada) and water. Cages and bedding were changed every 2 days. This work was approved and supervised by the INRS-Institut Armand-Frappier Animal Care Committee.
Fecal microbial population modulation following ingestion of LAB.
To investigate the safety and the impact on gut microbiota of the oral administration of bacteriocin-producing LAB, the methodology was adapted from O'Mahony et al. (25). Briefly, healthy mice received a daily dose of 100 μl of a solution containing 1010 CFU of viable bacteria (L. lactis MM19, P. acidilactici MM33, or P. acidilactici MM33A)/ml in 100 μl of PBS by the intragastric route by using a stainless steel feeding needle and a 1-ml syringe. A group of mice received PBS alone as a negative control. Mice were weighed on days 1, 9, 18, and 27 (day 27 being 9 days after the end of the feeding treatment), and any signs of physiological or behavioral perturbation during the experiment were noted. Stool samples were collected before the administration of PBS or LAB (day 1) and 9 and 18 days after the beginning of the feeding procedures. The final analysis was done 9 days after the end of the treatment (day 27 after the beginning of the treatment). After defecation, the stools were collected directly into a preweighted 2-ml sterile plastic tube. These tubes were kept on ice until microbial analysis (maximum, 1 h). Fecal populations of total culturable LAB, Lactobacillus spp., anaerobes, Enterobacteriaceae, Staphylococcus spp., and Enterococcus spp. were analyzed on selective media. This experiment was done twice using a total of 10 mice per experimental group.
VRE intestinal colonization experimental model.
The VRE intestinal colonization experimental model was adapted from Donskey et al. (7). Daily subcutaneous administration of clindamycin (1.4 mg/day) for 5 days was used to disrupt the intestinal microbiota as necessary to induce the VRE infection. Three days after the end of antimicrobial administration, gastric inoculation with 250 μl of an overnight culture of VRE in BHI broth was used to infect the mice. Approximately 108 CFU of VRE was administered to the mice. From the beginning of the antibiotic therapy, all groups of LAB-treated mice received once-daily doses of 100 μl of a 1010-CFU/ml concentration of L. lactis MM19, P. acidilactici MM33, or P. acidilactici MM33A bacteria, previously washed twice in sterile PBS. The mice received the LAB until the eighth day after infection. Bacitracin (Sigma) was administrated orally once daily at 600 U/day diluted in 100 μl of sterile PBS for 3 days following the infection of mice with VRE. This treatment was then discontinued and replaced by treatment with PBS alone. Stool samples were collected before the administration of antibiotics and 1, 3, 6, 8, and 12 days after the VRE infection. Fecal concentrations of total culturable VRE were analyzed using selective media. This experiment was done twice using a total of eight mice per experimental group.
Quantification of stool organisms.
All the feces were diluted in 1,000 μl of sterile saline, homogenized with a pestle, and serially diluted 10-fold in 0.1% peptone water. Finally, 100 μl of each dilution was inoculated onto the following media: MRS agar for the detection of total LAB, Rogosa SL agar for the selective detection of Lactobacillus spp., reinforced clostridium medium for the quantification of total anaerobic and mesophilic bacteria, Baird-Parker agar for the selective detection of Staphylococcus spp., MacConkey agar for the selective enumeration of Enterobacteriaceae, Enterococcosel agar for the selective quantification of total Enterococcus spp., and finally, Enterococcosel agar with 20 μg of vancomycin (Sigma)/ml for the detection and enumeration of VRE. A volume of 100 μl of the undiluted sample was also plated. MRS, Rogosa, and reinforced clostridium medium plates were incubated in anaerobic jars at 37°C for 72 h, while Baird-Parker agar, MacConkey agar, and both Enterococcosel agar plates were incubated under aerobic conditions at 37°C for 48 h. When negative results were obtained for Enterococcosel agar with 20 μg of vancomycin/ml after 48 h of incubation, the plates were allowed to incubate for another 24 h. Because the weights of fecal specimens varied, the lower limit of the assay for the detection of VRE also varied. For statistical purposes, a value in CFU per gram that was based upon the weights of individual specimens was assigned for feces without microorganisms.
Statistical analysis.
Acid resistance and bile salt tolerance experiments were carried out in triplicate (n = 3). For each independent replication, three independent samples were analyzed. Student's t test was done using the SPSS statistics program (version 10.0) to determine significant differences in viability between the LAB populations before and after acid treatment. Finally, an analysis of variance and a multiple-comparison Duncan test were used to compare microbial populations in the feces of mice fed various LAB to the populations at other time points and to those in the feces of the PBS control group. All significant differences were determined at a P value of ≤0.05.
RESULTS
Bacteriocin gene identification.
Two primers complementary to sequences occurring proximal to the 3′ and 5′ ends of the nisin A and pediocin PA-1 structural genes were used to amplify nisin and pediocin genes from the genomic DNA of L. lactis MM19 and P. acidilactici MM33, respectively. Results demonstrate that a 227-bp fragment was amplified from the genomic DNA of L. lactis MM19 and a 188-bp fragment was amplified from the genomic DNA of P. acidilactici MM33 when pediocin primers were used (data not shown). The amplified PCR products were sequenced and revealed 100% homology to the nisin Z gene of L. lactis MM19 and the pediocin PA-1/AcH gene of P. acidilactici MM33.
In vitro inhibitory activity.
The anti-VRE capacities of supernatants from L. lactis MM19 and P. acidilactici MM33 were observed following the well diffusion assay using the VRE strain (data not shown). No inhibition zone around the well filled with the supernatant from P. acidilactici MM33A was observed. In order to ensure that the inhibition of VRE was caused by bacteriocin, proteases were added to the agar. Following this treatment, no inhibition zones around wells containing the supernatants from L. lactis MM19 and P. acidilactici MM33 were observed (data not shown).
MICs for VRE.
The antibiotic resistance of the VanA-type VRE strain was also evaluated, and the results showed that the clinical isolate was resistant to all evaluated vancomycin concentrations between 0 and 800 μg/ml while, surprisingly, the VRE strain was very sensitive to clindamycin at a concentration of 2 μg/ml (data not shown). To our knowledge, this is the first report of a VanA-type VRE isolate's being sensitive to clindamycin.
Gastrointestinal resistance of the LAB.
The results showed that the bile salt mixture (Sigma) tolerance threshold was 4% for all bacteria. However, the tolerance to bile salts from Oxoid was 20% for the P. acidilactici MM33 and MM33A strains, compared to 16% for Lactobacillus rhamnosus GG and 12% for L. lactis MM19. Moreover, the results presented in Table 1 show that P. acidilactici MM33 and MM33A and Lactobacillus rhamnosus GG survived under an acidic environment for 30 min. No significant difference (P > 0.05) between the initial microbial population at 0 min and the population after 30 min at pH ≥2.5 was observed for Lactobacillus rhamnosus GG, while a slight reduction of viability was observed for both strains of P. acidilactici. However, a significant reduction of viability at pH 2 was observed for all bacteria tested. L. lactis MM19 did not tolerate the gastric simulation as well as the other bacteria tested, as seen by the high mortality rates at pH ≤2.5. A 4.2-log10 reduction at these pHs was observed. However, complete survival at pH 3 was observed.
TABLE 1.
Survival of LAB after an incubation of 30 min at 37°C in SGF (pH 1.5 to 3.0)
| Microorganism | Time (min) | pH | No. of surviving bacteria (log CFU)a |
|---|---|---|---|
| L. lactis MM19 | 0 | 8.91 ± 0.21B | |
| 30 | 1.5 | <1 | |
| 30 | 2.0 | 4.47 ± 0.46A | |
| 30 | 2.5 | 4.69 ± 0.71A | |
| 30 | 3.0 | 8.67 ± 0.26B | |
| P. acidilactici MM33 | 0 | 9.62 ± 0.10C | |
| 30 | 1.5 | <1 | |
| 30 | 2.0 | 4.7 ± 0.33A | |
| 30 | 2.5 | 8.62 ± 0.26B | |
| 30 | 3.0 | 9.54 ± 0.14C | |
| P. acidilactici MM33A | 0 | 9.47 ± 0.16C | |
| 30 | 1.5 | <1 | |
| 30 | 2.0 | 4.54 ± 0.42A | |
| 30 | 2.5 | 8.79 ± 0.38B | |
| 30 | 3.0 | 9.37 ± 0.22C | |
| Lactobacillus rhamnosus GG | 0 | 9.10 ± 0.13B | |
| 30 | 1.5 | <1 | |
| 30 | 2.0 | 5.33 ± 0.62A | |
| 30 | 2.5 | 9.08 ± 0.14B | |
| 30 | 3.0 | 9.01 ± 0.13B |
Data are means ± standard deviations. Superscript letters A, B, and C indicate a significant variation (P ≤ 0.05) compared to the control (0 min) for same LAB.
Changes in the intestinal microbiota as determined by plating onto selective agars.
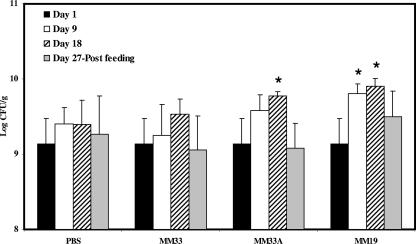
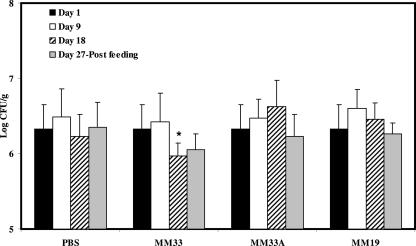
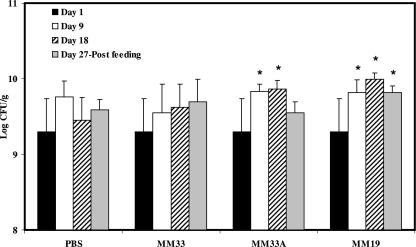
The influence of bacteriocin-producing LAB ingestion on the fecal microbial populations of healthy C57BL/6 mice is shown in Fig. 1, 2, and 3. The LAB counts in the feces of mice fed L. lactis MM19 were higher (P ≤ 0.05) than the starting levels after 9 and 18 days of feeding. However, after feeding interruption, the levels of LAB in these mice were similar to the initial counts (Fig. 1). The ingestion of P. acidilactici MM33A, the non-pediocin-producing mutant, led to an increase (P ≤ 0.05) of LAB after 18 days, while the pediocin-producing strain did not quantitatively influence the LAB population (Fig. 1). P. acidilactici MM33 is the only strain that significantly (P ≤ 0.05) reduced the Enterobacteriaceae population in mouse feces after 18 days of feeding (Fig. 2). Results shown in Fig. 3 indicate that MM19 feeding significantly (P ≤ 0.05) increased the culturable anaerobic populations after 9 and 18 days. This increase was maintained after the feeding treatment ended. The non-pediocin-producing strain increased the population of anaerobes as long as the feeding lasted, but after the feeding treatment ended, the level of anaerobes was similar to the initial count. No modification was observed for Lactobacillus spp., Staphylococcus spp., and Enterococcus spp. populations with either bacteriocin-producing bacterium assayed.
FIG. 1.
Total concentrations of culturable LAB in feces of C57BL/6 mice following the daily ingestion of L. lactis MM19, P. acidilactici MM33, or P. acidilactici MM33A. An asterisk indicates a significant variation (P ≤ 0.05) compared to the bacterial concentration on day 1 for the experimental group and that on the same day for the PBS control. Error bars represent the standard deviations.
FIG. 2.
Total concentrations of culturable Enterobacteriaceae in the feces of C57BL/6 mice following the daily ingestion of L. lactis MM19, P. acidilactici MM33, or P. acidilactici MM33A. An asterisk indicates a significant variation (P ≤ 0.05) compared to the bacterial concentration on day 1 for the experimental group and that on the same day for the PBS control. Error bars represent the standard deviations.
FIG. 3.
Total concentrations of culturable mesophilic anaerobes in feces of C57BL/6 mice following the daily ingestion of L. lactis MM19, P. acidilactici MM33, or P. acidilactici MM33A. An asterisk indicates a significant variation (P ≤ 0.05) compared to the bacterial concentration on day 1 for the experimental group and that on the same day for the PBS control. Error bars represent the standard deviations.
In vivo anti-VRE experiment.
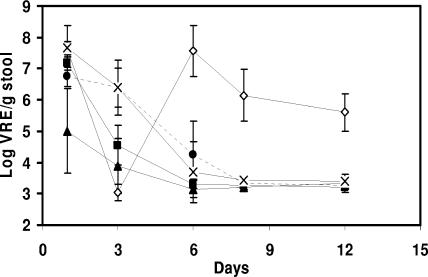
Results presented in Fig. 4 show that the VRE densities observed 1 day postinfection were lower by 1.73 log10 CFU/g (P ≤ 0.05) for the group of L. lactis MM19-fed mice than for the PBS-fed group. Moreover, L. lactis MM19- and P. acidilactici MM33-treated mice had significantly lower VRE densities than the PBS controls 3 days after the infection (P ≤ 0.05). The VRE populations were reduced by 2.50 and 1.85 log10 CFU/g, respectively. Six days after the infection, levels of VRE in L. lactis MM19- and P. acidilactici MM33-treated mice were undetectable. VRE densities in the feces of mice in the group fed the non-pediocin-producing strain were similar to the levels measured for controls for the duration of the experiment. Mice treated with bacitracin for 3 days after the infection had undetectable levels of VRE at day 3. However, the VRE population reappeared when the bacitracin treatment was discontinued. Results presented in Table 2 show that 1 day after the VRE infection, all mice were colonized by the pathogen, with the exception that only 83% of the mice fed L. lactis MM19 showed detectable levels of VRE. At 3 days postinfection, the level of VRE-colonized mice was reduced by 29% after the ingestion of L. lactis MM19. Also, at 6 days postinfection, no mice that had received bacteriocin-producing strains were colonized with the nosocomial pathogen, while 60 and 50% of the PBS- and P. acidilactici MM33A-treated groups were colonized, respectively.
FIG. 4.
Changes in the densities of total vancomycin-resistant Enterococcus populations in VRE-colonized CF-1 mice treated with PBS (•), bacitracin (⋄), L. lactis MM19 (▴), P. acidilactici MM33 (▪), and P. acidilactici MM33A (×). The intragastric VRE infection was realized on day 0. The oral administration of bacitracin began the day after the infection and continued for 3 days, after which it was stopped and replaced by PBS feeding. The oral administration of LAB and PBS began 7 days before the infection and continued for 8 days after the infection. Error bars represent the standard deviations.
TABLE 2.
Percentages of CF-1 mice colonized with a detectable level of VRE
| Inoculum | % of micea colonized with VRE on postinfection day:
|
|||||
|---|---|---|---|---|---|---|
| 0 | 1 | 3 | 6 | 8 | 12 | |
| PBS | 0 | 100 | 100 | 60 | 0 | 0 |
| Bacitracin | 0 | 100 | 0 | 100 | 100 | 100 |
| L. lactis MM19 | 0 | 83 | 71 | 0 | 0 | 0 |
| P. acidilactici MM33 | 0 | 100 | 100 | 0 | 0 | 0 |
| P. acidilactici MM33A | 0 | 100 | 100 | 50 | 0 | 0 |
A total of eight mice for each group were used in two independent experiments.
DISCUSSION
As reported by Corr et al. (5), intestinal infectious diseases are a major cause of morbidity and mortality. Enterococci are indigenous inhabitants of the mammal gut, are the second most common nosocomial pathogen in the United States, and are responsible for 3 to 4 cases of nosocomial bloodstream infection per 10,000 hospital discharges (11). Glycopeptides are often the only group of antibiotics able to control infection with multidrug-resistant strains of staphylococci, streptococci, and enterococci. The problem is that enterococci can be considered to be an important reservoir of resistance genes and this glycopeptide resistance can be transferred to more-pathogenic bacteria, such as staphylococci and streptococci (4, 6, 30). To date, few effective therapies are available to prevent and control intestinal colonization by VRE. As a result, alternative prophylactic and therapeutic strategies are urgently required.
The results obtained in our study demonstrate the survival of potentially probiotic L. lactis MM19, a nisin Z producer, and P. acidilactici MM33, a pediocin PA-1/AcH producer, following an in vitro model simulating the gastrointestinal transit and also show significant probiotic-associated alteration of some populations of the gastrointestinal microbiota. Moreover, the consumption of the probiotic was associated with a trend toward reduced intestinal colonization by VRE. These results warrant a large-scale probiotic study in order to reliably assess the statistical significance of these observations.
It was also possible to demonstrate that the genes encoding nisin in L. lactis MM19 and pediocin in P. acidilactici MM33 are identical to the nisZ gene (21) and to the pedA gene, respectively (15). The hypothesis suggested by Millette et al. (17) that the bacteriocin was different from nisin based on an antimicrobial spectrum and on the pH and the temperature stability of the supernatant was not confirmed by the results of this study. This outcome may be explained by the fact that crude neutralized supernatant was used and it is known that nisin in culture medium is not as heat stable as pure nisin (32). Moreover, the nisin concentration to inhibit Listeria monocytogenes should be higher than the quantity produced in the supernatant of L. lactis MM19.
The anti-VRE capacities of nisin Z and pediocin PA-1/AcH demonstrated in this study are important since little information exists about the capacities of bacteriocin-producing bacteria or their purified metabolites to control infections caused by multidrug-resistant gram-positive bacteria. Severina et al. (29) demonstrated the capacity of pure nisin to decrease the viability of Streptococcus pneumoniae, Staphylococcus aureus, and vancomycin-resistant E. faecium and Enterococcus faecalis isolates. However, stable nisin-resistant mutants were isolated after a few passages in nisin solution. Giacometti et al. (9) studied the addition of nisin and various antibiotics against methicillin-resistant Staphylococcus aureus (MRSA). They showed that nisin activity is enhanced in the presence of other antibiotics, and no resistance was observed. Brumfitt et al. (2) observed a synergy between nisin and ramoplanin against MRSA and VRE. Finally, Nascimento et al. (23) tested four bacteriocins against many MRSA strains or coagulase-negative staphylococci. They concluded that bacteriocins may represent alternative agents to control important nosocomial pathogens. Although some in vitro experiments have demonstrated the potential efficacy of bacteriocins against antibiotic-resistant bacteria, few studies have examined the utilization of live strains of bacteriocin-producing LAB alone or in combination with other antimicrobial compounds to eliminate MRSA or VRE. Other experiments using VRE strains presenting other vancomycin resistance genes will be needed to ascertain the anti-VRE capacities of both bacteriocins. Moreover, a nonconventional VRE strain harboring the vanA vancomycin resistance gene was used in this study. It is known that every other VRE strain reported in the literature has demonstrated intrinsic clindamycin resistance (3, 22). However, the particular strain of VRE in this study was very sensitive to clindamycin. Thus, more research is needed in order to elucidate this particularity.
A number of mechanisms for the modulation of the gastrointestinal microbiota by LAB are proposed. L. lactis MM19 and P. acidilactici MM33 produce nisin Z and pediocin PA-1/AcH, respectively, and in vitro these factors are antagonistic to a wide range of gram-positive bacteria. The production of these antimicrobial factors in the gut may increase the competition with other microorganisms, conferring a survival advantage on the bacteriocin-producing LAB. Thus, the competitive exclusion of other microorganisms from this niche environment would affect the composition of the fecal microbiota.
Our study demonstrated that the ingestion of bacteriocin-producing bacteria was well tolerated by C57BL/6 mice over the course of a 3-week feeding trial and could alter quantitatively the balance of colonic bacterial populations. The ingestion of P. acidilactici MM33 led to a reduction of the Enterobacteriaceae population, probably resulting from a modification of the microbial balance in the gut. The nisin-producing strain increased the total culturable LAB contents and the total anaerobes in the feces of the mice as long as the mice were fed L. lactis MM19, while the pediocin-producing strain did not quantitatively influence these populations. It should be hypothesized that a reorganization of the intestinal microbiota was induced by the bacteria or the nisin production. However, a mutant defective in nisin production should be used in order to validate this point. The administered strains may replace or stimulate the growth of the indigenous Lactobacillus strains, leading to a variation of the bacterial species but not to a quantitative modification, as observed for P. acidilactici MM33. Manninen et al. (14) observed that the indigenous Lactobacillus acidophilus populations in the small intestines of dogs were stimulated following the ingestion of other non-bacteriocin-producing species of the Lactobacillus genus. However, Bernbom et al. (1) demonstrated that the ingestion of nisin-producing and also non-nisin-producing L. lactis strains over 2 days increased only the Bifidobacterium populations in the feces of rats harboring a human microbiota during the first 8 days but decreased the numbers of Enterococcus and Streptococcus species in duodenum, ileum, cecum, and colon samples. Our experiment demonstrated that a longer period of ingestion of nisin-producing bacteria could alter other microbial populations in the mouse feces. Culture-independent techniques such as denaturing gradient gel electrophoresis, fluorescent in situ hybridization, or real time-PCR are presently done to precisely determine the variations in fecal microbial species during bacteriocin-producing-bacterium feeding experiments.
Some antibiotics, such as bacitracin, may be used to manage VRE infections, but in this study and also the study by Donskey et al. (7), an increase in VRE densities 3 days after the end of the bacitracin treatment was reported. Donskey et al. hypothesized that the inhibition of the normal microbiota by bacitracin facilitates the recurrence of VRE infection after the treatment ends. Recently, Corr et al. (5) demonstrated that the bacteriocin production capacity of Lactobacillus salivarius UCC118 is an important characteristic to control murine Listeria monocytogenes infection. Our study demonstrated that pediocin production by LAB is an important trait to reduce the intestinal colonization by a VRE strain. It is interesting that when the oral administration of LAB to the mice was discontinued, no recurrence of the VRE was observed. To our knowledge, this is the first clear evidence that bacteriocin-producing strains of LAB reduce the intestinal colonization by VRE in an animal model. No recurrence was observed. Donskey et al. (7) showed that a strain of Bacillus was able to reduce the fecal VRE concentration, but they did not implicate the production of a bacteriocin. Although the bacteriocin-producing strains are able to reduce the densities of VRE populations following an infection facilitated by clindamycin, these strains have no effect on the total Enterococcus spp. populations in the mouse feces. It can be hypothesized that the production of bacteriocin is an important characteristic of a probiotic in order to displace a sensitive VRE strain from the gut. The bacteriocin produced locally in the immediate environment of the producing bacteria may eliminate particular strains of antibiotic-resistant bacteria such as VRE. Also, the antimicrobial peptide or the presence of a novel intestinal inhabitant may modify the indigenous microbial populations, leading to the elimination of the VRE.
This is the first study reporting the capabilities of nisin- and pediocin-producing strains to modulate the intestinal microbiota of healthy mice and to reduce the intestinal colonization of VRE-infected mice. These LAB strains were almost as resistant to acid and bile as Lactobacillus rhamnosus GG, a well-recognized probiotic bacterium. Both LAB supernatants inhibited the growth of a clinical isolate of VRE in vitro. Although additional studies are needed to determine if our results are reproducible in VRE-infected patients, L. lactis MM19 and P. acidilactici MM33 seem to be good candidates to eradicate or control intestinal infections caused by multidrug-resistant bacterial pathogens.
Acknowledgments
We thank the NSERC (Natural Science and Engineering Research Council of Canada) partnership program and Bio-K+ International Inc. for their financial support. M. Millette is the recipient of a scholarship from Foundation Armand-Frappier.
The Centre Hospitalier de l'Université de Montréal microbiology laboratory is acknowledged for providing us with the VRE strain.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Bernbom, N., T. R. Licht, C. H. Brogren, B. Jelle, A. H. Johansen, I. Badiola, F. K. Vogensen, and B. Norrung. 2006. Effects of Lactococcus lactis on composition of intestinal microbiota: role of nisin. Appl. Environ. Microbiol. 72:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brumfitt, W., M. R. Salton, and J. M. Hamilton-Miller. 2002. Nisin, alone and combined with peptidoglycan-modulating antibiotics: activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. J. Antimicrob. Chemother. 50:731-734. [DOI] [PubMed] [Google Scholar]
- 3.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conly, J. 2002. Antimicrobial resistance in Canada. Can. Med. Assoc. J. 167:885-891. [PMC free article] [PubMed] [Google Scholar]
- 5.Corr, S. C., Y. Li, C. U. Riedel, P. W. O'Toole, C. Hill, and C. G. M. Gahan. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA 104:7617-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courvalin, P. 2006. Vancomycin-resistance in Gram-positive cocci. Clin. Infect. Dis. 42(Suppl. 1):S25-S34. [DOI] [PubMed] [Google Scholar]
- 7.Donskey, C. J., C. K. Hoyen, S. M. Das, S. Farmer, M. Dery, and R. A. Bonomo. 2001. Effect of the oral Bacillus coagulans administration on the density of vancomycin-resistant enterococci in the stool of colonized mice. Lett. Appl. Microbiol. 33:84-88. [DOI] [PubMed] [Google Scholar]
- 8.Food and Agriculture Organization-World Health Organization. 2006. FAO/WHO working group report. Probiotics in food health and nutritional properties and guidelines for evaluation. Food and Agriculture Organization of the United Nations-World Health Organization, Rome, Italy.
- 9.Giacometti, A., O. Cirioni, F. Barchiesi, and G. Scalise. 2000. In-vitro activity and killing effect of polycationic peptides on methicillin-resistant Staphylococcus aureus and interactions with clinically used antibiotics. Diagn. Microbiol. Infect. Dis. 38:115-118. [DOI] [PubMed] [Google Scholar]
- 10.Hasper, H. E., B. de Kruijff, and E. Breubink. 2004. Assembly and stability of nisin-lipid II pores. Biochemistry 43:11567-11575. [DOI] [PubMed] [Google Scholar]
- 11.Koch, S., M. Hufnagel, C. Theilacker, and J. Huebner. 2004. Enterococcal infections: host response, therapeutic, and prophylactic possibilities. Vaccine 22:822-830. [DOI] [PubMed] [Google Scholar]
- 12.Levine, D. P. 2006. Vancomycin. Clin. Infect. Dis. 42:S5-S12. [DOI] [PubMed] [Google Scholar]
- 13.Manley, K. J., M. B. Fraenkel, B. C. Mayall, and D. A. Power. 2007. Probiotic treatment of vancomycin-resistant enterococci: a randomised controlled trial. Med. J. Aust. 186:454-457. [DOI] [PubMed] [Google Scholar]
- 14.Manninen, T. J. K., M. L. Rinkinen, S. S. Beasley, and P. E. J. Saris. 2006. Alteration of the canine small-intestinal lactic acid bacterium microbiota by feeding of potential probiotics. Appl. Environ. Microbiol. 72:6539-6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marugg, J. D., C. F. Gonzalez, B. S. Kunka, A. M. Ledeboer, M. J. Pucci, M. Y. Toonen, S. A. Walker, L. C. Zoetmulder, and P. A. Vendenbergh. 1992. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl. Environ. Microbiol. 58:2360-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millette, M., W. Smoragiewicz, and M. Lacroix. 2004. Antimicrobial potential of immobilized Lactococcus lactis subsp. lactis ATCC 11454 against selected bacteria. J. Food Prot. 67:1184-1189. [DOI] [PubMed] [Google Scholar]
- 17.Millette, M., C. Dupont, D. Archambault, and M. Lacroix. 2007. Partial characterization of bacteriocins produced by human Lactococcus lactis and Pediococcus acidilactici isolates. J. Appl. Microbiol. 102:274-282. [DOI] [PubMed] [Google Scholar]
- 18.Millette, M., C. Dupont, F. Shareck, M. T. Ruiz, D. Archambault, and M. Lacroix. 2008. Purification and identification of the pediocin produced by Pediococcus acidilactici MM33, a new human intestinal strain. J. Appl. Microbiol. 104:269-275. [DOI] [PubMed] [Google Scholar]
- 19.Morency, H., M. Mota-Meira, G. LaPointe, C. Lacroix, and M. C. Lavoie. 2001. Comparison of the activity spectra against pathogens of bacterial strains producing a mutacin or a lantibiotic. Can. J. Microbiol. 47:322-331. [PubMed] [Google Scholar]
- 20.Mota-Meira, M., G. Lapointe, C. Lacroix, and M. C. Lavoie. 2000. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob. Agents Chemother. 44:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulders, J. W., I. J. Boerrigter, H. S. Rollema, R. J. Siezen, and W. M. de Vos. 1991. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 201:581-584. [DOI] [PubMed] [Google Scholar]
- 22.Murray, B. E. 1997. Vancomycin-resistant enterococci. Am. J. Med. 102:284-293. [DOI] [PubMed] [Google Scholar]
- 23.Nascimento, J. S., H. Ceotto, S. B. Nascimento, M. Giambiagi-Demarval, K. R. Santos, and M. C. Bastos. 2006. Bacteriocins as alternative agents for control of multiresistant staphylococcal strains. Lett. Appl. Microbiol. 42:215-221. [DOI] [PubMed] [Google Scholar]
- 24.Noonpakdee, W., C. Santivarangkna, P. Jumriangrit, K. Sonomoto, and S. Panyim. 2003. Isolation of nisin-producing Lactococcus lactis WNC 20 strain from nham, a traditional Thai fermented sausage. Int. J. Food Microbiol. 81:137-145. [DOI] [PubMed] [Google Scholar]
- 25.O'Mahony, L., M. Feeney, S. O'Halloran, L. Murphy, B. Kiely, J. Fitzgibbon, G. Lee, G. O'Sullivan, F. Shanahan, and J. K. Collins. 2001. Probiotic impact on microbial flora, inflammation and tumour development in IL-10 knockout mice. Aliment. Pharmacol. Ther. 15:1219-1225. [DOI] [PubMed] [Google Scholar]
- 26.Patel, R., J. R. Uhl, P. Kohner, M. K. Hopkins, and F. R. Cockerill III. 1997. Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J. Clin. Microbiol. 35:703-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice, L. B. 2006. Antimicrobial resistance in Gram-positive bacteria. Am. J. Med. 119:S11-S19. [DOI] [PubMed] [Google Scholar]
- 28.Sahm, D. F., J. Kissinger, J. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Severina, E., A. Severin, and A. Tomasz. 1998. Antibacterial efficacy of nisin against multidrug-resistant Gram-positive pathogens. J. Antimicrob. Chemother. 41:341-347. [DOI] [PubMed] [Google Scholar]
- 30.Shadel, B. N., L. A. Puzniak, K. N. Gillespie, S. J. Lawrence, M. Kollef, and L. M. Mundy. 2006. Surveillance for vancomycin-resistant enterococci: type, rates, costs, and implications. Infect. Control Hosp. Epidemiol. 27:1068-1075. [DOI] [PubMed] [Google Scholar]
- 31.Succi, M., P. Tremonte, A. Reale, E. Sorrentino, L. Grazia, S. Pacifico, and R. Coppola. 2005. Bile salt and acid tolerance of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. FEMS Microbiol. Lett. 244:129-137. [DOI] [PubMed] [Google Scholar]
- 32.Thomas, L. V., M. R. Clarkson, and J. Delves-Broughton. 2000. Nisin, p. 463-524. In A. S. Naidu (ed.), Natural food antimicrobial systems. CRC Press, Boca Raton, FL.
- 33.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i:57-58. [DOI] [PubMed] [Google Scholar]